Abstract
Although improvement in early diagnosis and treatment ameliorated life expectancy of cancer patients, metastatic disease still lacks effective therapeutic approaches. Resistance to anticancer therapies stems from the refractoriness of a subpopulation of cancer cells—termed cancer stem cells (CSCs)—which is endowed with tumor initiation and metastasis formation potential. CSCs are heterogeneous and diverge by phenotypic, functional and metabolic perspectives. Intrinsic as well as extrinsic stimuli dictated by the tumor microenvironment (TME)have critical roles in determining cell metabolic reprogramming from glycolytic toward an oxidative phenotype and vice versa, allowing cancer cells to thrive in adverse milieus. Crosstalk between cancer cells and the surrounding microenvironment occurs through the interchange of metabolites, miRNAs and exosomes that drive cancer cells metabolic adaptation. Herein, we identify the metabolic nodes of CSCs and discuss the latest advances in targeting metabolic demands of both CSCs and stromal cells with the scope of improving current therapies and preventing cancer progression.
1. Introduction
Despite significant advances in cancer prevention and treatment, metastatic disease is mostly incurable and resistant to common therapeutics.
The development of cancer depends on a small pool of tumor cells owing a phenotype comparable to normal adult stem cells, called cancer stem cells (CSCs), which are characterized by self-renewal and multilineage differentiation capabilities, as well as a great ability to initiate and promote tumorigenesis, metastasis formation and anticancer therapy resistance [1]. CSC features are also dictated by paracrine interactions occurring between tumor cells and their neighboring tumor microenvironment (TME), mainly composed by adipocytes, cancer-associated fibroblasts (CAFs), endothelial and immune cells. CAFs constitute the predominant cellular portion of TME thereby encouraging tumor expansion and progression by providing cytokines, growth factors, metabolites and extracellular matrix remodeling proteins [2,3].
Altered metabolism is a common feature of cancer. Normally, non-cancerous cells mainly catabolize glucose by oxidative phosphorylation (OXPHOS) in the mitochondria to produce ATP [4]. However, proliferating cancer cells metabolize significant amounts of glucose into lactate, even in the presence of normal oxygen, a phenomenon which is known as “Warburg effect” [5]. The preference to utilize glycolysis offers several advantages to cancer cells including adaptation in hypoxic and acidic environments (lactate production), which help cancer cells to rapidly proliferate and invade and surrounding tissues [6].
However, cancer cells are not characterized by a unique metabolic profile. Both intrinsic and extrinsic factors, such as a nutrient-poor microenvironment, have critical roles in determining cell metabolic phenotypes.
Indeed, it is well-established that CSCs can make use of their fully proficient mitochondrial respiration capacity, to face scarce nutrients supply or a hostile microenvironment, with a process named “reverse Warburg effect” [7].
CSCs rely on the release of nutrients, such as amino acids and fatty acids (FAs), by TME stromal cells, to accomplish the biosynthetic and energy requirements necessary to proliferate and metastasize even in a low oxygen microenvironment. One conceivable mechanism of interchange between neoplastic and stromal cells is represented by the release of miRNAs and exosomes, small particles containing nucleic acids (DNA, mRNA and miRNA), metabolites and inflammatory factors [8]. Moreover, exosomes and their cargo are in charge for the establishment of the pre-metastatic niche and, importantly, may contribute to cancer treatment failure [9,10].
Increasing the knowledge about the heterogeneity of CSC’s metabolic demands may be an essential tool for designing novel personalized therapeutic approaches aimed at enhancing the efficacy of treatments by restricting tumor metabolic adaptation.
Here, we will focus on the molecular mechanisms associated with both establishment and maintenance of CSC metabolic phenotypes listed in Table 1, and the biological processes that influence cell metabolic choices including hypoxia, reactive oxygen species (ROS), epithelial tomesenchymal transition (EMT) and the administration of anticancer therapies.

Table 1.
Metabolic phenotypes of cancer stem cells in various cancer models.
2. How Foodie Can Be Cancer Stem Cells? Glucose as the Main Metabolic Fuel
One of the most appointed metabolic hallmark of cancer cells was postulated by Otto Warburg in the early twentieth century [11]. The ‘Warburg effect’ foresees the conversion of pyruvate to lactate, from glucose, under aerobic conditions with a consequent predominance of glycolysis on OXPHOS [12]. This metabolic reprogramming has been initially ascribed to defects on mitochondrial function [13]. According to Warburg an injury on mitochondria respiration is at the base of the oncogenic transformation and the shift from OXPHOS toward glycolysis is persistent in the cancer cell progeny [13]. Recently, in a model of colorectal cancer it has been proved that tumor initiation is sustained by an enhancement of the glycolytic program in a condition in which the mitochondrial pyruvate carrier is inactivated, causing low pyruvate import, necessary for oxidative metabolism, onto the mitochondria. In vivo loss of mitochondrial pyruvate carrier 1 increased high-grade adenoma formation and it was fundamental for the subsistence of tumor initiating cells at the base of colon crypt [14].
Stem and cancer cells prefer glycolysis because it perfectly meets their metabolic requirements [15,16]. Notwithstanding glycolysis is an inefficient process with respect to ATP production, cancer cells uptake high quantity of glucose that enters glycolysis pathway and generates a comparable number of ATP molecules as OXPHOS [17]. Glycolysis is fundamental for cell bioenergetics as well as for the generation of metabolic intermediates used to produce NADPH, lipid or glycogen molecules. For instance, ribose-5-phosphate groups derived from glucose-6-phosphate are used for nucleotide biosynthesis [18]. In this context an enhanced nucleotide production, as it is the case in cancer cells, leads to a decrease in the NAD+/NADH ratio, which in turn encourages the conversion of glucose to lactate [19].
Glycolysis counteracts the formation of reactive oxygen species (ROS) in proliferating cells [28]. Luo et al. demonstrated that the inhibition of glycolysis by 2-deoxyglucose (2-DG) induced the transition of mesenchymal breast CSCs harboring low levels of ROS toward a ROShigh phenotype of epithelial breast CSCs, which are sensitive to auranofin and L-buthionine-sulfoximine (BSO), two inhibitors of the antioxidant pathways thioredoxin (TXN) and glutathione (GSH), respectively [27,29]. The latter study clearly indicates that despite glycolysis plays a central metabolic role, cancer cells modulate their metabolic phenotype in response to metabolic stress, including the possibility to switch to OXPHOS and vice versa if needed [30].
Indeed, pancreatic CSCs harboring high level of the peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC-1α) rely on OXPHOS and diverge from the glycolytic phenotype of their differentiated counterpart. Mitochondrial inhibition by metformin causes the selection of a resistant subtype of MYC-expressing pancreatic CSCs showing an intermediate glycolytic/oxidative metabolic phenotype [24].
3. The Mitostemness: Fiction or Reality?
A conspicuous entity of studies shows that cancer cells are characterized by high energy demand. According to the “Warburg effect” cancer cell sustenance is solely glycolytic.Nowadays we know that although the Warburg phenotype represents an undisputed feature of many cancer cells, most tumor cells possess intact tricarboxylic acid (TCA) cycle and OXPHOS [31,32].
The oxidative metabolism allows CSCs to have selective advantages, greater resistance to the inhibition of glycolysis, enhanced degree of independence from the supply of macronutrients and also a more efficient source for energy production [20,33]. In order to emphasize the central role of mitochondria in the self-renewal of CSCs and in their resistance to differentiation, two years ago it was coined the term “mitostemness” [34,35].
Several studies have highlighted that CSCs are endowed with a plastic metabolic phenotype, both glycolytic [36] and oxidative through the OXPHOS [37]. The latter process is mediated by electron transport chain (ETC) and involves supply of reducing equivalents from TCA cycle or fatty acid oxidation (FAO) for ATP and NADPH generation.
Janiszewska M. et al. demonstrated that the subpopulation of tumor-initiating gliomaspheres depends on OXPHOS for energy production. In particular, the oncofetal insulin-like growth factor 2 mRNA-binding protein 2, which regulates the protein synthesis and contributes to assembly of the subunits of ETC, sustained glioblastoma cell clonogenicity [21].
Mitochondrial metabolism is fundamental in CSCs because it constitutes a source of intermediate metabolites such as acetyl-coA, NAD+, alpha-ketoglutarate, succinate, FAD, S-adenosylmethionine. The variation in the concentration of these intermediate metabolites could lead to important phenotypic changes because they can be used by enzymes capable of modifying the state of the chromatin [38] and thus the transcriptomic profile of cancer cells. There are various ways to modify chromatin, for example, acetyl-coA is used to transfer acetyl group to lysine residues of histones [39] and S-adenosylmethionine is a donor of methyl group for the methylation of DNA and histones [40]. Histone acetylation, mediated by histone acetyl transferases, increases gene expression [41]. The proteins involved in the cytosine methylation of CpG dinucleotides are the DNA methyl transferases DNMT1, DNMT3A and DNMT3B and they have been identified as factors driving the CSCs formation and maintenance [42]. Histone methylation occurs predominantly on lysine (K) and arginine (R) residues. Such modifications are commonly associated with gene activation or repression, depending on the target histone modification [43]. Interestingly, the enhancer of zeste homolog 2 (EZH2), which is the catalytic subunit of polycomb repressive complex 2, has been demonstrated to downregulate gene transcription through trimethylation of histone H3 on lysine 27 [44], while it upregulates NOTCH expression, being crucial in the expansion of the stem pool in breast cancer [45].
Given the discovery of functional mitochondria in CSCs, it was hypothesized that the capability to rapid switch from glycolytic metabolism toward oxidative metabolism [46] and vice versa, could represent an escape mechanism to standard anticancer therapies. Therefore, using drugs that selectively target mitochondria could make CSCs more sensitive to standard therapies.
PGC-1α is a stress sensor, activated following the presence of limited amount of nutrients, oxidative stress and chemotherapy, to increase mitochondrial biogenesis and therefore all mitochondrial activities (OXPHOS, FAO and detoxification ROS) [47]. The XCT790 is an inhibitor of estrogen-related receptor α, the latter being a transcription factor coupled with PGC-1α. In this context, De Luca et al. have shown that the use of XCT790 reduces the proliferation of CSCs in breast cancer [48].
One of the most studied ETC inhibitor to target CSCs is the antidiabetic drug metformin, which targets the complex I in ETC in mitochondria. Metformin acts as driver of apoptosis in CD133+ pancreatic ductal adenocarcinomas cells and CD44highCD24lowbreast cancer spheres [24,49]. Promising clinical data havebeen reported for the use of metformin in breast, endometrial, prostate and pancreatic cancers (NCT01266486; NCT02755844; NCT01620593; NCT02978547) [50,51]. For instance, a retrospective study conducted on 445 patients affected by neuroendocrine pancreatic tumors revealed that progression free survival was doubled in diabetic patients receiving metformin than patients without diabetes (median progression free survival, 32.0 months versus 15.1 months) [51].
Another important mitochondrial target is represented by mitochondrial ribosomes, which are the site of protein synthesis. The homology existing between mammalian mitochondrial ribosomes and prokaryotic ribosomes is well established, for this reason it is possible to use a wide variety of antibiotics already approved by the FDA [52]. Ribosomes are targeted by the two main families of antibiotics, tetracycline and macrolide [53,54]. Among the wide range of antibiotics, tested by Lamb et al. doxycycline was found to inhibit mammosphere formation in breast cancer and many different tumor types [52]. Therefore, this therapeutic approach could be applicable to many cancer types.
