Simple Summary
Prostate-specific antigen and digital rectal examination, used to guide prostate biopsy, often result in overdiagnosis of indolent prostate cancer (PCa) while missing clinically significant PCa (csPCa). The aim of this study was to evaluate the diagnostic accuracy of SelectMDx and its association with multiparametric magnetic resonance imaging (mpMRI) in predicting PCa in prostate biopsies. SelectMDx was revealed to be a good predictor of PCa, while with regards to csPCa detection, it was demonstrated to be less effective, showing results similar to mpMRI. The best diagnostic strategy to avoid unnecessary biopsy is performing SelectMDx after an initial negative mpMRI. Biopsy could be proposed for all cases of positive mpMRI and to those with a negative mpMRI followed by a positive SelectMDx.
Abstract
Prostate-specific antigen (PSA) testing as the sole indication for prostate biopsy lacks specificity, resulting in overdiagnosis of indolent prostate cancer (PCa) and missing clinically significant PCa (csPCa). SelectMDx is a biomarker-based risk score to assess urinary HOXC6 and DLX1 mRNA expression combined with traditional clinical risk factors. The aim of this prospective multi-institutional study was to evaluate the diagnostic accuracy of SelectMDx and its association with multiparametric magnetic resonance (mpMRI) when predicting PCa in prostate biopsies. Overall, 310 consecutive subjects were included. All patients underwent mpMRI and SelectMDx prior to prostate biopsy. SelectMDx and mpMRI showed sensitivity and specificity of 86.5% vs. 51.9%, and 73.8% vs. 88.3%, respectively, in predicting PCa at biopsy, and 87.1% vs. 61.3%, and 63.7% vs. 83.9%, respectively, in predicting csPCa at biopsy. SelectMDx was revealed to be a good predictor of PCa, while with regards to csPCa detection, it was demonstrated to be less effective, showing results similar to mpMRI. With analysis of strategies assessed to define the best diagnostic strategy to avoid unnecessary biopsy, SelectMDx appeared to be a reliable pathway after an initial negative mpMRI. Thus, biopsy could be proposed for all cases of mpMRI PI-RADS 4–5 score, and to those with Prostate Imaging-Reporting and Data System (PI-RADS) 1–3 score followed by a positive SelectMDx.
1. Introduction
The European Association of Urology (EAU) guidelines on prostate cancer (PCa) recommend an individualized risk-adapted strategy for early detection of PCa, offering a prostate-specific antigen (PSA) test and digital rectal examination (DRE) to a well-informed man, aware of the related potential risks and benefits with at least ten to fifteen years of life expectancy [1]. However, use of PSA testing as the sole indication for prostate biopsy lacks specificity, resulting in overdiagnosis and potentially over-treatment of indolent PCa (i.e., non-aggressive), and, at the same time, may result in missing clinically significant PCa (csPCa) diagnoses in men with PSA levels below the cut-off value [2,3,4].
Many tools have been developed in recent years to avoid unnecessary biopsies and to overcome PSA limits. Improvements in risk stratification and a decrease of indolent PCa diagnosis have been reached, with the increased use of multiparametric magnetic resonance imaging (mpMRI) as one of the main diagnostic exams for PCa and as a tool for target biopsy [5,6].
To classify patients’ risk between low, intermediate or high (csPCa), in recent years different biomarkers and risk calculators (combining PSA and other risk factors), have been developed; however, their clinical benefit still needs to be proven [1]. Their development is guided by an appropriate risk stratification in order to avoid unnecessary biopsies and over-treatment for low-risk patients and to plan the correct treatment strategy for csPCa [7,8].
To consider initial prostate biopsy, blood biomarkers PHI and 4K Score, as well as urine biomarkers PCA3, SelectMDx and ExoDx have been developed [9,10,11,12,13,14,15,16,17,18]. To consider the treatment in patients with confirmed PCa, OncotypeDx, Prolaris and Decipher are available [19,20,21,22,23,24,25]. In order to evaluate a re-biopsy after an initially negative one, 4K score, PCA3, ExoDx and ConfirmMDx can help in the decision-making process [26,27,28,29,30,31,32,33,34].
SelectMDx is a novel biomarker-based risk score for assessing urinary HOXC6 and DLX1 mRNA expression combined with traditional clinical risk factors. Although SelectMDx is available in clinical practice to improve patient selection for initial prostate biopsy, previous studies have shown its ability to reduce the number of unnecessary biopsies, with current data being too limited to implement its use into routine screening programs [1]. Large prospective studies of the available biomarkers are still needed to assess whether their implementation in clinical practice is useful in providing guidance on decision making.
Following a prior positive single-institutional experience with SelectMDx test [17], we assessed a prospective and multi-institutional study with the aim to evaluate in a large cohort, if SelectMDx, associated with mpMRI, is a reliable method to predict PCa and csPCa for patients undergoing prostate biopsy.
2. Material and Methods
2.1. Study Population
In this multi-institutional prospective study, men from five different sites in Italy were consecutively enrolled between March 2018 and September 2019. All patients were scheduled for first prostate biopsy, and inclusion criteria were: elevated total PSA level (>3 ng/mL confirmed) and/or abnormal DRE. Exclusion criteria were those with history of PCa or different neoplasm under treatment, any medical treatment that could alter PSA value, any invasive treatments for BPH or any prior prostatic biopsy. Patient characteristics are shown in Table S1.
2.2. mpMRI
All patients underwent mpMRI prior to biopsy using a 1.5 or 3.0 Tesla scanner (Achieva XR, Philips Medical System, Best, Netherlands; GE Discovery MR750, GE Healthcare, Chicago, IL, USA) with or without an endorectal coil. The functional technique of MRI was based on a combination of T2-weighted (T2W) images, diffusion-weighted imaging (DWI) and dynamic contrast-enhanced (DCE) studies. Lesions were characterized and graded using the Prostate Imaging-Reporting and Data System (PI-RADS) version 2.0 or 2.1, with a final grade from 1 to 5 indicating a greater probability of csPCa [35]. All mpMRI performed were analyzed by expert uro-radiologists (one to three per center, with minimum of five years of experience) blinded from patient characteristic, urine test score and biopsy outcome.
2.3. SelectMDx Sampling
Urine samples from the first-voided stream (approximately 30 mL) were collected after the DRE was conducted with a standard scheme of three strokes for each prostate lobe [36]. Samples have been shipped at room temperature to the central laboratory (MDxHealth Servicelab B.V., Nijmegen, The Netherlands) and stored at −80 °C. The SelectMDx score (MDx Health) is obtained combining different levels of expression of HOXC6 and DLX1 with clinical risk factors (age, DRE, total PSA, prostate volume) in a logistic regression model [37]. The results of the test are given as percentage of probability of positive or negative prostate biopsy for PCa (with two different probabilities for PCa and csPCa). Looking at previous experiences, in which a cut-off point was offered, here, only the probability is given [38].
