Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study
Simple Summary
Abstract
1. Introduction
2. Results
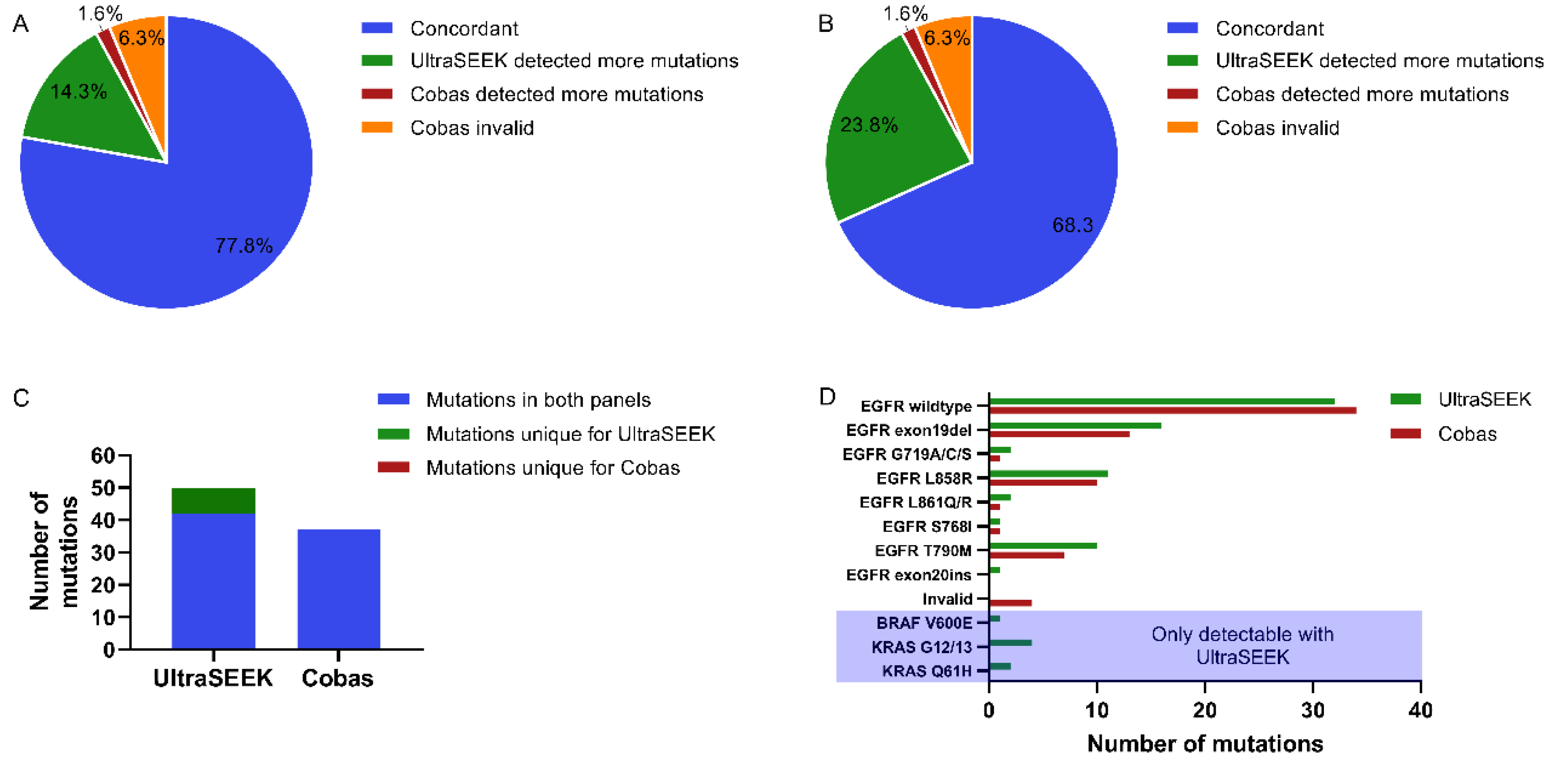
2.1. Comparison of Mutation Detection Using UltraSEEK™ and Cobas®
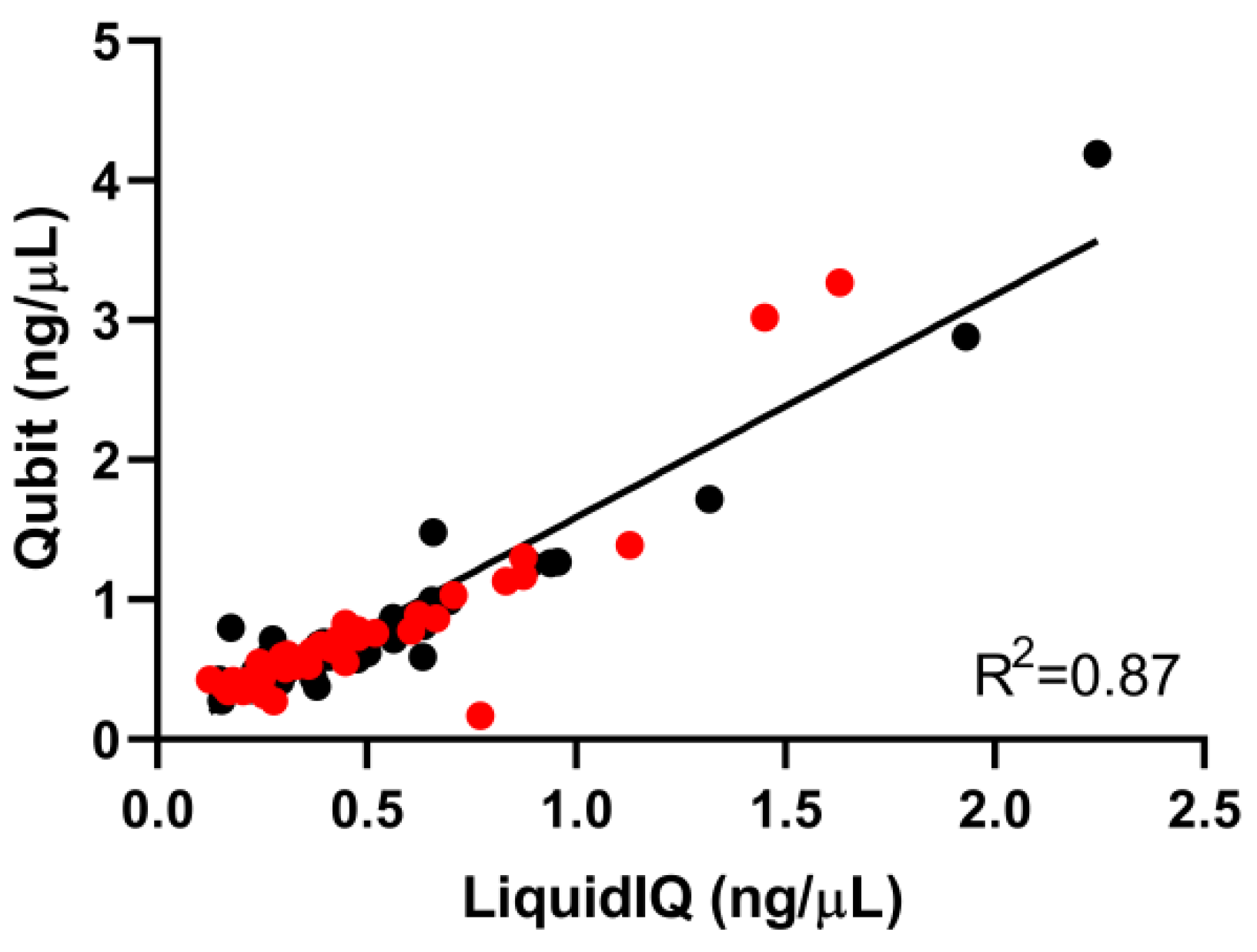
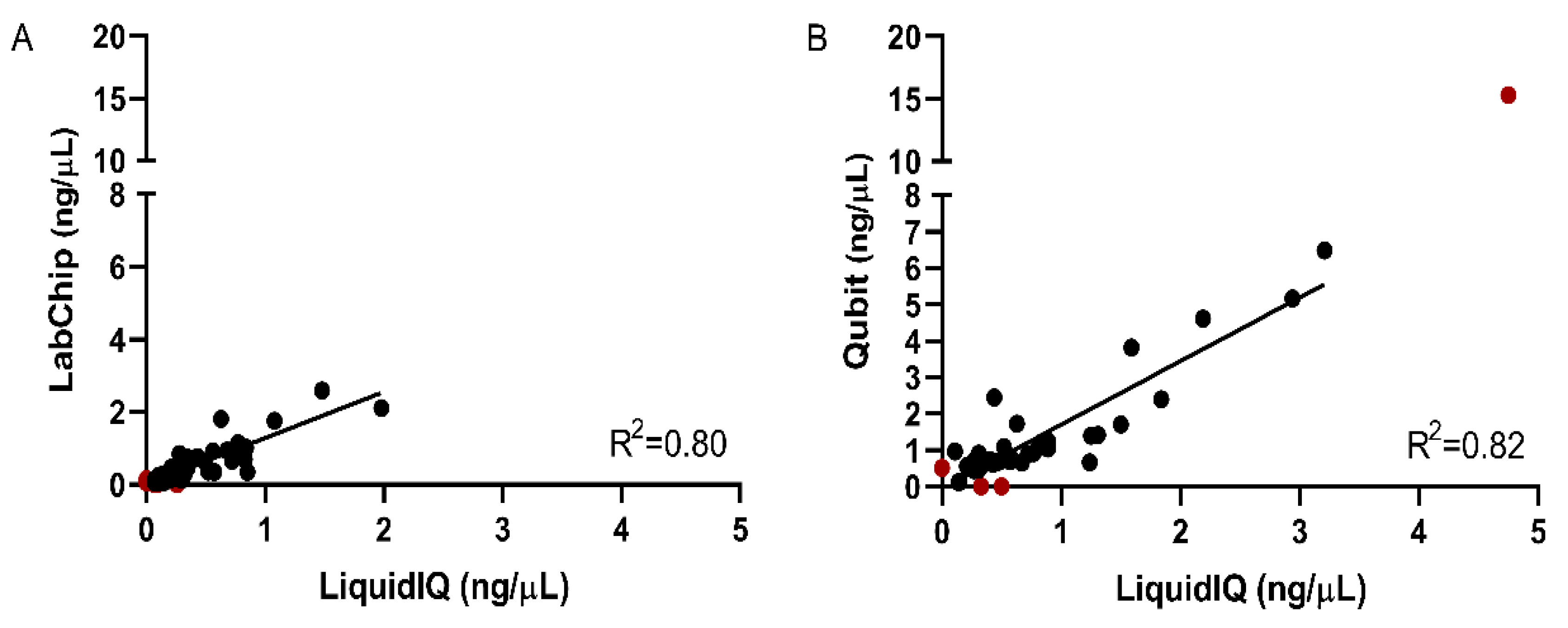
2.2. Validation of the Liquid IQ® Panel
2.3. Quantitative and Qualitative Analysis Using the Liquid IQ® Panel
2.4. Comparison of UltraSEEK™ and Cobas® with Harmonized Input
3. Discussion
4. Materials and Methods
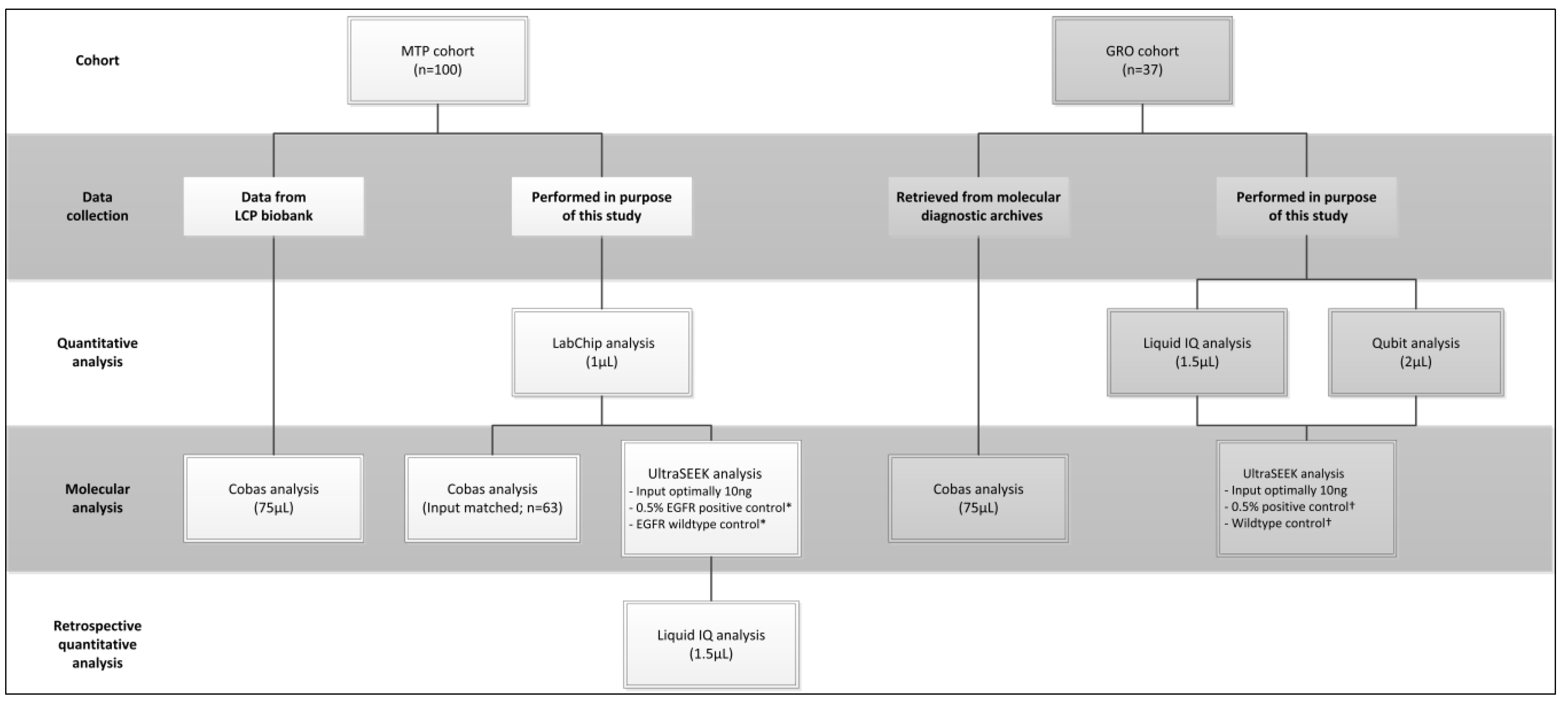
4.1. Patient Inclusion and Plasma Sample Processing
4.2. CcfDNA Extraction and Sample Quantity and Quality Assessment
4.3. Molecular Analysis
4.4. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Recondo, G.; Facchinetti, F.; Olaussen, K.A.; Besse, B.; Friboulet, L. Making the first move in EGFR-driven or ALK-driven NSCLC: First-generation or next-generation TKI? Nat. Rev. Clin. Oncol. 2018, 15, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar] [PubMed]