Notwithstanding inside the cell there are various production sites of ROS, mitochondrial ETC represents their primary endogenous source [55]. The intracellular increase of ROS may result from the activation of oncogenes, inactivation of tumor suppressor genes, enhanced oxidative metabolism and mitochondrial dysfunction [56]. The accumulation of ROS causes damage to proteins, lipids and DNA and therefore irrecoverable damage to the cell [27]. Notably, cells have protection mechanisms from oxidative stress, including enzymatic antioxidants such as superoxide dismutase (SOD), catalases, TXN, peroxiredoxins, glutathione (GSH) peroxidases, p38-MAPK and sirtuins [57,58,59] and non-enzymatic molecules such as GSH, vitamin C (ascorbate), vitamin E (tocopherols) and polyphenols [60]. Interestingly, the administration of SOD-1 inhibitor, disulfiram, strongly counteracted breast CSCs expansion by modulating MAPK, NF-kB and STAT3 pathways [61,62].
Although it has been shown that there is an increase in mitochondrial functionality in CSCs compared to other cancer cells [48], it appears that there are low concentrations of ROS within CSCs [63,64] as there is a high production of antioxidant agents that maintain the disposal of ROS, protecting CSCs from oxidative damage [65]. The excessive increase in ROS levels reduced clonogenicity and mediated death of CSCs [66]. Moreover, the use of paraquat, an inducer of ROS, increased ROS production and significantly reduced tumor spheroid formation of CSCs [16].
Albeit several authors have shown that CSCs harbor high ROS levels, further studies clarified their role in cancer onset and progression. In particular, ROS facilitates CSCs self-renewal, expansion, invasiveness [67,68,69] and potentiated tumor progression [70,71].
Myant et al. highlighted the connection between Rac1, NF-kB and ROS in CSCs, in which ROS act as connecting molecules between Rac1 and NF-kB. Rac1 is required for NF-kB activation and initiation of colon tumorigenesis. Rac1 is also part of a protein complex containing NADPH oxidase, which generates superoxide, a major constituent of the pool of intracellular ROS. On the other hand, NF-kB is a redox-sensitive transcription factor that is activated by increased levels of ROS. In this study, the authors showed that increased NADPH oxidase activity, ROS intracellular concentration and NF-kB signaling in the APC-inactivated intestinal cells are all dependent on Rac1 expression [68,72].
Within tumor bulk, CSCs represent the subpopulation with the highest degree of adaptability to redox stress. The GSH system, adopted by CSCs, is the major antioxidant defense mechanism that reduces the intracellular levels of ROS. Gln-derived glutamate (Glu) is a necessary substrate for the synthesis of GSH and an exchange ion for the cellular import of cystine, which cooperates with Glu for the production of GSH, through the CD44v-xCT transporter [73]. Cells showing stem-like features strongly rely on mitochondrial OXPHOS sustained by glutamine (Gln) catabolism [23]. Of note, cancer cells are metabolically heterogeneous, since they can also retain the OXPHOS capacity and replace glucose with Gln or fatty acids (FAs), as energy supply. Despite being a non-essential amino acid, Gln is considered as a ‘conditionally essential’ amino acid since its de novo synthesis does not satisfy the demands of cancer cell, which become reliant on its exogenous uptake. Gln is also required for TCA cycle intermediate replenishment, protein translation and the biosynthesis of amino acids and nucleotides [74].
Interestingly, metformin resistance can be mediated by Gln compensation. In human cancer organoids and in in vivo model it was described that the administration of metformin and a glutaminase inhibitor (GLSi), which blocks Gln consumption, efficiently overcome metformin resistance [75]. Glutaminase activity also regulates head and neck cancer metabolism though the expression of the stemness marker ALDH [25].
It has been shown that survival of lapatinib-resistant breast cancer cells is supported by Gln metabolism [76], whose inhibition could be exploited for sensitizing cancer cells to standard therapies. Notably, GLSi have been recently launched to Phase II in combination with paclitaxel for the treatment of triple negative breast cancer patients (NCT03057600).
Recently, different clinical studies demonstrated that leukemia stem cells are greedy of amino acids and possess enhanced catabolic activity [26]. Some amino acids, such as methionine, influence the epigenetic state of cancer cells and fuel tumor initiation [77]. In addition, glycine decarboxylase, belonging to the serine–glycine pathway, is the metabolic driving force of tumor-initiating cells in non-small cell lung cancer [22]. Metabolic targeting of amino acids resulted often in neoplasia regression [77]. However, phenomena of cancer metabolic compensation, mediated by FA oxidation, have been observed [26] (Figure 1).
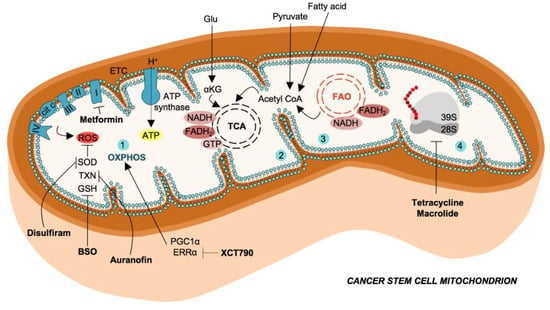
Figure 1.
Mitochondrial metabolism in cancer stem cells and therapeutic strategies.Cancer stem cells exhibit enhanced catabolism of glutamate, pyruvate and fatty acids (FAs) to generate intermediate metabolites, which converge in tricarboxylic acid (TCA) cycle. The oxidative phosphorylation (OXPHOS) (1), mediated by the electron transport chain (ETC), use the reducing equivalents (FADH2 and NADH) produced by tricarboxylic acid (TCA) cycle (2) and fatty acids oxidation (FAO) (3), with the scope of producing ATP molecules. Mitochondrial ETC is also the main endogenous source of reactive oxygen species (ROS), which possess several scavenger enzymes and molecules in CSCs such as superoxide dismutase (SOD), thioredoxin (TXN) and glutathione (GSH). These antioxidant mechanisms are targeted by Disulfiram, auranofin and L-buthionine-sulfoximine (BSO), respectively. Cancer stem cells possess a high number of mitochondria, mitochondrial ribosomes, necessary for protein synthesis are targetable with antibiotics such as tetracyclines and macrolides (4).
4. Fatty Acids as a Metabolic Reservoir for Cancer Cell Plasticity
As we have already extensively discussed in this review, the ability to adjust cell metabolism is crucial in cancer cells, because it supports the neoplastic proliferation and survival during the different steps of cancer progression [78]. Given the high plasticity of CSCs, this phenomenon is highly pronounced in this cell subset, in particular regarding FA metabolism, which is fundamental to sustain their boosted growth, division and survival during cancer progression. FAs play an important role in different aspects of cancer cell life, being crucial for cell membranes assembly, acting as structural components of the extracellular matrix, secondary messengers and energy source [79]. FAs abundance is regulated by two different processes, de novo biosynthesis mediated by nutrients as glucose and amino acids, and exogenous uptake thanks to the presence of specific transporters, both mechanisms are finely regulated at different levels during cancer progression by the dynamic TME components and dysregulation of oncogenic signaling pathways (reviewed in [80]). Importantly, in 1984, Ookhtens and colleagues demonstrated in vivo that most of the esterified FAs in tumors were derived from de novobiosynthesis [81], as confirmed few years later by Kuhajdaet al., who showed for the first time that tumor cells (breast cancer in this study) over-express fatty acid synthase (FASN) [82]. These findings were also demonstrated in glioma, breast and pancreatic CSCs, where FASN expression dictated the acquisition of stemness features [83,84,85,86].
Lipid droplets (LDs) are dynamic and multifunctional cytoplasmic organelles involved in the storage of FAs, which are fundamental to avoid lipotoxicity when cells start accumulating an excess of FAs, in particular under hypoxic conditions that induce an upregulation of FAs uptake, but also to provide an important source of ATP and NADH under stress. The generation of energy from LDs is achieved by β-oxidation of stored lipids that is sufficient to provide ATP during the metastatic cascade and essential for production of NADPH useful as detox agent against ROS [87,88]. The role of LDs biogenesis and their regulated mechanisms in cancer cells has been widely described over the last decade, and observed in all the phases of cancer development, including initiation, promotion and progression [89,90] in particular in CSCs from different organs, including colon cancer [91,92]. These findings suggest that LD biogenesis could be considered a promising target for designing innovative antitumor therapeutic strategies [93,94].
In addition to de novo synthesis, FAs can be collected from exogenous environment through the expression of specialized transporters that facilitate the efficient movement across the plasma membrane. The best characterized include CD36, also known as fatty acid translocase (FAT), the fatty acid transport protein family (FATPs) and the fatty acid-binding protein (FABPs), which have been shown to be upregulated in tumors [95,96]. In particular, CD36 has been demonstrated to be crucial for metastatic dissemination of cancer cell in different tumor type, with its high expression being correlated with poor prognosis [87]. Accordingly, targeting of CD36 has shown pre-clinical evidence that its inhibition is sufficient to impair metastases [87]. Interestingly, the expression of CD36 in cancer cells is also able to regulate the metabolic crosstalk with TME by increasing their dependency on exogenous lipid uptake. Lipid uptake from cancer cells becomes crucial mostly under condition of metabolic stress. Indeed, it has been shown that in hypoxia, the glucose to acetyl-CoA flux is downregulated, as well as unsaturation of FA that is driven by stearoyl-CoA desaturase-1 SCD-1, which is an oxygen-consuming enzyme. To compensate this prohibitive microenvironment, during hypoxia cancer cells upregulate FABP4 [97], which is a target of the hypoxia-induced factor 1 alpha (HIF1α) [98]. Notably, FABP4 negatively correlated with progression-free survival [99].
Adipose tissue cells establish symbiotic relationship with cancer cells. Interestingly, some cancers show preferential homing to adipose tissue [100]. Adipocytes sustain cancer cell growth by activating lipolytic processes with consequent release of free FAs that are up taken by cancer cells over-expressing FABP4, which in turn release exosomes containing pro-lipolytic factors (i.e., miR-144 and miR-126) [101,102]. We can distinguish two different populations of cancer adipocytes, the “peritumoral adipocytes” that do not penetrate tumor tissue (surrounding tumor tissue, tumor-educated adipose cells can be also found distant from tumor growth [103]), and the “metastasis-associated adipocytes”, which arise from the transient or prolonged contact between metastatic cancer cells and resident adipose cells. The most important mechanism involved in cancer cell metabolic reprogramming, is driven by cancer cell-driven lipolysis in adipose cells [104]. Indeed, it has been demonstrated that following prolonged exposure of cancer cells to adipose cells, the latter start losing their lipid content (responsible for tumor cachexia), gaining fibroblast-like phenotype, which further promotes the metastatic behavior of cancer cells. Recently, Ye et al. demonstrated that leukemic stem cells use gonadal adipose tissue as niche to support their growth and to protect themselves from chemotherapy, by inducing an inflammatory microenvironment and adipose tissue lipolysis [105]. Interestingly, the most beneficial effect was gained by leukemic stem cells over-expressing the FA transporter CD36 [105].
Another important source of heterogeneity in the metabolic signature of cancer cells is due to the genetic background since several metabolic processes are directly regulated by constitutive activation of oncogenes and deactivation of tumor suppressor genes [106]. In particular, oncogenes as RAS and PI3K are usually associated with glycolysis over OXPHOS, while tumor suppressor genes, as p53, have opposite effects [107].
One of the most important oncogenes in cancers is RAS. Indeed, constitutive activation of KRAS is frequent in colon and non-small cell lung cancer [108]. KRAS by activating ERK1/2 reprogramscancer cell lipid metabolism, leading to increased synthesis of glycerophospholipids and FAs [109,110]. G12V mutated KRAS is also able to boost de novo lipogenesis [111].