2.4. Prostatic Biopsy
The trans-rectal ultrasound (TRUS) guided prostate biopsies have been performed by a single uro-radiologist for every center, all with more than 20 years of experience. The standardized biopsy scheme was: 12 random systematic cores from the peripheral zone of the prostate at the base, mid gland, and apex. In all patients where a mpMRI PI-RADS 3–5 lesion has been described, additional targeted samples (2 to 3 biopsy cores per lesion) have been obtained using an imaging fusion technique. All samples have been evaluated by experienced genitourinary pathologists and histological grading have been assessed following the Gleason grading system and Gleason Grade Groups (International Society of Urological Pathologist (ISUP) 2014) [39]. A csPCa was defined as ISUP score ≥2 (Gleason score ≥7) [1].
2.5. Statistical Analysis
Descriptive statistics were used to compare patients’ characteristics. A Pearson Chi-Square test was used to test association between categorical variables. Non-parametric Mann-Whitney U tests were used for comparisons of continuous covariates among groups. Sensitivity, specificity, and areas under the curves (AUC) were evaluated by computing receiver operating characteristic (ROC) curves. To compare the clinical utility of each tool alone or in association decision curve analysis were used (DCA). The SelectMDx scores were divided between positive (with the percentage of probability for PCa and csPCa) or negative for the suspicious of PCa by manufacturers’ report. PI-RADS 4–5 for mpMRI were considered positive while PI-RADS 1–3 negative. SelectMDx and positive mpMRI were used to evaluate the performance of tests together. To evaluate discordant cases, we carried out a simulated analysis, determining the number of avoided biopsies and missed PCa by SelectMDx results and PSAD values. Cut-off levels were >3 ng/mL for PSA, and ≥0.15 ng/mL/mL for PSAD.
Additionally, a multivariable stepwise logistic regression model (forward selection) was generated to assess the relative influence of those predictive variables that were significant upon univariate analysis on both the outcome of PCa and csPCa detection. Entering and removing limits were set at p = 0.05 and p = 0.10, respectively. In particular, age (continuous; <65 vs. ≥65 years), prostate volume (continuous; <56 vs. ≥56 mL), PCa familiarity (no vs. yes), DRE (negative vs. suspicious), total PSA (continuous, ng/mL), PSAD (<0.15 vs. ≥0.15), mpMRI score (negative 1–3 vs. positive 4–5) and SelectMDx (negative vs. positive) were included into the model. Finally, a locally weighted scatter plot smoother (LOWESS) function was used to graphically depict the relationship between the predicted probability of PCa or csPCa and the SelectMDx score provided within the ‘manufacturers’ report. Statistical analysis was performed using Stata (version 16.1) and SPSS (version 21.0) statistical programs, having set p-values <0.05 as statistically significant.
3. Results
Overall, 310 consecutive subjects have been included in our prospective analysis. Table S1 reports patients characteristics. SelectMDx was positive in 144 (46.5%) cases and a PI-RADS score 4–5 in 78 (25.2%) cases. A concordance between SelectMDx and mpMRI was found in 63.3% of cases. Out of 104 PCa (33.5%) detected at biopsy, 62 (20.0%) were csPCa. Table 1 reports stratification of subjects according to pathologic results at biopsy (i.e., PCa negative, all PCa, and csPCa). There was a significantly difference between PSA levels between the groups (p < 0.0001, and p = 0.001) (Table 1 and Figure S1a).

Table 1.
Patient characteristics stratified according to prostatic biopsy results (number, %, mean ± SD, median, range).
SelectMDx score was positive in 86.5% and 87.1% of PCa and csPCa, respectively, and in 26.2% of cases with no PCa at biopsy (Table 1 and Figure 1a). The probability for a csPCa at the SelectMDx score was higher in csPCa (27.7 ± 18.1) than in PCa cases (25.9 ± 18.2) at biopsy (Table 1).
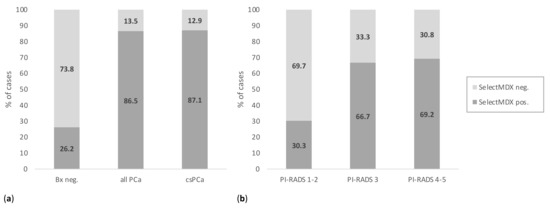
Figure 1.
SelectMDx positive and negative results according to (a) histologic diagnosis for PCa at biopsy; and (b) to PI-RADS score at mpMRI. (PCa = prostate cancer; PI-RADS = Prostate Imaging-Reporting and Data System; mpMRI = multiparametric magnetic resonance imaging; Bx = biopsy; csPCa = clinically significant PCa).
3.1. SelectMDx Performance for PCa and csPCa at Biopsy
The performance of SelectMDx compared to that of mpMRI PI-RADS score, and of the association between mpMRI and SelectMDx, PSA or PSAD to predict PCa and csPCa at biopsy is reported in Table 2.

Table 2.
Performance of individual parameters and their combination to predict PCa and csPCa on biopsy (% and 95% CI).
SelectMDx and mpMRI PI-RADS scores showed sensitivity and specificity of 86.5% (95% CI 78.5–91.9) vs. 51.9% (95% CI 42.4–61.3), and 73.8% (95% CI 64.7–79.3) vs. 88.3% (95% CI 83.2–92.1), respectively, in predicting PCa at biopsy, and 87.1% (95% CI 76.2–93.5) vs. 61.3% (95% CI 48.8–72.4), and 63.7% (95% CI 57.5–69.4) vs. 83.9% (95% CI 78.7–87.9), respectively, in predicting csPCa at biopsy. Negative predictive value (NPV) and positive predictive value (PPV) were 91.6% vs. 78.4%, and 62.5% vs. 69.2%, respectively, for PCa and 95.2% vs. 89.7%, and 37.5% vs. 48.7%, respectively, for csPCa (Table 2). mpMRI sensitivity and specificity-considering positive those cases with PI-RADS score 3–5-were 82.7% (95% CI 74.2–88.8) and 77.7% (95% CI 71.5–82.8), respectively.