PI3K is one of the most frequently dysregulated pathways in cancers. Constitutive activation of this signaling regulates de novo lipid biosynthesis by increasing acetyl-CoA synthesis [112], and promoting NADPH production to fuel lipogenesis [113]. Several metabolic pathways are also regulated by mTOR, which is strictly linked to dysregulation of PI3K/Akt pathway. mTOR can affect cancer cell lipid metabolism by activating OXPHOS that in turn promote lipogenesis [114], as recently demonstrated by the evidence of downregulation of lipogenic enzymes, including FASN, acetyl-CoA carboxylase 1 and ATP citrate lyase, following treatment with mTOR inhibitor or by raptor genetic knockdown [111,115]. PI3K can be activated by stimulation of growth factor receptor tyrosine kinases, such as HER2. HER2 positive tumors are characterized by enhanced de novo lipogenesis, which contributes to the aggressiveness of these tumors [116]. The overexpression of HER2 is sufficient to prompt a FASN-dependent lipogenic phenotype in non-transformed epithelial cells that recapitulate the cancer cell metabolism. Conversely, HER2 inhibition—or de novo lipogenesis—inhibit oncogenic potential and induce apoptosis [117].
Among the others, p53 can directly or indirectly regulate lipid metabolism-related gene expression [118]. p53 downregulates FAs synthesis by inhibiting the pentose phosphate pathway and downregulating SREBP1 [119]. On the other hand, p53 is able to increase FAs oxidation and prevent lipid accumulation, by inhibiting the pyruvate dehydrogenase kinase at transcriptional level, thus leading to increase in pyruvate dehydrogenase and to the conversion of pyruvate to acetyl-CoA [120,121].
The complex intra-tumor regulation of metabolic activities is further enriched at cellular level, as best represented by breast cancer, which includes multiple cellular subtypes characterized by specific hormone/growth factor receptor and genetic profile. A recent study has highlighted how triple-negative breast cancers over-express genes involved in exogenous lipid uptake (high FABP5 and FABP7 expression), while the receptor-positive breast cancers are associated with de novo lipogenesis [122]. For these reasons, a better characterization of lipid-associated signatures in cancers could help to guide therapeutic interventions.
In additionto fuel the boosted proliferation, aberrant FA metabolism protect cancer cell from surrounding stress of different nature. It has been demonstrated that an excess of saturated FAs is responsible of mitochondrial/ER stress and dysfunction [123]. To prevent accumulation of saturated FAs, cancer cells overexpress SCD1 and SCD5, which convert saturated into monounsaturated FAs [124]. To cope with hypoxic microenvironment that often characterizes tumor progression, and which inhibits the activity of SCD enzymes, cancer cells upregulate the uptake of exogenous unsaturated FAs in order to maintain lipid homeostasis [123]. Importantly, to increase FAs uptake, it has been demonstrated that cancer cells over-express lipoprotein–lipase, which are involved in lipolysis of extracellular TG-rich lipoproteins, thus facilitating the hypoxia-induced FA uptake directly from TME [125]. However, hypoxia-induced regulation of FAs is highly cell-specific [126]. The major difference among the available studies is due to the cell culture conditions, and in particular, by the abundance of nutrients in cell culture media. Indeed, it is now clear that hypoxia or serum starving alone induce different metabolic phenotype in cancer cells, compared to their combination. Importantly, hypoxic regions are characterized by both oxygen and nutrients deprivation. In such condition, cancer cells are dependent on Gln and acetate as carbon source for the production of acetyl-CoA [127,128]. In line with these findings, when both oxygen and serum are limited, cancer cells upregulate nuclear acyl-coenzyme A synthetase that plays a dual role, aiding the use of extracellular acetate as carbon source and maintaining high histone acetylation levels, to prevent induction of apoptosis and promoting cancer cell growth [129].
Another important consequence of hypoxia microenvironment is represented by the excess in ROS production. This phenomenon results from FA oxidation process, activated by patatin like phospholipase domain containing 2 (PNPLA2) and mitochondrial electron leakage, which both lead to oxidative stress. To counteract this phenomenon, cancer cell downregulate PNPLA2 through the hypoxia-inducible protein 2, to promotes cancer cell survival [130,131]. The resistance to oxidative stress in hypoxic condition is also driven by a deregulated membrane lipid saturation, with increased saturated and monounsaturated FAs, which are less susceptible to peroxidation [132].
Dysregulation of membrane lipids saturation, as well as cholesterol content, could also affect cell membrane fluidity and dynamics, both mechanisms associated with the acquisition of a mesenchymal phenotype, which is crucial in metastatic cancer cells. Indeed, cholesterol synthesis is finely regulated during metastatic dissemination of cancer cells. Ehmsen, et al. recently demonstrated that elevated cholesterol biosynthesis identifies the subpopulation of breast CSCs. The administration of simvastatin, by inhibiting the 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase, hampered mammosphere formation [133].
All these pre-clinical findings regarding the role of FA metabolism in cancer progression arouse clinical interest for development of innovative antitumor strategies targeting FA metabolism, each one directed against specific phases of lipid metabolism: de novo synthesis, modification, uptake, activation, storage, mobilization and degradation [134].
Most of the enzymes involved in FA metabolism are regulated by sterol regulatory element binding protein-1SREBP-1 at transcriptional level. However, despite targeting SREBP-1 could potentially represent a promising strategy to regulate lipid metabolism in cancer cells, transcription factors are mostly considered undruggable. For this reason, several pre-clinical studies are now considering the idea to target its protein partner, SREBP-cleavage activating protein, which drive SREBP-1 to the Golgi for activation, by treating cancer cells with fatostatin and betulin [135,136]. Another important druggable player in FA biosynthesis is FASN. Although several pre-clinical studies showed efficacy in inhibiting cancer growth, the pronounced side effects, mostly on neuronal stem cells, impeded its use in clinical settings [137,138]. FA modification is an important step in lipid metabolism. Among the enzymes driving this step we find the elongases (seven members, ELOVL1-7) and desaturases (two members, SCD-1 and SCD-5). Importantly, our group has recently shown that inhibition of SCD-1, using the betulinic acid, in vitro is sufficient to efficiently kill colon cancer cells, in particular the CSC subset, leading to mitochondria-dependent cell death [139,140]. Similarly, the SCD-1 inhibitor, CAY10566, inhibits glioblastoma stem cells expansion and in vivo tumor growth [141]. As previously mentioned, one of the most important transporters of FAs is CD36. Targeting this membrane protein has been demonstrated to impair metastases in breast cancer and melanoma, by inhibiting the metastasis-initiating cell compartment [87].
5. The Role of miRNAs in Cancer Stem Cells Metabolism
Recent studies shed light on the role of microRNAs (miRNAs) in regulating stemness features of cancer cells, such as self-renewal, differentiation, metabolic reprogramming, metastasis formation and anticancer therapy resistance, associated with the pathogenesis of various types of human cancer [142].
miRNAs are single strand short non-coding RNA molecules (21–23 nucleotides) transcribed as precursor molecules, which are subsequently cleaved by the endoribonucleases Drosha and Dicer. Mature miRNAs are bound by a member of the Argonaute(AGO) protein family to form the RNA- induced silencing complex (RISC) in a process termed RISC loading. The miRNA guides RISC to complementary sequences located mainly in the region of its target mRNAs, silencing gene expression [143]. Following nuclear transcription, miRNAs are processed within the "multi-vesicular bodies", that consist of phospholipid membranes and lipoprotein microvesicles of endocytic origin, known as exosomes, of about 30–100 nm. After the binding of bodies to the plasma membrane, miRNAs are released into the bloodstream [144].
miRNAs have the role of coordinating gene expression programs at the base of physiological and pathologic cellular processes, including cancer [145]. Interestingly, miRNAs can act as both oncosuppressors, inhibiting proliferation and oncogenes, promoting tumor initiation, growth and metastasis formation [146]. Each miRNA can bind and regulate the expression of multiple coding or non-coding mRNAs. Therefore, the aberrant expression of a single miRNA can deleteriously influence the translation of multiple genes within a cell, leading to profound phenotypic changes [146].
To better understand the interaction between miRNAs and CSCs, recent studies evaluated how miRNAs can play a key role in maintaining and regulating the functioning of CSCs by targeting various oncogenic signaling pathways, such as Notch, WNT/β-catenin, JAK/STAT, PI3K/AKT and NF-kB [142].
Some miRNAs, such as miR-145, miR-200c, miR-494, miR-195-5p, miR-34, miR-519d, miR-128, miR-99a inhibit CSCs expansion by inhibiting of ADAM, BMI, Notch, caspases and mTOR, respectively, while others, such as miR-19, miR-501-5p, miR-21, miR221/222, miR-483-5p, miR-196b-5p, miR-494-3p stimulate stemness through the activation of WNT/β-catenin, PTEN, cyclin D1 and MMP-2, STAT3 and Notch 1 pathways [142].
The deregulation of Drosha and Dicer has been observed in different types of cancer [147,148]. For instance, the downregulation of Dicer led to decreased miR-130b and tumorigenesis [149]. In addition to Drosha and Dicer, other enzymes involved in the miRNAs biogenesis pathway, are the trans-activation responsive RNA-binding protein (TARBP2) and the Argonaute RISC catalytic component 2 (AGO2). In sporadic and hereditary carcinomas, downregulation of TARBP2 protein expression in CSCs was shown to be important for pro-metastasis signaling [150].
A key role in the miRNAs biogenesis pathway is played by Exportin-5 (XPO5) protein. It mediates the nuclear export of miRNAs precursors to the cytoplasm. Genetic mutations of XPO5 leads to an entrapment and loss of precursor miRNAs in the nucleus promoting tumor initiation through an increase in the expression of stemness-related genes such as EZH2 and MYC [151].
Notably, different miRNAs have been associated with the regulation of cancer cell metabolism [152].
The transport of glucose inside the cells is an important event for the correct cellular functioning. This mechanism is mediated by tissue-specific glucose transporters known as GLUTs, which are directly or indirectly targeted by several miRNAs. Presumably, deregulation of GLUTs can increase glucose uptake, satisfying the high glucose requirement and accelerating metabolism in cancer cells. However, the direct links between miRNAs deregulation and glucose transport in cancer are largely unknown therefore further studies are needed. A key enzyme for aerobic glycolysis is the hexokinase 2 (HK2), which is overexpressed in tumors. miR-143 is inversely correlated to the expression of HK2 in the head and neck squamous cell carcinoma and in lung cancer [153]. Likewise, the loss of miR-143, in glioma tissues and glioblastoma stem-like cells, promoted the expression of HK2, resulting in enhanced aerobic glycolysis and inhibition of cell differentiation [154]. The connection between miR-143 and HK2 has been studied also in colorectal cancer, where it has been shown that it downregulates HK2 expression and that its reintroduction leads to a decrease in lactate secretion by impairing the rate of glycolysis [155]. Another miRNA-regulated glycolytic enzyme is phosphofructokinase 1 (PFK1). PFK1 catalyzes the phosphorylation reaction of fructose-6-phosphate to convert it into fructose-1,6-bisphosphate. Yang et al. showed that miR-135 targets PFK1 and inhibits aerobic glycolysis and suppresses tumor growth [146].
Chong et al. studied the role of miRNAs in maintaining the glycolytic metabolism of CSCs [156]. In their study, they demonstrated a critical role of the LIN28B in stemness. In breast cancer cell lines, the inhibition of LIN28B suppresses MYC expression and increased miR-34a-5p levels expression, correlating with inhibition of glucose uptake/lactate production and a better patient’s prognosis. Therefore, blocking of the LIN28B/MYC/miR-34a-5p signaling pathway, by the LIN28B specific inhibitor, causes a dramatic inhibition of tumor growth and metastatic potential in orthotopic immunodeficient mouse models of human breast cancer cells [156].
A class of miRNAs that regulates the choice between self-renewal and differentiation of breast CSC is represented by miR-600. In breast CSCs, the miR-600 silencing caused cancer cell expansion, while its overexpression reduced CSC self-renewal, leading to a decreased in vivo tumorigenicity. Interestingly, the main target of miR-600 is SCD1, an enzyme required to produce active lipid-modified WNT proteins, which are involved in cell fate determination during tissue development and oncogenic processes [157]. Indeed, the authors showed that low levels of miR-600 are correlated with active WNT signaling and a poor prognosis. These findings highlighted that miR-600 is involved in breast cancer cells-fate decisions and influences tumor progression [157].
In addition to glucose, cancer cells need Gln for their growth. This adaptive metabolism by cancer cells appears to provide substrates for an increase in lipogenesis and biosynthesis of nucleic acids which are fundamental for the proliferative phenotype of the cancer cell. Cell transformation has been shown to stimulate glutaminolysis and many cancer cells are closely dependent on this metabolic path [158]. One of the main regulators of glutaminolysis is MYC, which promotes not only cell proliferation, but also the generation of macromolecules and antioxidants necessary for the growth of cancer cells. Interestingly, MYC’s inhibition of miR-23A/B improves mitochondrial expression of glutaminase and Gln metabolism and correlates with the onset of a neoplastic phenotype [158]. Similarly, Wang et al. have demonstrated that MYC, via miR-33b induction, supports glioblastoma CSCs via the activation of mevalonate metabolism [159]. Pyruvate dehydrogenase kinase 4 is target of miR-122, a liver-specific miRNA. mir-122 is able to hamper the glycolytic metabolism in the CD133+ CSC compartment, decreasing spheroids formation and sensitizing to standard anticancer therapy [160].
An increased expression of miR-210-3p correlates with colorectal cancer progression [161]. In fact, the stable overexpression of miR-210 in colorectal cancer spheroid culturesresulted in significantly enhanced CSC self-renewal activity [162]. In colon CSCs miR-210 inhibits mitochondrial TCA cycle activity to enhance lactate production [163]. The lactate stimulation leads to an increased self-renewal capacity of different colon CSC cultures [163].
The rapid proliferation of cancer cells within a tumor mass leads to the formation of hypoxic regions caused by the absence of an efficient vascular network. Cancer cell survival in hypoxia requires the activation of adaptive pathways [164] and the downregulation of miRNA through a reduction of Dicer and Drosha [165,166].
Within tumor is possible that miRNAs processed in the normoxic portion of a tumor diffuse toward hypoxic zones to promote tumor growth and regulate gene expression [149]. In colon CSCs, miR-210 promotes the self-renewal and reprograms cell metabolism toward a prompted glycolytic and lactate yield. In hypoxic conditions HIF1α induces miR-210-3p expression, this determines a reduced oxidative metabolism (TCA cycle and OXPHOS) [161]. miR-210 is an oncogenic miRNA and a target of HIF-1 and HIF-2 [167]. It has been shown that during hypoxia, miR-210 targets the mRNA that encodes the mitochondrial electron transport chain component protein succinate dehydrogenase complex subunit D (SDHD). Downregulation of SDHD results in an increased stabilization of HIF1α and cancer cell survival [168,169]. In the hypoxic microenvironment, miRNAs contribute to metabolic activities, to the maintenance of stemness in cancer cells and to therapy resistance. miRNAs, hypoxia and CSC are part of a complex signaling network that promotes tumor aggressiveness [170].
Several miRNAs target important cancer cell regulatory molecules and are involved in an intricate network of signaling between cancer cells and the TME. In addition to their involvement in direct cell-to-cell signaling, several miRNAs are secreted through micro vesicles or exosomes and affect cancer cell growth and metastasis [165]. Broniszeet al. studied the role of miRNAs in the transformation of normal fibroblasts into CAFs, focusing on PTEN-regulated miR-320. Downregulation of miR-320 and upregulation of one of its direct targets, ETS Proto-Oncogene 2, concomitantly with loss of PTEN, is a key event in oncogenic process. In fact, this event leads to increased angiogenesis and tumor formation [171]. miR-320 was found to regulate CAF-secreted proteins, including MMP9, MMP2, lysyl oxidase homolog 2 and elastin microfibril interfacer 2, which are known to enhance tumor metastasis by programming the TME via degradation of extracellular matrices [165]. Hua Y et al., identified an important chemokine in the TME, CCL5, as target of miR-214, the latter being downregulated in CAFs. These data support the idea that miRNAs could alter the TME by changing protein production in CAFs, such as chemokines, sustaining tumor growth [172].
Invasion and metastasis of cancer cells have been associated with phosphoglucose isomerase, thedownregulation of which by mir-200 in breast cancer cells impaired metastasis spread [173]. Indeed, well-known miRNAs that are downregulated in cancer belong to miR-200 family. These miRNAs are involved in many different functions, such as induction of EMT via downregulation of E-cadherin and consequent increases in zinc finger E-Box (ZEB) proteins. It was demonstrated that miR-200 influences angiogenesis indirectly via downregulation of chemokine and interleukin such as CXCL1 and IL-8, which are major players in the TME [174].
Knowledge about the role of miRNAs in development and diseases, particularly in cancer, encouraged the use of miRNAs as tools or as targets for novel therapeutic approaches. For instance, several approaches are based on the use of miRNA mimics to restore the expression of tumor suppressors or antimiR to target oncogenes [169]. miRNAs mimics are synthetic double-stranded small RNA molecules that match the corresponding miRNA sequence. In human disease this therapy is used to replace the lost miRNA expression and their function. In contrast, antimiRs are single stranded and based on first-generation antisense oligonucleotides, which had been designed to target mRNAs or modified with locked nucleic acids.
For example, a mimic of miR-34, whose target is lactate dehydrogenase A, is used like a tumor suppressor in Phase I clinical trials (NCT01829971).
On the other hand, in Phase II clinical trials for thetreatment of hepatitis, it was used antimiRs targeting miR-122 (NCT01200420; NCT01872936; NCT02031133; NCT02508090) [169].
Studies have shown that miRNAs such as let-7, miR-34, miR-451 and miR-200 can sensitize cancer cells to chemotherapy, and mimics of these miRNAs could be rationally combined with chemotherapeutic drugs [175,176,177].
MRX34 was the first miRNA-based therapy undergoing clinical trial for treatment of lymphoma, melanoma, multiple myeloma, liver, lung and renal carcinoma (NCT01829971) [178]. The use of MRX34 in the aggressive KRAS TP53 mutated non-small cell lung cancer mouse model, led to significant tumor reduction. Moreover, combination treatment with the epidermal growth factor receptor (EGFR) inhibitor, erlotinib and miR-34 mimic and let-7 showed synergistic effects in inhibiting the growth of non-small cell lung cancer cell lines in vitro [177].
A similar example is the MesomiR-1 drug, which reintroduces miR-16, a miRNA that regulates aldolase A in glycolysis process [179,180].
Another therapy approach valuated in clinical trial is the locked oligonucleotide acid-modified inhibitor for miR-155 (MRG-106) (NCT03713320) [181]. MRG-106 replaces both miR-155 and miR-143, that negatively regulates HK2 and counteracts glycolytic phenotype [182].
Given that miR-29 is frequently lost in cancer and has been reported to negatively regulates monocarboxylate transporter 1 (MCT1), a lactate transporter [183,184] it has been tempted a novel approach that involves the use of miR-29 mimic (MRG-201).
In conclusion, although numerous studies have been conducted to understand the role of miRNA in the CSC metabolism, further studies are needed to define their role more precisely.
6. The Influence of Microenvironment in Cancer Stem Cells Metabolism
The TME is characterized by a high degree of metabolic heterogeneity and dynamics that involves both cancer and microenvironmental cells. Based on the metabolic activity it is possible to classify tumors in “glycolytic” (lung, liver, colorectal, leukemia) versus “oxidative” (melanoma, glioblastoma) [185]. The complexity of this scenario is even increased by the evidence that tumors originated from the same organ—or characterized by the same genetic background—are not metabolically uniform [186,187]. This intra-tumor metabolic variability is mainly dictated by different access to oxygen and glucose, which occurs in proximity of blood vessels and by the different cell population co-existing in TME. Indeed, cancer and stromal cells can compete and/or cooperate for nutrients [188].
The neoplastic tissue is constituted not only by the tumor cells, but also by the stromal cells; together they constitute the TME. CAFs, a cellular component of the TME, influence tumor growth by supplying nutrients or directly by cell-to-cell communication. In addition, CAFs provide a stromal framework to cancer cells during early growth and development, leading to malignant transformation [149].
The metabolic coupling between cancer and stromal cells, which is responsible for the ‘reverse Warburg effect’, is mediated by an interchange of cytokines, metabolites and miRNAs. Stromal cells allow the creation a permissive soil that sustains cancer cell growth and fuel cancer cell energy by supplying metabolic substrates, including Gln and lactate, which enhance a specific metabolic pathway, such as OXPHOS [105,189,190] (Figure 2).
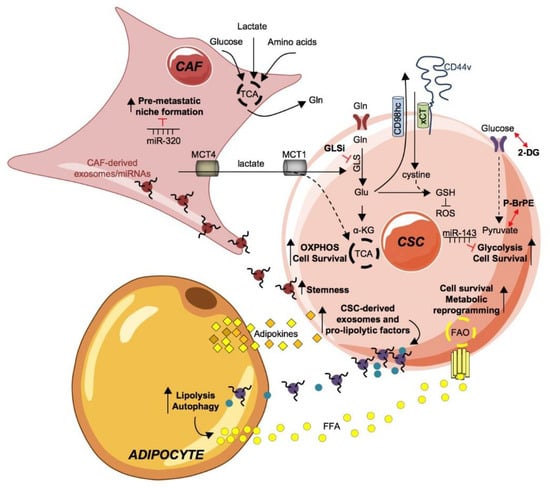
Figure 2.
Metabolic fingerprint of cancer stem cells and their coupling with tumor microenvironment. The tumor microenvironment and every single part of it is an important element to be taken into consideration in tumor development. Through the production and excretion by the cancer associated fibroblast (CAF) of intermediate metabolites, such as lactate and glutamine (Gln), they are able to meet the continuous and demanding energy needs of CSCs. Gln is internalized by its transporter ASCT2 and converted in glutamate (Glu), by the enzyme glutaminase (GLS), finally inducing OXPHOS and cell survival of CSCs. GLS activity is blocked by GLU inhibitors. Glu contributes to the formation of reactive oxygen species (ROS) inhibitor molecule glutathione (GSH) by acting as an exchange ion, through the CD44v-xCT transporter, for the import of cystine, which is necessary for the production of GSH together with Glu. The uptake of glucose through the GLUT foster a glycolytic phenotype in CSCs boosting cell survival. The administration of the glycolysis inhibitors 2-deoxyglucose (2-DG), 3-bromopyruvate ester (pBr-PE) or miR-143 proved to effective in dampening the expansion of the CSC subpopulation.The crosstalk of CAF and CSC is possible through CAF-derived exosomes/miRNAs, including miR-21, miR-378e and miR-143. CAF-derived exosomes/miRNAs are internalized by CSC, sustaining stemness and positively influences tumor progression. Conversely, miR-320 counteracts pre-metastatic niche formation by CAFs.Adipocyte-secreted adipokines are taken upby CSC and stimulate the production and release of exosomes and pro-lipolytic factors, including miRNA-144 and miRNA-126, which in turn active lipolysis and autophagy pathways in adipocytes. Consequently, free fatty acids (FFA) are released in the tumor microenvironment (TME) that, upon absorption by CSCs through the fatty acid transporters (fatty acid translocase, FAT/CD36; fatty acid transport protein, FATP and fatty acid-binding protein, FABP), undergo fatty acid oxidation (FAO) to sustain CSCs expansion and metabolic reprogramming.