Sensitivity and specificity for both positive tests were 46.2% (95% CI 36.9–55.7) and 97.1% (95% CI 93.6–98.8), respectively, in predicting PCa at biopsy, and 54.8% (95% CI 42.5–66.6) and 91.9% (95% CI 87.8–94.8), respectively, in predicting csPCa at biopsy. Compared to the association of mpMRI and SelectMDx, that of mpMRI PI-RADS score and PSA showed slightly higher sensitivity and lower specificity in predicting both PCa and csPCa at biopsy, while the association of mpMRI PI-RADS score and PSAD showed lower sensitivity and specificity in predicting both PCa and csPCa at biopsy (Table 2).
SelectMDx score performance in predicting PCa and csPCa at biopsy was evaluated as area under the curve (AUC) of the receiver operating characteristics (ROC) in Figure 2. The AUC was 0.80 (95% CI 0.76–0.85) for PCa and 0.75 (95% CI 0.70–0.81) for csPCa; mpMRI PI-RADS score AUC was 0.70 (95% CI 0.65–0.75) for PCa and 0.73 (95% CI 0.66–0.79) for csPCa (Figure 2a,b). The AUC of using mpMRI and SelectMDx test together was 0.72 (95% CI 0.67–0.77) to detect PCa and 0.73 (95% CI 0.67–0.80) for csPCa, compared to 0.70 (95% CI 0.65–0.75) and 0.72 (95% CI 0.65–0.78), respectively, for the association mpMRI and PSA, and 0.67 (95% CI 0.62–0.71) and 0.68 (95% CI 0.62–0.75), respectively, for the association mpMRI and PSAD (Figure S2a,b).
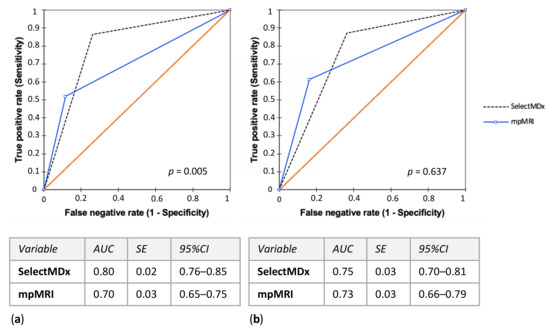
Figure 2.
SelectMDx score and mpMRI PI-RADS score performance evaluated as area under the curve (AUC) of the receiver operating characteristics (ROC) in predicting (a) PCa and (b) csPCa histological diagnosis at biopsy. (mpMRI = multiparametric magnetic resonance imaging; PI-RADS = Prostate Imaging-Reporting and Data System; PCa = prostate cancer; csPCa = clinically significant PCa; AUC = area under the curve; SE = standard error; CI = confidence interval).
Decision curve analyses (DCA) were depicted to evaluate the clinical net benefit of SelectMDx and mpMRI (alone or in combination). The best combination for the detection of PCa and csPCa, found at DCA, was SelectMDX + mpMRI if compared to the association of mpMRI with other diagnostic tools (i.e., PSA and PSAD) (Figure 3a,b).
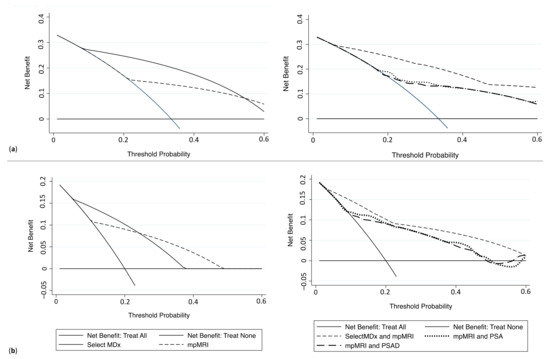
Figure 3.
Decision curve analysis comparing clinical utility of SelectMDx score, mpMRI, and the associations mpMRI+ SelectMDx, mpMRI + PSA and mpMRI + PSAD for detecting (a) PCa and (b) csPCa. (mpMRI = multiparametric magnetic resonance imaging; PSA = prostatic-specific antigen; PCa = prostate cancer; csPCa = clinically significant PCa).
Moreover, at multivariable logistic regression analysis, mpMRI and SelectMDx were confirmed to be independently and strongly associated with both the outcomes of PCa and csPCa diagnosis at prostate biopsy (Odds Ratio [OR] PCa: 7.14, 95% CI: 3.31–15.39 and 25.57, 95% CI: 11.05–59.16 respectively; OR csPCa, 6.10, 95% CI: 2.94–12.64 and 9.49, 95% CI: 4.01–22.49, respectively; Table S2). Finally, at LOWESS function analysis, the SelectMDx score given by the ‘manufacturers’ report (percentage (%) indicating the probability of PCa and/or csPCa from the test) showed an almost linear raising correlation when plotted against the multivariable adjusted predicted probability for the prostate biopsy detection of PCa (Figure 4a) and csPCa respectively (Figure 4b).
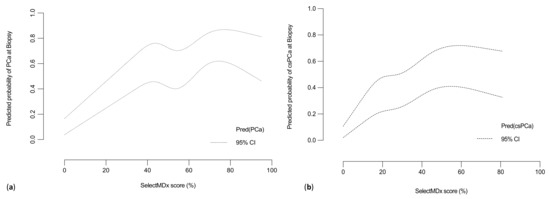
Figure 4.
Multivariable adjusted Locally Weighted Scatter Plot Smoother (LOWESS) function depicting the predicted probability of (a) PCa or (b) csPCa detection rate and SelectMDx score (%). (PCa = prostate cancer; csPCa = clinically significant PCa).
3.2. Impact of Implementation of SelectMDx Versus PSAD into the mpMRI Pathway to Select Patients Candidate for Prostate Biopsy
The distribution of SelectMDx scores according to PI-RADS findings in mpMRI is reported in Figure 1b and compared to those for total PSA in Figure S1b. Interestingly, SelectMDx positivity increased from 30.3% for PI-RADS 1–2 cases, to 66.7% and 69.2% for PI-RADS 3 and 4–5 cases (p < 0.01); additionally, total PSA values were differently distributed among PI-RADS score groups (p < 0.01) (Figure 1b and Figure S1b). With regards to PI-RADS 3 lesions, 59.3% (32/54) showed PCa at biopsy, and 14 (25.9%) were csPCa; SelectMDx score was positive in 81.3% of PI-RADS score 3 associated with PCa diagnosis and positive in 45.4% of those negative for PCa at biopsy. In PI-RADS 4–5 lesions, SelectMDx score was positive in 88.9% of PCa cases, and in 25.0% of those with no PCa at biopsy.
Cases with discordant tests were investigated to analyze the potential added value of implementing SelectMDx in the mpMRI diagnostic pathway. If we look at PI-RADS 1–2 cases, according to SelectMDx results, 30.3% (54/178) of patients would undergo biopsy with the detection of 16 (15.4%) PCa and 8 (12.9%) csPCa. Avoiding biopsy in patients with a PI-RADS score 4–5 and a negative SelecMDx test would result in 24 (30.8%) being spared biopsies within this category, while missing 6 (5.8%) PCa and 4 (6.5%) csPC. Regarding PI-RADS 3 cases, performing prostate biopsy only in those with a positive SelectMDx would result in 81.3% (26/32) of PCa detected, while avoiding biopsy in those with a negative SelectMDx would result in 18.8% (6/32) of PCa and 14.3% (2/14) of csPCa missed.
Performing prostate biopsy in patients with a PI-RADS score 1–2 and PSAD ≥ 0.15, would result in 48/178 (27.0%) biopsies performed in this category, with the detection of 6 (5.8%) PCa and 4 (6.5%) csPC. Avoiding biopsy in patients with a PI-RADS score 4–5 and PSAD < 0.15, would result in 28 (35.9%) spared biopsies within this category, yet missing 14 (13.5%) PCa and 10 (16.1%) csPC. If we perform prostate biopsy among PI-RADS score 3 cases only in those with PSAD ≥ 0.15, this would result in the detection of 8/32 (25.0%) PCa and 4/14 (28.6%) csPCa, while avoiding biopsy in those with PSAD < 0.15 would miss 24/32 (75.5%) PCa and 10/14 (71.4%) csPCa diagnosed within this category.
3.3. Impact of Different Screening Strategies to Select Patients Candidate for Prostate Biopsy
Several strategies of combining and sequencing SelectMDx and mpMRI have been simulated to investigate their impact in terms of number of avoided biopsies, missed PCa and csPCa (Table 3). Limiting biopsy to men with a positive SelectMDx would result in avoiding 53.5% (166/310) of biopsies, while missing 13.5% (14/104) of PCa and 12.9% (8/62) of csPCa; performing a biopsy only in those men with a positive mpMRI (PI-RADS 4–5) would avoid 74.8% (232/310) of biopsies and miss 48.1% (50/104) of PCa and 38.7% (24/62) of csPCa. Initial SelectMDx test followed by mpMRI if the test was positive and a subsequent biopsy if the mpMRI showed PI-RADS 4–5 findings, would result in 82.6% (256/310) of biopsies avoided, yet with 53.9% (116/104) of PCa and 45.2% (28/62) of csPCa missed. Initial mpMRI followed by biopsy for positive mpMRI cases (PI-RADS 4–5) and negative mpMRI cases (PI-RADS 1–3) (only if SelectMDx was positive), would result in avoiding 45.8% (142/310) of biopsies, while only missing 7.7% (8/104) of PCa and 6.5% (6/62) of csPCa.

Table 3.
Prostate cancer (PCa) and clinically significant prostate cancer (csPCa) detection rate, avoided biopsies and missed PC and csPC among the study population according to different strategies.
4. Discussion
In order to tailor risk stratification and improve PCa detection, and particularly to reduce unnecessary prostate biopsies and diagnosis of indolent PCa, several tissues based urine and blood tests have been introduced to overcome PSA’s well-known limits [7,40,41,42,43,44,45,46,47]. Avoiding unnecessary prostate biopsies would allow for a reduction in the risk of side effects inherent to prostate biopsies, as well as associated health costs. This is especially relevant when a trans-rectal (TR) route is chosen, since evidence has suggested significantly higher infectious complications following TR biopsies, compared to a transperineal approach [48,49]. Among these tests, a 3-gene urinary panel using HOXC6, TRD1 and DLX1 were proposed in 2015 by Leyten et al. [50]. One year later, Van Neste et al. [38] reported that TRD1 was not implementing the panel and that HOXC6 and DLX1 were sufficient for prediction of positive prostate biopsy and csPCa, with a sensitivity of 91%, specificity of 36% and NPV of 93%. Recently, a large multicenter trial including 1955 patients prior to initial prostate biopsy compared performance of urinary HOXC6 and DLX1 mRNA (combined with other risk factors) with Prostate Cancer Prevention Trial Risk Calculator (PCPTRC). AUC for molecular test was 0.85 and 0.76 for PCPTRC, demonstrating high sensitivity and NPV to detect csPCa [51]. The study by Rubio-Briones et al., analyzing 492 men with PSA 3–10 ng/mL, compared 2-gene urine-based molecular test targeting mRNAs with Prostate Cancer Antigen 3 (PCA3), European Randomized Screening in Prostate Cancer (ERSPC) and Prostate Biopsy Collaborative Group (PBCG) risk calculators. Focusing on patients with a Grading Group ≥2, the test avoided 37.2% of unnecessary biopsies, while delaying the diagnosis in 1.6% of cases between all patients. The authors concluded that the test could be useful to avoid unnecessary biopsies and to identify patients most likely to benefit from prostate biopsy [52].
To date, SelectMDx is not recommended by EAU guidelines since only few clinical trials prospectively investigated its performance in patients with an initial suspicious of PCa [1].
Initial prostate biopsy, in our prospective study, was scheduled for patients selected on the basis of PSA values or DRE results, and SelectMDx score was positive in 86.5% of PCa, 87.1% of csPCa, and in 26.2% of cases with no PCa at biopsy. Compared to mpMRI, SelectMDx had the best performance in predicting PCa and csPCa after biopsy. The association of mpMRI and SelectMDx compared to the association of mpMRI and other tools (i.e., PSA and PSAD), additionally, had the best performance. Moreover, SelectMDx showed a significant association with mpMRI results in terms of PI-RADS score, as test positivity significantly increased according to PI-RADS score (p < 0.001). It should be noted, however, that mpMRI results are particularly affected by the strategy used with PI-RADS 3 lesions (if considered as positive or negative cases). Indeed, considering PI-RADS 3 score as a positive test, with respect to PCa outcome, mpMRI sensitivity and specificity increased from 51.9% (if considered as a negative test) to 82.7%, and decreased from 88.3% (if considered as a negative test) to 77.7%, respectively.
In our personal clinical experience SelectMDx demonstrated to be a good predictor of PCa. We reported that in patients before initial biopsy it could reach high levels of sensitivity and specificity for the diagnosis of all PCa (AUC 0.80), while it seems slightly less effective in detecting csPCa (AUC 0.75). Moreover, with regards to csPCa detection, SelectMDx results were similar to mpMRI. On the contrary, mpMRI demonstrated a better diagnostic performance with respect to csPCa outcome (AUC 0.73), than to PCa outcome (AUC 0.70).