In nutrient-deprived conditions, CAFs can undergo metabolic adaptation and synthesize Gln that is released in TME for cancer cell consumption, thus fostering tumor growth and metastasis formation. CAF-derived lactate, in turn, promotes OXPHOS in cancer cells, favors Gln uptake and catabolism and is responsible for the resistance to glutaminase inhibitors [191]. Monocarboxylate transporter (MCT) 4 exports lactate from CAFs, while MCT1 allows lactate uptake in CSCs [192].
Another example is the alanine uptake by pancreatic cancer cells. This essential amino acid is released by pancreatic stellate cells by autophagy and fuel TCA cycle in pancreatic ductal adenocarcinoma cells for the production of essential amino acids and lipids [193].
On the contrary, it seems that CSCs prefer glycolysis as source of ATP in a low oxygen microenvironment. Inhibition of glycolysis with a derivate of 3-bromopyruvate ester (pBr-PE) sensitized chemotherapy-resistant glioblastoma stem-like cells to standard therapeutic agents and counteracted tumor growth in vivo [194]. Accordingly, the activation of an EMT program repressed fructose-1,6-bisphosphatase through a Snail-G9a-Dnmt1 complex causing the production of ROS and enhancement of glycolysis under hypoxia in breast CSCs [15].
Cancer metabolic reprogramming is indeed influenced by proximity to blood vessels and thus the availability of oxygen. By using a methodology capable of sorting of stroma-free glioblastoma cells and based on the distance of cells from vasculature, Kumar et al. demonstrated that glioblastoma cells positioned in close proximity to blood vessels were highly chemo- and radio-resistant and possessed an OXPHOS metabolism [195]. These observations shed light on the presence of metabolic zonation, which means that cancer cell metabolism is highly heterogeneous within the same tumor due to the presence of different metabolic niches that favor the expansion of CSCs [105,196].
Besides being directly involved in nutrients exchange, CAFs secreted cytokines are involved in the activation of EMT, which is associated with metabolic reprogramming of CSCs [197]. In particular, several studies demonstrated that EMT prompts glycolytic flux in CSCs, at the expense of mitochondrial respiration [3]. Interestingly, glucose consumption by cancer cells can in turn can suppress the immune response [198] and create an acidic microenvironment that favors cancer progression [199].
Beyond the cytokine-driven effects, cancer and TME cells can communicate and benefit from the exchange of vesicles-contained messengers. Exosomes are nanovesicles (40–100 nm in diameter) that are released from most cell types into the extracellular space after fusion with the plasma membrane [200,201]. Many reports have highlighted that exosomes play important roles in immune response and tumor progression [200]. It was demonstrated that cancer cells secrete a larger number of exosomes than normal cells. Tumor-derived exosomes are necessary for cell-to-cell communication. They transport a cargo of growth factors, chemokines, miRNAs and other small molecules [202,203]. Moreover, it has been demonstrated in multiple cancer models that the exposure of cancer cells to the conditioned media of CAFs promotes cancer cell growth. Besides the essential role played by CAFs in secreting factors that shape TME to allow the expansion of cancer cell population, CAFs secrete micro vesicles that act as cargo for nutrients, including intermediate metabolites, lipids and amino acids, which are ultimately absorbed by cancer cells [204]. Microvesicles released in the TME by stromal cells and loaded with several metabolites, following the uptake by cancer cells, directly promote cell proliferation and fuel tumor progression [204].
Moreover, CAF-derived exosomes loaded with miRNAs impact metabolism, migration, invasion and metastasis in CSCs. Exosomal miR-21, miR-378e and miR-143, horizontally transferred from CAFs to breast CSC, encouraged the formation of mammospheres concomitantly with the acquisition EMT and stemness markers [205].
Interestingly, in metastatic breast cancer, extracellular vesicles released by CAFs contain mitochondrial DNA that is internalized by CSCs and used to sustain OXPHOS and exit from quiescence induced by hormonal therapy [206].
7. Concluding Remarks and Future Perspectives
Herein we show an overview on the metabolic demand of CSCs highlighting the importance of looking at the ‘whole picture’ of tumor metabolism. CSCs possess a heterogeneous metabolism that can vary in different zones of the tumor in response to nutrients and oxygen supply.
Novel frontiers of cancer treatment are focused on the use of biocompatible particles, usually conjugated with anticancer compounds, which are capable of releasing the drug directly at the tumor site. For instance, gold nanoparticles conjugated with salinomycin, caused negligible damage to healthy cells while inducing cell death of the subpopulation of CD44+/CD24− breast CSCs by inducing ferroptosis, consisting in the accumulation of iron and ROS derived from lipid oxidation [207].
Thus, the identification of metabolic traits of CSCs and the adaptation of CSCs metabolism in response to a hostile or supportive TME led to the development of promising therapeutic treatments, summarized in Table 2, which interfere with CSCs metabolism and thus, disease progression.

Table 2.
Metabolic Therapeutic Targets of Cancer Stem Cells.
Author Contributions
A.T., G.P., C.D. and G.S. conceived and wrote the manuscript. S.D.F., F.V., F.C., S.F., D.G., L.M. and M.T. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work wassupported by grants from AIRC (14415) to M.T. and AIRC 5x1000 (9979), AIRC IG (16746) and PRIN (2017WNKSLR) to G.S.
Acknowledgments
A.T. is a research fellow funded by European Union- FESR FSE, PON Ricerca e Innovazione 2014–2020 (AIM line 1).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 2DG | 2-deoxyglucose |
| AGO | Argonaute |
| AGO2 | Argonaute RISC catalytic component 2 |
| BSO | L-buthionine-sulfoximine |
| CAFs | Cancer associated fibroblasts |
| CSCs | Cancer Stem Cells |
| DNMT | DNA-methyltransferases |
| EMT | Epithelial mesenchymal transition |
| ETC | Electron transport chain |
| EZH2 | Enhancer of zeste homolog 2 |
| FA | Fatty acid |
| FABP | Fatty acid-binding protein |
| FAO | Fatty acid oxidation |
| FASN | Fatty acid synthase |
| FAT | Fatty acid translocase |
| FATP | Fatty acid transport protein |
| GLSi | Glutaminase inhibitors |
| GLU | Glutamate |
| GSH | Glutathione |
| HIF1α | Hypoxia-inducible factor 1 alpha |
| HK2 | Hexokinase |
| MCT1 | Monocarboxylate transporter 1 |
| MCT4 | Monocarboxylate transporter 4 |
| miRNA | microRNA |
| OXPHOS | Oxidative phosphorylation |
| pBr-PE | 3-bromopyruvate ester |
| PFK1 | Phosphofructokinase 1 |
| PGC-1α | Peroxisome proliferator activated receptor gamma coactivator 1 α |
| PNPLA2 | Patatin like phospholipase domain containing 2 |
| RISC | RNA-induced silencing complex |
| ROS | Reactive oxygen species |
| SCD-1 | Stearoyl-CoA desaturase-1 |
| SDHD | Succinate dehydrogenase complex subunit D |
| SOD | Superoxide dismutase |
| SREBP | Sterol regulatory element binding protein |
| TARBP2 | Trans-activation responsive RNA-binding protein |
| TCA | Tricarboxylic acid |
| TXN | thioredoxin |
| TME | Tumor microenvironment |
| XPO5 | Exportin 5 |
| ZEB | Zinc finger E-box-binding homeobox |
References
- Turdo, A.; Veschi, V.; Gaggianesi, M.; Chinnici, A.; Bianca, P.; Todaro, M.; Stassi, G. Meeting the Challenge of Targeting Cancer Stem Cells. Front. Cell Dev. Biol. 2019, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Tu, G.; Liu, Z.; Liu, M. Cancer-associated fibroblasts: A multifaceted driver of breast cancer progression. Cancer Lett. 2015, 361, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Escalona, R.; Leung, D.; Chan, E.; Kannourakis, G. Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: Perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin. Cancer Biol. 2018, 53, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Brand, R.E.; Mehla, K. MicroRNAs in pancreatic cancer metabolism. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef]
- Zong, W.X.; Rabinowitz, J.D.; White, E. Mitochondria and Cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef]
- Lacina, L.; Plzak, J.; Kodet, O.; Szabo, P.; Chovanec, M.; Dvorankova, B.; Smetana, K., Jr. Cancer Microenvironment: What Can We Learn from the Stem Cell Niche. Int. J. Mol. Sci. 2015, 16, 24094–24110. [Google Scholar] [CrossRef]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef]
- Mohammed, H.R.; Bayraktar, E.; Gouda, K.H.; Abd-Ellah, M.F.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On respiratory impairment in cancer cells. Science 1956, 124, 269–270. [Google Scholar] [PubMed]
- Bensard, C.L.; Wisidagama, D.R.; Olson, K.A.; Berg, J.A.; Krah, N.M.; Schell, J.C.; Nowinski, S.M.; Fogarty, S.; Bott, A.J.; Wei, P.; et al. Regulation of Tumor Initiation by the Mitochondrial Pyruvate Carrier. Cell Metab. 2020, 31, 284–300.e7. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Yuan, T.; Wu, Y.; Wang, Y.; Fan, T.W.; Miriyala, S.; Lin, Y.; Yao, J.; Shi, J.; Kang, T.; et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2013, 23, 316–331. [Google Scholar] [CrossRef]
- Chen, C.L.; Uthaya Kumar, D.B.; Punj, V.; Xu, J.; Sher, L.; Tahara, S.M.; Hess, S.; Machida, K. NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab. 2016, 23, 206–219. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Lane, A.N.; Fan, T.W. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Ye, X.Q.; Li, Q.; Wang, G.H.; Sun, F.F.; Huang, G.J.; Bian, X.W.; Yu, S.C.; Qian, G.S. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int. J. Cancer 2011, 129, 820–831. [Google Scholar] [CrossRef]
- Janiszewska, M.; Suva, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clement-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Viale, A.; Pettazzoni, P.; Lyssiotis, C.A.; Ying, H.; Sanchez, N.; Marchesini, M.; Carugo, A.; Green, T.; Seth, S.; Giuliani, V.; et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Sancho, P.; Burgos-Ramos, E.; Tavera, A.; Bou Kheir, T.; Jagust, P.; Schoenhals, M.; Barneda, D.; Sellers, K.; Campos-Olivas, R.; Grana, O.; et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015, 22, 590–605. [Google Scholar] [CrossRef] [PubMed]
- Kamarajan, P.; Rajendiran, T.M.; Kinchen, J.; Bermudez, M.; Danciu, T.; Kapila, Y.L. Head and Neck Squamous Cell Carcinoma Metabolism Draws on Glutaminolysis, and Stemness Is Specifically Regulated by Glutaminolysis via Aldehyde Dehydrogenase. J. Proteome Res. 2017, 16, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.L.; Stevens, B.M.; D’Alessandro, A.; Reisz, J.A.; Culp-Hill, R.; Nemkov, T.; Pei, S.; Khan, N.; Adane, B.; Ye, H.; et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018, 34, 724–740.e4. [Google Scholar] [CrossRef]