According to EAU guidelines on PCa, mpMRI is recommended before performing prostate biopsy in biopsy-naïve patients with clinical suspicion of PCa [1]. In the era of mpMRI pathway, to aid clinicians in decision making (i.e., to decide whether a prostate biopsy can be omitted, in an effort to avoid unnecessary biopsy), and to improve the detection of csPCa while limiting the detection of indolent cases, combining mpMRI with serum biomarkers would be of clinical value, yet the optimal sequence and timing remains to be determined. According to our simulated strategies, limiting biopsy to men with a mpMRI PI-RADS score 4–5 would have resulted in avoiding 74.8% (232/310) of biopsies, yet missing 38.7% (24/62) of csPCa. Interestingly, implementing SelectMDx test after a mpMRI PI-RADS score 1–3, and performing a biopsy in mpMRI PI-RADS score 4–5 and in those with PI-RADS score 1–3 yet with a positive SelectMDx, would have resulted in missing only 6.5% (6/62) of csPCa, still avoiding 45.8% (142/310) of biopsies. SelectMDx could potentially lower the number of mpMRI scans and biopsies performed without increasing the risk of missing csPCa. However, our results suggest that upfront SelectMDx, although in 82.6% of avoided biopsies, is associated with a high risk of missing csPCa (45.2%).
Cases with discordant tests were analyzed to better investigate the potential added value of implementing SelectMDx in the mpMRI diagnostic pathway. With regards to PI-RADS 3 lesions, which are equivocal by nature, several possible clinical factors have been evaluated as predictors of positive biopsy [53]. In this context, SelectMDx might aid the decision-making scenario (i.e., whether a prostate biopsy versus observation should be advised). In our experience, performing a biopsy only in those with a positive SelectMDx, would result in the detection of 81.3% of PCa and 85.7% of csPCa diagnosed within this category. Differing from data reported by some authors, in our analysis, PSAD with a cut-off value of ≥0.15 to decide on the need for biopsy, did not show a clinical benefit for the detection of PCa and csPCa.
In conclusion, our analysis suggests several points of interest: (I) SelectMDx demonstrated a valid diagnostic accuracy for the detection of PCa. (II) With regards to csPCa detection, SelectMDx showed a reliable diagnostic performance, and comparable to that of mpMRI. (III) In the era of mpMRI pathway, the association of mpMRI and SelectMDx showed the best performance, compared to the association of mpMRI and other tools (i.e., PSA and PSAD). (IV) Upfront mpMRI followed by SelectMDx in cases with PI-RADS 1–3 scores to decide the need to undergo biopsy, appeared a reliable strategy to avoid unnecessary biopsies, with a reasonable csPCa detection rate. (V) Regarding the management of equivocal PI-RADS 3 lesions, SelectMDx performed better than PSAD in selecting patients for biopsy.
Our study warrants certain limitations. Due to the multicentric nature of our study, diagnostic performance data for mpMRI could be at least partially affected by variability in mpMRI-related factors (i.e., inter-reader variability, readers experience and technical performance) among the different centers included in the study. In our study all mpMRI performed have been analyzed by expert uro-radiologists, 1 to 3 per center, with minimum 5 years of experience, adopting high standards of image quality. However, a central review was not assessed. Moreover, we did not perform a cost-effectiveness analysis of SelectMDx compared to that of mpMRI, which is a main issue that should be covered before its routinely implementation in clinical practice for an extended population. To this end, although elevated costs represent a main limit, recent studies suggest that saving healthcare costs for PCa diagnosis is possible and could be done because SelectMDx quality-adjusted life years (QALYs) increase. If comparing standard of care for PCa diagnosis with SelectMDx, it could be a potentially correct cost-effective strategy [54,55,56]. On the contrary, comparing the performance of mpMRI with biomarkers, it seems that minimizing cost and maximizing effectiveness, would be the optimal strategy [48]. Future research should address the cost-effectiveness of SelectMDx compared with mpMRI and other biomarkers in order to offer to the clinicians the best diagnostic toll at the lowest price.
5. Conclusions
In our clinical experience, SelectMDx was revealed to be a good predictor of PCa, while, with regards to csPCa detection, it was demonstrated to be less effective, showing results similar to mpMRI. With analysis of strategies assessed to define the best diagnostic strategy to avoid unnecessary biopsy without missing csPCa, SelectMDx appeared after an initial negative mpMRI to be a reliable pathway. Thus, biopsy could be proposed to all cases of mpMRI PI-RADS 4–5 score, and to those with mpMRI PI-RADS 1–3 score, followed by a positive SelectMDx.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13092047/s1. Table S1: Patients’ characteristics; Figure S1: Total PSA in negative and positive PCa cases at biopsy; Total PSA distribution according to PI-RADS score at mpMRI; Figure S2: Performance of the association of mpMRI PI-RADS score with SelectMDx, PSA or PSAD evaluated as area under the curve (AUC) of the receiver operating characteristics (ROC) in predicting PCa and csPCa histological diagnosis at biopsy. Table S2: Uni- and Multivariable Logistic Regression Model predicting variables influencing detection rate of PCa and csPCa.
Author Contributions
Conceptualization, M.M., A.S. and G.M.B.; methodology, F.G.; software, M.F. and G.L.; validation, U.G.F., G.I.R.; formal analysis, M.M. and F.D.G.; investigation, R.L., G.I.R., G.S.S. and A.P.; resources, M.D.M., R.L. and G.I.; data curation, F.D.G. and F.G.; writing—original draft preparation, G.M.B.; writing—review and editing, E.D.B. and G.S.S.; visualization, F.D.G. and A.C.; supervision, P.R.C. and M.R.C.; project administration, D.T. and L.C.; funding acquisition, G.M.B. and G.C. All authors have read and agreed to the published version of the manuscript.
Funding
This paper has been published with the financial support of the Department of Medical and Surgical Sciences of the University of Foggia.
Institutional Review Board Statement
This study was conducted in line with European Urology and Good Clinical Practice guidelines, with ethical principles laid down in the latest version of the Declaration of Helsinki and in accordance with the ethical standards of the institutional research committee (Sapienza Rome University, reference number RM11916B70C64901, 07/01/2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Draisma, G.; Etzioni, R.; Tsodikov, A.; Mariotto, A.; Wever, E.; Gulati, R.; Feuer, E.; de Koning, H. Lead time and overdiagnosis in prostate-specific antigen screening: Importance of methods and context. J. Natl. Cancer Inst. 2009, 101, 374–383. [Google Scholar] [CrossRef]
- Logozzi, M.; Mizzoni, D.; Capasso, C.; Del Prete, S.; Di Raimo, R.; Falchi, M.; Angelini, D.F.; Sciarra, A.; Maggi, M.; Supuran, C.T.; et al. Plasmatic exosomes from prostate cancer patients show increased carbonic anhydrase IX expression and activity and low pH. J. Enzyme Inhib. Med. Chem. 2020, 35, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Maggi, M.; Gentilucci, A.; Salciccia, S.; Gatto, A.; Gentile, V.; Colarieti, A.; Von Heland, M.; Busetto, G.M.; Del Giudice, F.; Sciarra, A. Psychological impact of different primary treatments for prostate cancer: A critical analysis. Andrologia 2019, 51, e13157. [Google Scholar] [CrossRef] [PubMed]
- Panebianco, V.; Barchetti, F.; Sciarra, A.; Ciardi, A.; Indino, E.L.; Papalia, R.; Gallucci, M.; Tombolini, V.; Gentile, V.; Catalano, C. Multiparametric magnetic resonance imaging vs. standard care in men being evaluated for prostate cancer: A randomized study. Urol. Oncol. 2015, 33, 17.e1–17.e7. [Google Scholar] [CrossRef]
- Kasivisvanathan, V.; Rannikko, A.S.; Borghi, M.; Panebianco, V.; Mynderse, L.A.; Vaarala, M.H.; Briganti, A.; Budäus, L.; Hellawell, G.; Hindley, R.G.; et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N. Engl. J. Med. 2018, 378, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Cooperberg, M.R.; Carroll, P.R.; Dall’Era, M.A.; Davies, B.J.; Davis, J.W.; Eggener, S.E.; Feng, F.Y.; Lin, D.W.; Morgan, T.M.; Morgans, A.K.; et al. The State of the Science on Prostate Cancer Biomarkers: The San Francisco Consensus Statement. Eur. Urol. 2019, 76, 268–272. [Google Scholar] [CrossRef]
- Sciarra, A.; Maggi, M.; Salciccia, S.; Nicolai, A.; Tortorella, E.; Giantulli, S.; Magliocca, F.M.; Silvestri, I.; Taglieri, L.; Cattarino, S.; et al. Tissue Expression of Androgen Receptor Splice Variant 7 at Radical Prostatectomy Predicts Risk of Progression in Untreated Nonmetastatic Prostate Cancer. Oncology 2021, 18, 1–5. [Google Scholar]
- Catalona, W.J.; Partin, A.W.; Sanda, M.G.; Wei, J.T.; Klee, G.G.; Bangma, C.H.; Slawin, K.M.; Marks, L.S.; Loeb, S.; Broyles, D.L.; et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/mL prostate specific antigen range. J. Urol. 2011, 185, 1650–1655. [Google Scholar] [CrossRef]
- Loeb, S.; Sanda, M.G.; Broyles, D.L.; Shin, S.S.; Bangma, C.H.; Wei, J.T.; Partin, A.W.; Klee, G.G.; Slawin, K.M.; Marks, L.S.; et al. The prostate health index selectively identifies clinically significant prostate cancer. J. Urol. 2015, 193, 1163–1169. [Google Scholar] [CrossRef]
- Ferro, M.; Bruzzese, D.; Perdonà, S.; Marino, A.; Mazzarella, C.; Perruolo, G.; D’Esposito, V.; Cosimato, V.; Buonerba, C.; Di Lorenzo, G.; et al. Prostate Health Index (Phi) and Prostate Cancer Antigen 3 (PCA3) significantly improve prostate cancer detection at initial biopsy in a total PSA range of 2–10 ng/mL. PLoS ONE 2013, 8, e67687. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.; Sjoberg, D.D.; Vickers, A.J.; Lilja, H.; Bjartell, A.S. A Four-kallikrein Panel Predicts High-grade Cancer on Biopsy: Independent Validation in a Community Cohort. Eur. Urol. 2016, 69, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Nordström, T.; Vickers, A.; Assel, M.; Lilja, H.; Grönberg, H.; Eklund, M. Comparison Between the Four-kallikrein Panel and Prostate Health Index for Predicting Prostate Cancer. Eur. Urol. 2015, 68, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Cronin, A.M.; Aus, G.; Pihl, C.; Becker, C.; Pettersson, K.; Scardino, P.T.; Hugosson, J.; Lilja, H. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: Data from the European Randomized Study of Prostate Cancer Screening in Göteborg, Sweden. BMC Med. 2008, 6, 19. [Google Scholar] [CrossRef]
- Leyten, G.H.; Hessels, D.; Jannink, S.A.; Smit, F.P.; de Jong, H.; Cornel, E.B.; de Reijke, T.M.; Vergunst, H.; Kil, P.; Knipscheer, B.C.; et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur. Urol. 2014, 65, 534–542. [Google Scholar] [CrossRef]
- Ochiai, A.; Okihara, K.; Kamoi, K.; Oikawa, T.; Shimazui, T.; Murayama, S.; Tomita, K.; Umekawa, T.; Uemura, H.; Miki, T. Clinical utility of the prostate cancer gene 3 (PCA3) urine assay in Japanese men undergoing prostate biopsy. BJU Int. 2013, 111, 928–933. [Google Scholar] [CrossRef]
- Busetto, G.M.; Del Giudice, F.; Maggi, M.; De Marco, F.; Porreca, A.; Sperduti, I.; Magliocca, F.M.; Salciccia, S.; Chung, B.I.; De Berardinis, E.; et al. Prospective assessment of two-gene urinary test with multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy. World J. Urol. 2020. [Google Scholar] [CrossRef]
- De la Calle, C.M.; Fasulo, V.; Cowan, J.E.; Lonergan, P.E.; Maggi, M.; Gadzinski, A.J.; Yeung, R.A.; Saita, A.; Cooperberg, M.R.; Shinohara, K.; et al. Clinical Utility of 4Kscore®, ExosomeDx™ and Magnetic Resonance Imaging for the Early Detection of High Grade Prostate Cancer. J. Urol. 2021, 205, 452–460. [Google Scholar] [CrossRef]
- Leapman, M.S.; Nguyen, H.G.; Cooperberg, M.R. Clinical Utility of Biomarkers in Localized Prostate Cancer. Curr. Oncol. Rep. 2016, 18, 30. [Google Scholar] [CrossRef]