- Luo, M.; Shang, L.; Brooks, M.D.; Jiagge, E.; Zhu, Y.; Buschhaus, J.M.; Conley, S.; Fath, M.A.; Davis, A.; Gheordunescu, E.; et al. Targeting Breast Cancer Stem Cell State Equilibrium through Modulation of Redox Signaling. Cell Metab. 2018, 28, 69–86.e6. [Google Scholar] [CrossRef]
- Brand, K.A.; Hermfisse, U. Aerobic glycolysis by proliferating cells: A protective strategy against reactive oxygen species. FASEB J. 1997, 11, 388–395. [Google Scholar] [CrossRef]
- Harris, I.S.; Treloar, A.E.; Inoue, S.; Sasaki, M.; Gorrini, C.; Lee, K.C.; Yung, K.Y.; Brenner, D.; Knobbe-Thomsen, C.B.; Cox, M.A.; et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 2015, 27, 211–222. [Google Scholar] [CrossRef]
- Smolkova, K.; Plecita-Hlavata, L.; Bellance, N.; Benard, G.; Rossignol, R.; Jezek, P. Waves of gene regulation suppress and then restore oxidative phosphorylation in cancer cells. Int. J. Biochem. Cell Biol. 2011, 43, 950–968. [Google Scholar] [CrossRef]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, E.C.; Van Houten, B. Metabolic symbiosis in cancer: Refocusing the Warburg lens. Mol. Carcinog 2013, 52, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Cuyas, E.; Verdura, S.; Folguera-Blasco, N.; Bastidas-Velez, C.; Martin, A.G.; Alarcon, T.; Menendez, J.A. Mitostemness. Cell Cycle 2018, 17, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Bezuidenhout, N.; Shoshan, M. A Shifty Target: Tumor-Initiating Cells and Their Metabolism. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef]
- Chae, Y.C.; Kim, J.H. Cancer stem cell metabolism: Target for cancer therapy. BMB Rep. 2018, 51, 319–326. [Google Scholar] [CrossRef]
- Reid, M.A.; Dai, Z.; Locasale, J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017, 19, 1298–1306. [Google Scholar] [CrossRef]
- Racey, L.A.; Byvoet, P. Histone acetyltransferase in chromatin. Evidence for in vitro enzymatic transfer of acetate from acetyl-coenzyme A to histones. Exp. Cell Res. 1971, 64, 366–370. [Google Scholar] [CrossRef]
- Wongtrakoongate, P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J. Stem Cells 2015, 7, 137–148. [Google Scholar] [CrossRef]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.B.; Lim, J.J.; Chow, E.K. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 29. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.E.; Moore, H.M.; Li, X.; Toy, K.A.; Huang, W.; Sabel, M.S.; Kidwell, K.M.; Kleer, C.G. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc. Natl. Acad Sci. USA 2014, 111, 3098–3103. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Wicha, M.S. Metabolic plasticity of cancer stem cells. Oncotarget 2015, 6, 35141–35142. [Google Scholar] [CrossRef]
- Shin, M.K.; Cheong, J.H. Mitochondria-centric bioenergetic characteristics in cancer stem-like cells. Arch. Pharm. Res. 2019, 42, 113–127. [Google Scholar] [CrossRef]
- De Luca, A.; Fiorillo, M.; Peiris-Pages, M.; Ozsvari, B.; Smith, D.L.; Sanchez-Alvarez, R.; Martinez-Outschoorn, U.E.; Cappello, A.R.; Pezzi, V.; Lisanti, M.P.; et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget 2015, 6, 14777–14795. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res. 2011, 71, 3196–3201. [Google Scholar] [CrossRef]
- Jagust, P.; de Luxan-Delgado, B.; Parejo-Alonso, B.; Sancho, P. Metabolism-Based Therapeutic Strategies Targeting Cancer Stem Cells. Front. Pharmacol. 2019, 10, 203. [Google Scholar] [CrossRef]
- Pusceddu, S.; Vernieri, C.; Di Maio, M.; Marconcini, R.; Spada, F.; Massironi, S.; Ibrahim, T.; Brizzi, M.P.; Campana, D.; Faggiano, A.; et al. Metformin Use Is Associated With Longer Progression-Free Survival of Patients With Diabetes and Pancreatic Neuroendocrine Tumors Receiving Everolimus and/or Somatostatin Analogues. Gastroenterology 2018, 155, 479–489.e7. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.; Ozsvari, B.; Lisanti, C.L.; Tanowitz, H.B.; Howell, A.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget 2015, 6, 4569–4584. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, E.M.; Baquero, P.; Michie, A.M.; Dunn, K.; Tardito, S.; Holyoake, T.L.; Helgason, G.V.; Gottlieb, E. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 2017, 23, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, F.; Ozsvari, B.; Fiorillo, M.; De Francesco, E.M.; Bonuccelli, G.; Lisanti, M.P. A mitochondrial based oncology platform for targeting cancer stem cells (CSCs): MITO-ONC-RX. Cell Cycle 2018, 17, 2091–2100. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.W.L.; Ghode, P.; Ong, D.S.T. Redox regulation of cell state and fate. Redox. Biol. 2019, 25, 101056. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef]
- Chandimali, N.; Jeong, D.K.; Kwon, T. Peroxiredoxin II Regulates Cancer Stem Cells and Stemness-Associated Properties of Cancers. Cancers 2018, 10. [Google Scholar] [CrossRef]
- Heusch, P.; Canton, M.; Aker, S.; van de Sand, A.; Konietzka, I.; Rassaf, T.; Menazza, S.; Brodde, O.E.; Di Lisa, F.; Heusch, G.; et al. The contribution of reactive oxygen species and p38 mitogen-activated protein kinase to myofilament oxidation and progression of heart failure in rabbits. Br. J. Pharmacol. 2010, 160, 1408–1416. [Google Scholar] [CrossRef]
- Hwang, J.W.; Yao, H.; Caito, S.; Sundar, I.K.; Rahman, I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic. Biol. Med. 2013, 61, 95–110. [Google Scholar] [CrossRef]
- Ahmadinejad, F.; Geir Moller, S.; Hashemzadeh-Chaleshtori, M.; Bidkhori, G.; Jami, M.S. Molecular Mechanisms behind Free Radical Scavengers Function against Oxidative Stress. Antioxidants 2017, 6. [Google Scholar] [CrossRef]
- Yip, N.C.; Fombon, I.S.; Liu, P.; Brown, S.; Kannappan, V.; Armesilla, A.L.; Xu, B.; Cassidy, J.; Darling, J.L.; Wang, W. Disulfiram modulated ROS-MAPK and NFkappaB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br. J. Cancer 2011, 104, 1564–1574. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, J.Y.; Lee, N.; Oh, E.; Sung, D.; Cho, T.M.; Seo, J.H. Disulfiram suppresses cancer stem-like properties and STAT3 signaling in triple-negative breast cancer cells. Biochem. Biophys Res. Commun. 2017, 486, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, P.H.; Wiesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Achuthan, S.; Santhoshkumar, T.R.; Prabhakar, J.; Nair, S.A.; Pillai, M.R. Drug-induced senescence generates chemoresistant stemlike cells with low reactive oxygen species. J. Biol. Chem. 2011, 286, 37813–37829. [Google Scholar] [CrossRef] [PubMed]
- Sitwala, K.V.; Hess, J.L. Structural basis of multimer-mediated mayhem. Cancer Cell 2006, 9, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, H.; Iwata, K.; Ogonuki, N.; Inoue, K.; Atsuo, O.; Kanatsu-Shinohara, M.; Morimoto, T.; Yabe-Nishimura, C.; Shinohara, T. ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell 2013, 12, 774–786. [Google Scholar] [CrossRef]
- Pelicci, P.G.; Dalton, P.; Giorgio, M. The other face of ROS: A driver of stem cell expansion in colorectal cancer. Cell Stem Cell 2013, 12, 635–636. [Google Scholar] [CrossRef]
- Mori, K.; Shibanuma, M.; Nose, K. Invasive potential induced under long-term oxidative stress in mammary epithelial cells. Cancer Res. 2004, 64, 7464–7472. [Google Scholar] [CrossRef]
- Radisky, D.C.; Levy, D.D.; Littlepage, L.E.; Liu, H.; Nelson, C.M.; Fata, J.E.; Leake, D.; Godden, E.L.; Albertson, D.G.; Nieto, M.A.; et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005, 436, 123–127. [Google Scholar] [CrossRef]
- Droge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Myant, K.B.; Cammareri, P.; McGhee, E.J.; Ridgway, R.A.; Huels, D.J.; Cordero, J.B.; Schwitalla, S.; Kalna, G.; Ogg, E.L.; Athineos, D.; et al. ROS production and NF-kappaB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell 2013, 12, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, T.; Nagano, O.; Yae, T.; Tamada, M.; Motohara, T.; Oshima, H.; Oshima, M.; Ikeda, T.; Asaba, R.; Yagi, H.; et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell 2011, 19, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, K.J.; Seo, Y.; Kwon, J.H.; Yoon, J.P.; Kang, J.Y.; Lee, H.J.; Park, S.J.; Hong, S.P.; Cheon, J.H.; et al. Effects of metformin on colorectal cancer stem cells depend on alterations in glutamine metabolism. Sci. Rep. 2018, 8, 409. [Google Scholar] [CrossRef]
- Deblois, G.; Smith, H.W.; Tam, I.S.; Gravel, S.P.; Caron, M.; Savage, P.; Labbe, D.P.; Begin, L.R.; Tremblay, M.L.; Park, M.; et al. ERRalpha mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat. Commun. 2016, 7, 12156. [Google Scholar] [CrossRef]
- Wang, Z.; Yip, L.Y.; Lee, J.H.J.; Wu, Z.; Chew, H.Y.; Chong, P.K.W.; Teo, C.C.; Ang, H.Y.; Peh, K.L.E.; Yuan, J.; et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019, 25, 825–837. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fagone, P.; Jackowski, S. Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res. 2009, 50, S311–S316. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Ookhtens, M.; Kannan, R.; Lyon, I.; Baker, N. Liver and adipose tissue contributions to newly formed fatty acids in an ascites tumor. Am. J. Physiol. 1984, 247, R146–R153. [Google Scholar] [CrossRef] [PubMed]
- Kuhajda, F.P.; Jenner, K.; Wood, F.D.; Hennigar, R.A.; Jacobs, L.B.; Dick, J.D.; Pasternack, G.R. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc. Natl. Acad. Sci. USA 1994, 91, 6379–6383. [Google Scholar] [CrossRef] [PubMed]
- Yasumoto, Y.; Miyazaki, H.; Vaidyan, L.K.; Kagawa, Y.; Ebrahimi, M.; Yamamoto, Y.; Ogata, M.; Katsuyama, Y.; Sadahiro, H.; Suzuki, M.; et al. Inhibition of Fatty Acid Synthase Decreases Expression of Stemness Markers in Glioma Stem Cells. PLoS ONE 2016, 11, e0147717. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.R.; Xing, F.; Sharma, S.; Watabe, M.; Pai, S.K.; Iiizumi-Gairani, M.; Fukuda, K.; Hirota, S.; Mo, Y.Y.; Watabe, K. Elevated lipogenesis in epithelial stem-like cell confers survival advantage in ductal carcinoma in situ of breast cancer. Oncogene 2013, 32, 5111–5122. [Google Scholar] [CrossRef] [PubMed]
- Brandi, J.; Dando, I.; Pozza, E.D.; Biondani, G.; Jenkins, R.; Elliott, V.; Park, K.; Fanelli, G.; Zolla, L.; Costello, E.; et al. Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J. Proteom. 2017, 150, 310–322. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Wong, J.; Conklin, D.S. PPARgamma maintains ERBB2-positive breast cancer stem cells. Oncogene 2013, 32, 5512–5521. [Google Scholar] [CrossRef]