- Klein, E.A.; Cooperberg, M.R.; Magi-Galluzzi, C.; Simko, J.P.; Falzarano, S.M.; Maddala, T.; Chan, J.M.; Li, J.; Cowan, J.E.; Tsiatis, A.C.; et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy under sampling. Eur. Urol. 2014, 66, 550–560. [Google Scholar] [CrossRef]
- Tosoian, J.J.; Chappidi, M.R.; Bishoff, J.T.; Freedland, S.J.; Reid, J.; Brawer, M.; Stone, S.; Schlomm, T.; Ross, A.E. Prognostic utility of biopsy-derived cell cycle progression score in patients with National Comprehensive Cancer Network low-risk prostate cancer undergoing radical prostatectomy: Implications for treatment guidance. BJU Int. 2017, 120, 808–814. [Google Scholar] [CrossRef]
- Roach, M., 3rd; Hanks, G.; Thames, H., Jr.; Schellhammer, P.; Shipley, W.U.; Sokol, G.H.; Sandler, H. Defining biochemical failure following radiotherapy with or without hormonal therapy in men with clinically localized prostate cancer: Recommendations of the RTOG-ASTRO Phoenix Consensus Conference. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Cooperberg, M.R.; Simko, J.P.; Cowan, J.E.; Reid, J.E.; Djalilvand, A.; Bhatnagar, S.; Gutin, A.; Lanchbury, J.S.; Swanson, G.P.; Stone, S.; et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J. Clin. Oncol. 2013, 31, 1428–1434. [Google Scholar] [CrossRef] [PubMed]
- Erho, N.; Crisan, A.; Vergara, I.A.; Mitra, A.P.; Ghadessi, M.; Buerki, C.; Bergstralh, E.J.; Kollmeyer, T.; Fink, S.; Haddad, Z.; et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2013, 8, e66855. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.G.; Chang, S.L.; Spratt, D.E.; Erho, N.; Yu, M.; Ashab, H.A.; Alshalalfa, M.; Speers, C.; Tomlins, S.A.; Davicioni, E.; et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: A matched, retrospective analysis. Lancet Oncol. 2016, 17, 1612–1620. [Google Scholar] [CrossRef]
- Scattoni, V.; Lazzeri, M.; Lughezzani, G.; De Luca, S.; Passera, R.; Bollito, E.; Randone, D.; Abdollah, F.; Capitanio, U.; Larcher, A.; et al. Head-to-head comparison of prostate health index and urinary PCA3 for predicting cancer at initial or repeat biopsy. J. Urol. 2013, 190, 496–501. [Google Scholar] [CrossRef]
- Boegemann, M.; Stephan, C.; Cammann, H.; Vincendeau, S.; Houlgatte, A.; Jung, K.; Blanchet, J.; Semjonow, A. The percentage of prostate-specific antigen (PSA) isoform [-2]proPSA and the Prostate Health Index improve the diagnostic accuracy for clinically relevant prostate cancer at initial and repeat biopsy compared with total PSA and percentage free PSA in men aged ≤65 years. BJU Int. 2016, 117, 72–79. [Google Scholar]
- Stephan, C.; Jung, K.; Semjonow, A.; Schulze-Forster, K.; Cammann, H.; Hu, X.; Meyer, H.; Bögemann, M.; Miller, K.; Friedersdorff, F. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin. Chem. 2013, 59, 280–288. [Google Scholar] [CrossRef]
- Gupta, A.; Roobol, M.J.; Savage, C.J.; Peltola, M.; Pettersson, K.; Scardino, P.T.; Vickers, A.J.; Schröder, F.H.; Lilja, H. A four-kallikrein panel for the prediction of repeat prostate biopsy: Data from the European Randomized Study of Prostate Cancer screening in Rotterdam, Netherlands. Br. J. Cancer 2010, 103, 708–714. [Google Scholar] [CrossRef]
- Auprich, M.; Augustin, H.; Budäus, L.; Kluth, L.; Mannweiler, S.; Shariat, S.F.; Fisch, M.; Graefen, M.; Pummer, K.; Chun, F.K.H. A comparative performance analysis of total prostate-specific antigen, percentage free prostate-specific antigen, prostate-specific antigen velocity and urinary prostate cancer gene 3 in the first, second and third repeat prostate biopsy. BJU Int. 2012, 109, 1627–1635. [Google Scholar] [CrossRef]
- Haese, A.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.A.; Huland, H.; Abbou, A.; Remzi, M.; Tinzl, M.; et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur. Urol. 2008, 54, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.K.; Reese, A.C.; Cooperberg, M.R.; Sadetsky, N.; Shinohara, K. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis. 2012, 15, 100–105. [Google Scholar] [CrossRef]
- Stewart, G.D.; Van Neste, L.; Delvenne, P.; Delrée, P.; Delga, A.; McNeill, S.A.; O’Donnell, M.; Clark, J.; Van Criekinge, W.; Bigley, J.; et al. Clinical utility of an epigenetic assay to detect occult prostate cancer in histopathologically negative biopsies: Results of the MATLOC study. J. Urol. 2013, 189, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Uhr, A.; Glick, L.; Gomella, L.G. An overview of biomarkers in the diagnosis and management of prostate cancer. Can. J. Urol. 2020, 27, 24–27. [Google Scholar]
- Weinreb, J.C.; Barentsz, J.O.; Choyke, P.L.; Cornud, F.; Haider, M.A.; Macura, K.J.; Margolis, D.; Schnall, M.D.; Shtern, F.; Tempany, C.M.; et al. PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2. Eur. Urol. 2016, 69, 16–40. [Google Scholar] [CrossRef]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, R.J.; van der Leest, M.M.G.; Dijkstra, S.; Barentsz, J.O.; Criekinge, W.V.; Hulsbergen-van de Kaa, C.A.; Schalken, J.A.; Mulders, P.F.A.; van Oort, I.M. A urinary biomarker-based risk score correlates with multiparametric MRI for prostate cancer detection. Prostate 2017, 77, 1401–1407. [Google Scholar] [CrossRef]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker-Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Carlsson, S.V.; Roobol, M.J. Improving the evaluation and diagnosis of clinically significant prostate cancer in 2017. Curr. Opin. Urol. 2017, 27, 198–204. [Google Scholar] [CrossRef]
- Sciarra, A.; Gentilucci, A.; Silvestri, I.; Salciccia, S.; Cattarino, S.; Scarpa, S.; Gatto, A.; Frantellizzi, V.; Von Heland, M.; Ricciuti, G.P.; et al. Androgen receptor variant 7 (AR-V7) in sequencing therapeutic agents for castratrion resistant prostate cancer: A critical review. Medicine 2019, 98, e15608. [Google Scholar] [CrossRef] [PubMed]
- Busetto, G.M.; Giovannone, R.; Antonini, G.; Rossi, A.; Del Giudice, F.; Tricarico, S.; Ragonesi, G.; Gentile, V.; De Berardinis, E. Short-term pretreatment with a dual 5α-reductase inhibitor before bipolar transurethral resection of the prostate (B-TURP): Evaluation of prostate vascularity and decreased surgical blood loss in large prostates. BJU Int. 2015, 116, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Flammia, S.; Frisenda, M.; Maggi, M.; Magliocca, F.M.; Ciardi, A.; Panebianco, V.; De Berardinis, E.; Salciccia, S.; Di Pierro, G.B.; Gentilucci, A.; et al. Cribriform pattern does not have a significant impact in Gleason Score ≥7/ISUP Grade ≥2 prostate cancers submitted to radical prostatectomy. Medicine 2020, 99, e22156. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Musi, G.; Matei, D.V.; Mistretta, A.F.; Luzzago, S.; Cozzi, G.; Bianchi, R.; Di Trapani, E.; Cioffi, A.; Lucarelli, G.; et al. Assessment of PSIM (Prostatic Systemic Inflammatory Markers) Score in Predicting Pathologic Features at Robotic Radical Prostatectomy in Patients with Low-Risk Prostate Cancer Who Met the Inclusion Criteria for Active Surveillance. Diagnostics 2021, 11, 355. [Google Scholar] [CrossRef] [PubMed]
- Ferro, M.; Lucarelli, G.; de Cobelli, O.; Del Giudice, F.; Musi, G.; Mistretta, F.A.; Luzzago, S.; Busetto, G.M.; Buonerba, C.; Sciarra, A.; et al. The emerging landscape of tumor marker panels for the identification of aggressive prostate cancer: The perspective through bibliometric analysis of an Italian translational working group in uro-oncology. Minerva Urol. Nephrol. 2021. [Google Scholar] [CrossRef]
- Sciarra, A.; Maggi, M.; Del Proposto, A.; Magliocca, F.M.; Ciardi, A.; Panebianco, V.; De Berardinis, E.; Salciccia, S.; Di Pierro, G.B.; Gentilucci, A.; et al. Impact of uni- or multifocal perineural invasion in prostate cancer at radical prostatectomy. Transl. Androl. Urol. 2021, 10, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Cerrato, A.; Bedia, C.; Capriotti, A.L.; Cavaliere, C.; Gentile, V.; Maggi, M.; Montone, C.M.; Piovesana, S.; Sciarra, A.; Tauler, R.; et al. Untargeted metabolomics of prostate cancer zwitterionic and positively charged compounds in urine. Anal. Chim. Acta 2021, 1158, 338381. [Google Scholar] [CrossRef]
- Busetto, G.M.; Giovannone, R.; Ferro, M.; Tricarico, S.; Del Giudice, F.; Matei, D.V.; De Cobelli, O.; Gentile, V.; De Berardinis, E. Chronic bacterial prostatitis: Efficacy of short-lasting antibiotic therapy with prulifloxacin (Unidrox®) in association with saw palmetto extract, lactobacillus sporogens and arbutin (Lactorepens®). BMC Urol. 2014, 14, 53. [Google Scholar] [CrossRef]
- Porreca, A.; D’Agostino, D.; Romagnoli, D.; Del Giudice, F.; Maggi, M.; Palmer, K.; Falabella, R.; De Berardinis, E.; Sciarra, A.; Ferro, M.; et al. The Clinical Efficacy of Nitrofurantoin for Treating Uncomplicated Urinary Tract Infection in Adults: A Systematic Review of Randomized Control Trials. Urol. Int. 2021, 1–10. [Google Scholar] [CrossRef]
- Leyten, G.H.; Hessels, D.; Smit, F.P.; Jannink, S.A.; de Jong, H.; Melchers, W.J.; Cornel, E.B.; de Reijke, T.M.; Vergunst, H.; Kil, P.; et al. Identification of a Candidate Gene Panel for the Early Diagnosis of Prostate Cancer. Clin. Cancer Res. 2015, 21, 3061–3070. [Google Scholar] [CrossRef]
- Haese, A.; Trooskens, G.; Steyaert, S.; Hessels, D.; Brawer, M.; Vlaeminck-Guillem, V.; Ruffion, A.; Tilki, D.; Schalken, J.; Groskopf, J.; et al. Multicenter Optimization and Validation of a 2-Gene mRNA Urine Test for Detection of Clinically Significant Prostate Cancer before Initial Prostate Biopsy. J. Urol. 2019, 202, 256–263. [Google Scholar] [CrossRef]
- Rubio-Briones, J.; Borque-Fernando, A.; Esteban, L.M.; Mascarós, J.M.; Ramírez-Backhaus, M.; Casanova, J.; Collado, A.; Mir, C.; Gómez-Ferrer, A.; Wong, A.; et al. Validation of a 2-gene mRNA urine test for the detection of ≥GG2 prostate cancer in an opportunistic screening population. Prostate 2020, 80, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Maggi, M.; Panebianco, V.; Mosca, A.; Salciccia, S.; Gentilucci, A.; Di Pierro, G.; Busetto, G.M.; Barchetti, G.; Campa, R.; Sperduti, I.; et al. Prostate Imaging Reporting and Data System 3 Category Cases at Multiparametric Magnetic Resonance for Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Focus 2020, 6, 463–478. [Google Scholar] [CrossRef]
- Dijkstra, S.; Govers, T.M.; Hendriks, R.J.; Schalken, J.A.; Van Criekinge, W.; Van Neste, L.; Grutters, J.P.C.; Sedelaar, J.P.M.; van Oort, I.M. Cost-effectiveness of a new urinary biomarker-based risk score compared to standard of care in prostate cancer diagnostics—A decision analytical model. BJU Int. 2017, 120, 659–665. [Google Scholar] [CrossRef]
- Govers, T.M.; Hessels, D.; Vlaeminck-Guillem, V.; Schmitz-Dräger, B.J.; Stief, C.G.; Martinez-Ballesteros, C.; Ferro, M.; Borque-Fernando, A.; Rubio-Briones, J.; Sedelaar, J.P.M.; et al. Cost-effectiveness of SelectMDx for prostate cancer in four European countries: A comparative modeling study. Prostate Cancer Prostatic Dis. 2019, 22, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Sathianathen, N.J.; Kuntz, K.M.; Alarid-Escudero, F.; Lawrentschuk, N.L.; Bolton, D.M.; Murphy, D.G.; Weight, C.J.; Konety, B.R. Incorporating Biomarkers into the Primary Prostate Biopsy Setting: A Cost-Effectiveness Analysis. J. Urol. 2018, 200, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).