- Pascual, G.; Avgustinova, A.; Mejetta, S.; Martin, M.; Castellanos, A.; Attolini, C.S.; Berenguer, A.; Prats, N.; Toll, A.; Hueto, J.A.; et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 2017, 541, 41–45. [Google Scholar] [CrossRef]
- Mitsuishi, Y.; Taguchi, K.; Kawatani, Y.; Shibata, T.; Nukiwa, T.; Aburatani, H.; Yamamoto, M.; Motohashi, H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012, 22, 66–79. [Google Scholar] [CrossRef]
- Cruz, A.L.S.; Barreto, E.A.; Fazolini, N.P.B.; Viola, J.P.B.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef]
- Petan, T.; Jarc, E.; Jusovic, M. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23. [Google Scholar] [CrossRef]
- Tirinato, L.; Pagliari, F.; Limongi, T.; Marini, M.; Falqui, A.; Seco, J.; Candeloro, P.; Liberale, C.; Di Fabrizio, E. An Overview of Lipid Droplets in Cancer and Cancer Stem Cells. Stem Cells Int. 2017, 2017, 1656053. [Google Scholar] [CrossRef] [PubMed]
- Tirinato, L.; Liberale, C.; Di Franco, S.; Candeloro, P.; Benfante, A.; La Rocca, R.; Potze, L.; Marotta, R.; Ruffilli, R.; Rajamanickam, V.P.; et al. Lipid droplets: A new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells 2015, 33, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Favaro, E.; Lewis, C.A.; Peck, B.; Lord, S.; Collins, J.M.; Pinnick, K.E.; Wigfield, S.; Buffa, F.M.; Li, J.L.; et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014, 9, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Cotte, A.K.; Aires, V.; Fredon, M.; Limagne, E.; Derangere, V.; Thibaudin, M.; Humblin, E.; Scagliarini, A.; de Barros, J.P.; Hillon, P.; et al. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat. Commun. 2018, 9, 322. [Google Scholar] [CrossRef]
- Glatz, J.F.; Luiken, J.J.; Bonen, A. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol. Rev. 2010, 90, 367–417. [Google Scholar] [CrossRef]
- Su, X.; Abumrad, N.A. Cellular fatty acid uptake: A pathway under construction. Trends Endocrinol. Metab. 2009, 20, 72–77. [Google Scholar] [CrossRef]
- Gharpure, K.M.; Pradeep, S.; Sans, M.; Rupaimoole, R.; Ivan, C.; Wu, S.Y.; Bayraktar, E.; Nagaraja, A.S.; Mangala, L.S.; Zhang, X.; et al. FABP4 as a key determinant of metastatic potential of ovarian cancer. Nat. Commun. 2018, 9, 2923. [Google Scholar] [CrossRef]
- Hu, B.; Guo, Y.; Garbacz, W.G.; Jiang, M.; Xu, M.; Huang, H.; Tsung, A.; Billiar, T.R.; Ramakrishnan, S.K.; Shah, Y.M.; et al. Fatty acid binding protein-4 (FABP4) is a hypoxia inducible gene that sensitizes mice to liver ischemia/reperfusion injury. J. Hepatol. 2015, 63, 855–862. [Google Scholar] [CrossRef]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef]
- Lengyel, E.; Makowski, L.; DiGiovanni, J.; Kolonin, M.G. Cancer as a Matter of Fat: The Crosstalk between Adipose Tissue and Tumors. Trends Cancer 2018, 4, 374–384. [Google Scholar] [CrossRef]
- Balaban, S.; Shearer, R.F.; Lee, L.S.; van Geldermalsen, M.; Schreuder, M.; Shtein, H.C.; Cairns, R.; Thomas, K.C.; Fazakerley, D.J.; Grewal, T.; et al. Adipocyte lipolysis links obesity to breast cancer growth: Adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer Metab. 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Li, Z.; Sun, S.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Zhang, Y.; Sun, S.; et al. Exosomes from the tumour-adipocyte interplay stimulate beige/brown differentiation and reprogram metabolism in stromal adipocytes to promote tumour progression. J. Exp. Clin. Cancer Res. 2019, 38, 223. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.M.; Stemmler, B.; Lopez-Soriano, F.J.; Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol. 2018, 15, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef]
- Ye, H.; Adane, B.; Khan, N.; Sullivan, T.; Minhajuddin, M.; Gasparetto, M.; Stevens, B.; Pei, S.; Balys, M.; Ashton, J.M.; et al. Leukemic Stem Cells Evade Chemotherapy by Metabolic Adaptation to an Adipose Tissue Niche. Cell Stem Cell 2016, 19, 23–37. [Google Scholar] [CrossRef]
- Tarrado-Castellarnau, M.; de Atauri, P.; Cascante, M. Oncogenic regulation of tumor metabolic reprogramming. Oncotarget 2016, 7, 62726–62753. [Google Scholar] [CrossRef]
- Iurlaro, R.; Leon-Annicchiarico, C.L.; Munoz-Pinedo, C. Regulation of cancer metabolism by oncogenes and tumor suppressors. Methods Enzymol. 2014, 542, 59–80. [Google Scholar] [CrossRef]
- Downward, J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 2003, 3, 11–22. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 2005, 121, 179–193. [Google Scholar] [CrossRef]
- Gouw, A.M.; Eberlin, L.S.; Margulis, K.; Sullivan, D.K.; Toal, G.G.; Tong, L.; Zare, R.N.; Felsher, D.W. Oncogene KRAS activates fatty acid synthase, resulting in specific ERK and lipid signatures associated with lung adenocarcinoma. Proc. Natl. Acad. Sci. USA 2017, 114, 4300–4305. [Google Scholar] [CrossRef]
- Ricoult, S.J.; Yecies, J.L.; Ben-Sahra, I.; Manning, B.D. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene 2016, 35, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Berwick, D.C.; Hers, I.; Heesom, K.J.; Moule, S.K.; Tavare, J.M. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J. Biol. Chem. 2002, 277, 33895–33900. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect Biol. 2012, 4, a006783. [Google Scholar] [CrossRef] [PubMed]
- Porstmann, T.; Santos, C.R.; Griffiths, B.; Cully, M.; Wu, M.; Leevers, S.; Griffiths, J.R.; Chung, Y.L.; Schulze, A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008, 8, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Zheng, Y.; Cho, S.; Jang, C.; England, C.; Dempsey, J.M.; Yu, Y.; Liu, X.; He, L.; Cavaliere, P.M.; et al. Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell 2017, 171, 1545–1558.e18. [Google Scholar] [CrossRef]
- Menendez, J.A. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: Molecular mechanisms and therapeutic perspectives. Biochim. Biophys. Acta 2010, 1801, 381–391. [Google Scholar] [CrossRef]
- Menendez, J.A.; Vellon, L.; Mehmi, I.; Oza, B.P.; Ropero, S.; Colomer, R.; Lupu, R. Inhibition of fatty acid synthase (FAS) suppresses HER2/neu (erbB-2) oncogene overexpression in cancer cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10715–10720. [Google Scholar] [CrossRef]
- Puzio-Kuter, A.M. The Role of p53 in Metabolic Regulation. Genes Cancer 2011, 2, 385–391. [Google Scholar] [CrossRef]
- Yahagi, N.; Shimano, H.; Matsuzaka, T.; Najima, Y.; Sekiya, M.; Nakagawa, Y.; Ide, T.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. p53 Activation in adipocytes of obese mice. J. Biol. Chem. 2003, 278, 25395–25400. [Google Scholar] [CrossRef]
- Liu, Y.; He, Y.; Jin, A.; Tikunov, A.P.; Zhou, L.; Tollini, L.A.; Leslie, P.; Kim, T.H.; Li, L.O.; Coleman, R.A.; et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. USA 2014, 111, E2414–E2422. [Google Scholar] [CrossRef]
- Contractor, T.; Harris, C.R. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 2012, 72, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.E. Fatty acid metabolism in breast cancer subtypes. Oncotarget 2017, 8, 29487–29500. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, D.; Simon, M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014, 24, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Tracz-Gaszewska, Z.; Dobrzyn, P. Stearoyl-CoA Desaturase 1 as a Therapeutic Target for the Treatment of Cancer. Cancers 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 2019, 120, 1090–1098. [Google Scholar] [CrossRef]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef]
- Schug, Z.T.; Peck, B.; Jones, D.T.; Zhang, Q.; Grosskurth, S.; Alam, I.S.; Goodwin, L.M.; Smethurst, E.; Mason, S.; Blyth, K.; et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 2015, 27, 57–71. [Google Scholar] [CrossRef]
- Bulusu, V.; Tumanov, S.; Michalopoulou, E.; van den Broek, N.J.; MacKay, G.; Nixon, C.; Dhayade, S.; Schug, Z.T.; Vande Voorde, J.; Blyth, K.; et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017, 18, 647–658. [Google Scholar] [CrossRef]
- Zhang, X.; Saarinen, A.M.; Hitosugi, T.; Wang, Z.; Wang, L.; Ho, T.H.; Liu, J. Inhibition of intracellular lipolysis promotes human cancer cell adaptation to hypoxia. Elife 2017, 6. [Google Scholar] [CrossRef]
- VandeKopple, M.J.; Wu, J.; Auer, E.N.; Giaccia, A.J.; Denko, N.C.; Papandreou, I. HILPDA Regulates Lipid Metabolism, Lipid Droplet Abundance, and Response to Microenvironmental Stress in Solid Tumors. Mol. Cancer Res. 2019, 17, 2089–2101. [Google Scholar] [CrossRef] [PubMed]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniels, V.W.; Machiels, J.; et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef] [PubMed]
- Ehmsen, S.; Pedersen, M.H.; Wang, G.; Terp, M.G.; Arslanagic, A.; Hood, B.L.; Conrads, T.P.; Leth-Larsen, R.; Ditzel, H.J. Increased Cholesterol Biosynthesis Is a Key Characteristic of Breast Cancer Stem Cells Influencing Patient Outcome. Cell Rep. 2019, 27, 3927–3938.e6. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Huang, J. The expanded role of fatty acid metabolism in cancer: New aspects and targets. Precis. Clin. Med. 2019, 2, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Kamisuki, S.; Mao, Q.; Abu-Elheiga, L.; Gu, Z.; Kugimiya, A.; Kwon, Y.; Shinohara, T.; Kawazoe, Y.; Sato, S.; Asakura, K.; et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 2009, 16, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Krol, S.K.; Kielbus, M.; Rivero-Muller, A.; Stepulak, A. Comprehensive review on betulin as a potent anticancer agent. Biomed. Res. Int. 2015, 2015, 584189. [Google Scholar] [CrossRef]
- Knobloch, M.; Braun, S.M.; Zurkirchen, L.; von Schoultz, C.; Zamboni, N.; Arauzo-Bravo, M.J.; Kovacs, W.J.; Karalay, O.; Suter, U.; Machado, R.A.; et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 2013, 493, 226–230. [Google Scholar] [CrossRef]
- Loftus, T.M.; Jaworsky, D.E.; Frehywot, G.L.; Townsend, C.A.; Ronnett, G.V.; Lane, M.D.; Kuhajda, F.P. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science 2000, 288, 2379–2381. [Google Scholar] [CrossRef]
- Potze, L.; Di Franco, S.; Grandela, C.; Pras-Raves, M.L.; Picavet, D.I.; van Veen, H.A.; van Lenthe, H.; Mullauer, F.B.; van der Wel, N.N.; Luyf, A.; et al. Betulinic acid induces a novel cell death pathway that depends on cardiolipin modification. Oncogene 2016, 35, 427–437. [Google Scholar] [CrossRef]
- Potze, L.; di Franco, S.; Kessler, J.H.; Stassi, G.; Medema, J.P. Betulinic Acid Kills Colon Cancer Stem Cells. Curr. Stem Cell Res. Ther. 2016, 11, 427–433. [Google Scholar] [CrossRef]
- Pinkham, K.; Park, D.J.; Hashemiaghdam, A.; Kirov, A.B.; Adam, I.; Rosiak, K.; da Hora, C.C.; Teng, J.; Cheah, P.S.; Carvalho, L.; et al. Stearoyl CoA Desaturase Is Essential for Regulation of Endoplasmic Reticulum Homeostasis and Tumor Growth in Glioblastoma Cancer Stem Cells. Stem Cell Rep. 2019, 12, 712–727. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef]
- Adams, B.D.; Kasinski, A.L.; Slack, F.J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 2014, 24, R762–R776. [Google Scholar] [CrossRef]
- Rakheja, D.; Chen, K.S.; Liu, Y.; Shukla, A.A.; Schmid, V.; Chang, T.C.; Khokhar, S.; Wickiser, J.E.; Karandikar, N.J.; Malter, J.S.; et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat. Commun. 2014, 2, 4802. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Priest, J.R.; Duchaine, T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef]
- De Vito, C.; Riggi, N.; Cornaz, S.; Suva, M.L.; Baumer, K.; Provero, P.; Stamenkovic, I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell 2012, 21, 807–821. [Google Scholar] [CrossRef]
- Melo, S.A.; Moutinho, C.; Ropero, S.; Calin, G.A.; Rossi, S.; Spizzo, R.; Fernandez, A.F.; Davalos, V.; Villanueva, A.; Montoya, G.; et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010, 18, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.; Manley, J.; Lee, J.; Singh, S.R. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015, 356, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Xiao, T.; Fang, Z.; Sun, Y.; Li, F.; Gao, Y.; Feng, Y.; Li, L.; Wang, Y.; Liu, X.; et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 2012, 287, 23227–23235. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, H.; Liu, Y.; Wu, J.; Wang, C.; Hou, X.; Chen, X.; Yang, G.; Zhao, L.; Che, H.; et al. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013, 333, 253–260. [Google Scholar] [CrossRef]
- Gregersen, L.H.; Jacobsen, A.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer 2012, 12, 232. [Google Scholar] [CrossRef]
- Chen, C.; Bai, L.; Cao, F.; Wang, S.; He, H.; Song, M.; Chen, H.; Liu, Y.; Guo, J.; Si, Q.; et al. Targeting LIN28B reprograms tumor glucose metabolism and acidic microenvironment to suppress cancer stemness and metastasis. Oncogene 2019, 38, 4527–4539. [Google Scholar] [CrossRef]
- El Helou, R.; Pinna, G.; Cabaud, O.; Wicinski, J.; Bhajun, R.; Guyon, L.; Rioualen, C.; Finetti, P.; Gros, A.; Mari, B.; et al. miR-600 Acts as a Bimodal Switch that Regulates Breast Cancer Stem Cell Fate through WNT Signaling. Cell Rep. 2017, 18, 2256–2268. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Wang, X.; Huang, Z.; Wu, Q.; Prager, B.C.; Mack, S.C.; Yang, K.; Kim, L.J.Y.; Gimple, R.C.; Shi, Y.; Lai, S.; et al. MYC-Regulated Mevalonate Metabolism Maintains Brain Tumor-Initiating Cells. Cancer Res. 2017, 77, 4947–4960. [Google Scholar] [CrossRef]
- Song, K.; Kwon, H.; Han, C.; Zhang, J.; Dash, S.; Lim, K.; Wu, T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: Regulation by MIR-122. Oncotarget 2015, 6, 40822–40835. [Google Scholar] [CrossRef]
- Ullmann, P.; Nurmik, M.; Begaj, R.; Haan, S.; Letellier, E. Hypoxia- and MicroRNA-Induced Metabolic Reprogramming of Tumor-Initiating Cells. Cells 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Qureshi-Baig, K.; Ullmann, P.; Rodriguez, F.; Frasquilho, S.; Nazarov, P.V.; Haan, S.; Letellier, E. What Do We Learn from Spheroid Culture Systems? Insights from Tumorspheres Derived from Primary Colon Cancer Tissue. PLoS ONE 2016, 11, e0146052. [Google Scholar] [CrossRef] [PubMed]
- Ullmann, P.; Qureshi-Baig, K.; Rodriguez, F.; Ginolhac, A.; Nonnenmacher, Y.; Ternes, D.; Weiler, J.; Gabler, K.; Bahlawane, C.; Hiller, K.; et al. Hypoxia-responsive miR-210 promotes self-renewal capacity of colon tumor-initiating cells by repressing ISCU and by inducing lactate production. Oncotarget 2016, 7, 65454–65470. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.S.; Keith, B.; Simon, M.C. Oxygen availability and metabolic adaptations. Nat. Rev. Cancer 2016, 16, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Wu, S.Y.; Pradeep, S.; Ivan, C.; Pecot, C.V.; Gharpure, K.M.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; Zand, B.; et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat. Commun. 2014, 5, 5202. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 2020. [Google Scholar] [CrossRef]
- Gee, H.E.; Ivan, C.; Calin, G.A.; Ivan, M. HypoxamiRs and cancer: From biology to targeted therapy. Antioxid Redox. Signal. 2014, 21, 1220–1238. [Google Scholar] [CrossRef] [PubMed]
- Puissegur, M.P.; Mazure, N.M.; Bertero, T.; Pradelli, L.; Grosso, S.; Robbe-Sermesant, K.; Maurin, T.; Lebrigand, K.; Cardinaud, B.; Hofman, V.; et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011, 18, 465–478. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Macharia, L.W.; Wanjiru, C.M.; Mureithi, M.W.; Pereira, C.M.; Ferrer, V.P.; Moura-Neto, V. MicroRNAs, Hypoxia and the Stem-Like State as Contributors to Cancer Aggressiveness. Front. Genet. 2019, 10, 125. [Google Scholar] [CrossRef]
- Bronisz, A.; Godlewski, J.; Wallace, J.A.; Merchant, A.S.; Nowicki, M.O.; Mathsyaraja, H.; Srinivasan, R.; Trimboli, A.J.; Martin, C.K.; Li, F.; et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat. Cell Biol. 2011, 14, 159–167. [Google Scholar] [CrossRef]
- Mitra, A.K.; Zillhardt, M.; Hua, Y.; Tiwari, P.; Murmann, A.E.; Peter, M.E.; Lengyel, E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012, 2, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Aboukameel, A.; Kong, D.; Wang, Z.; Sethi, S.; Chen, W.; Sarkar, F.H.; Raz, A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011, 71, 3400–3409. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, O.; Filkowski, J.; Meservy, J.; Ilnytskyy, Y.; Tryndyak, V.P.; Chekhun, V.F.; Pogribny, I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008, 7, 2152–2159. [Google Scholar] [CrossRef]
- Sun, L.; Yao, Y.; Liu, B.; Lin, Z.; Lin, L.; Yang, M.; Zhang, W.; Chen, W.; Pan, C.; Liu, Q.; et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene 2012, 31, 432–445. [Google Scholar] [CrossRef]
- Stahlhut, C.; Slack, F.J. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle 2015, 14, 2171–2180. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085. [Google Scholar] [CrossRef]
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, L.F.; Zhang, H.W.; Hu, S.; Lu, M.H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.D.; et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Pullen, T.J.; da Silva Xavier, G.; Kelsey, G.; Rutter, G.A. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol. Cell Biol. 2011, 31, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Zhang, Y.; Han, J.; Wang, W.; Liu, Y.; Li, J.; Jiang, X. Embryonic stem cell-derived pancreatic endoderm transplant with MCT1-suppressing miR-495 attenuates type II diabetes in mice. Endocr. J. 2015, 62, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Lehuede, C.; Dupuy, F.; Rabinovitch, R.; Jones, R.G.; Siegel, P.M. Metabolic Plasticity as a Determinant of Tumor Growth and Metastasis. Cancer Res. 2016, 76, 5201–5208. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Moscat, J.; Diaz-Meco, M. Metabolism shapes the tumor microenvironment. Curr. Opin. Cell Biol. 2017, 48, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Tasdogan, A.; Huang, F.; Hu, Z.; Morrison, S.J.; DeBerardinis, R.J. The abundance of metabolites related to protein methylation correlates with the metastatic capacity of human melanoma xenografts. Sci. Adv. 2017, 3, eaao5268. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef]
- Yang, L.; Achreja, A.; Yeung, T.L.; Mangala, L.S.; Jiang, D.; Han, C.; Baddour, J.; Marini, J.C.; Ni, J.; Nakahara, R.; et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016, 24, 685–700. [Google Scholar] [CrossRef]
- Anderson, A.S.; Roberts, P.C.; Frisard, M.I.; Hulver, M.W.; Schmelz, E.M. Ovarian tumor-initiating cells display a flexible metabolism. Exp. Cell Res. 2014, 328, 44–57. [Google Scholar] [CrossRef]
- Perez-Escuredo, J.; Dadhich, R.K.; Dhup, S.; Cacace, A.; Van Hee, V.F.; De Saedeleer, C.J.; Sboarina, M.; Rodriguez, F.; Fontenille, M.J.; Brisson, L.; et al. Lactate promotes glutamine uptake and metabolism in oxidative cancer cells. Cell Cycle 2016, 15, 72–83. [Google Scholar] [CrossRef]
- Fiaschi, T.; Marini, A.; Giannoni, E.; Taddei, M.L.; Gandellini, P.; De Donatis, A.; Lanciotti, M.; Serni, S.; Cirri, P.; Chiarugi, P. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012, 72, 5130–5140. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.M.; Biancur, D.E.; Wang, X.; Halbrook, C.J.; Sherman, M.H.; Zhang, L.; Kremer, D.; Hwang, R.F.; Witkiewicz, A.K.; Ying, H.; et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016, 536, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, Y.; Shingu, T.; Feng, L.; Chen, Z.; Ogasawara, M.; Keating, M.J.; Kondo, S.; Huang, P. Metabolic alterations in highly tumorigenic glioblastoma cells: Preference for hypoxia and high dependency on glycolysis. J. Biol. Chem. 2011, 286, 32843–32853. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharife, H.; Kreisel, T.; Mogilevsky, M.; Bar-Lev, L.; Grunewald, M.; Aizenshtein, E.; Karni, R.; Paldor, I.; Shlomi, T.; et al. Intra-Tumoral Metabolic Zonation and Resultant Phenotypic Diversification Are Dictated by Blood Vessel Proximity. Cell Metab. 2019, 30, 201–211.e6. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Chapple, R.H.; Lin, A.; Kitano, A.; Nakada, D. AMPK Protects Leukemia-Initiating Cells in Myeloid Leukemias from Metabolic Stress in the Bone Marrow. Cell Stem Cell 2015, 17, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jeong, E.K.; Ju, M.K.; Jeon, H.M.; Kim, M.Y.; Kim, C.H.; Park, H.G.; Han, S.I.; Kang, H.S. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol. Cancer 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241. [Google Scholar] [CrossRef]
- Prager, B.C.; Xie, Q.; Bao, S.; Rich, J.N. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 2019, 24, 41–53. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Gross, J.C.; Chaudhary, V.; Bartscherer, K.; Boutros, M. Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 2012, 14, 1036–1045. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharm. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, X.; Gong, S.; Yuan, H.; Jiang, Y.; Huang, W.; Sun, X.; Dang, X. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018, 25, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Donnarumma, E.; Fiore, D.; Nappa, M.; Roscigno, G.; Adamo, A.; Iaboni, M.; Russo, V.; Affinito, A.; Puoti, I.; Quintavalle, C.; et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget 2017, 8, 19592–19608. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, W.; Lim, Y.C.; Liu, T. Salinomycin-Loaded Gold Nanoparticles for Treating Cancer Stem Cells by Ferroptosis-Induced Cell Death. Mol. Pharm. 2019, 16, 2532–2539. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).