- Steeghs, E.M.P.; Kroeze, L.I.; Tops, B.B.J.; van Kempen, L.C.; Ter Elst, A.; Kastner-van Raaij, A.W.M.; Hendriks-Cornelissen, S.J.B.; Hermsen, M.J.W.; Jansen, E.A.M.; Nederlof, P.M.; et al. Comprehensive routine diagnostic screening to identify predictive mutations, gene amplifications, and microsatellite instability in FFPE tumor material. BMC Cancer 2020, 20, 291. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Maruvka, Y.E.; Michor, F.; Pao, W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J. Clin. Oncol. 2013, 31, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.S.; Wu, Y.-L.; Ahn, M.-J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.M.E.; et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Tagrisso Summary of Product Characteristics. Available online: https://www.ema.europa.eu/en/glossary/summary-product-characteristics (accessed on 6 September 2019).
- Rangachari, D.; To, C.; Shpilsky, J.E.; VanderLaan, P.A.; Kobayashi, S.S.; Mushajiang, M.; Lau, C.J.; Paweletz, C.P.; Oxnard, G.R.; Jänne, P.A.; et al. EGFR-Mutated Lung Cancers Resistant to Osimertinib through EGFR C797S Respond to First-Generation Reversible EGFR Inhibitors but Eventually Acquire EGFR T790M/C797S in Preclinical Models and Clinical Samples. J. Thorac. Oncol. 2019, 14, 1995–2002. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef]
- Engelman, J.A.; Mukohara, T.; Zejnullahu, K.; Lifshits, E.; Borrás, A.M.; Gale, C.-M.; Naumov, G.N.; Yeap, B.Y.; Jarrell, E.; Sun, J.; et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Investig. 2006, 116, 2695–2706. [Google Scholar] [CrossRef]
- Keppens, C.; Dequeker, E.M.C.; Rouleau, E.; ’t Hart, N.; Bubendorf, L.; Dufraing, K.; Garrec, C.; Guéguen, P.; Lamy, A.; Marchetti, A.; et al. Sensitive detection methods are key to identify secondary EGFR c.2369C>T p.(Thr790Met) in non-small cell lung cancer tissue samples. BMC Cancer 2020, 20, 366. [Google Scholar] [CrossRef]
- Douillard, J.-Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: Circulating-free tumor DNA as a surrogate for determination of EGFR status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef]
- Diao, Z.; Han, Y.; Zhang, R.; Li, J. Circulating tumour DNA: A new biomarker to monitor resistance in NSCLC patients treated with EGFR-TKIs. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188363. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Lampignano, R.; Neumann, M.H.D.; Weber, S.; Kloten, V.; Herdean, A.; Voss, T.; Groelz, D.; Babayan, A.; Tibbesma, M.; Schlumpberger, M.; et al. Multicenter Evaluation of Circulating Cell-Free DNA Extraction and Downstream Analyses for the Development of Standardized (Pre)analytical Work Flows. Clin. Chem. 2019, 66, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Spiegl, B.; Perakis, S.O.; Ulz, C.M.; Abuja, P.M.; Kashofer, K.; van der Leest, P.; Azpurua, M.A.; Tamminga, M.; Brudzewsky, D.; et al. Technical Evaluation of Commercial Mutation Analysis Platforms and Reference Materials for Liquid Biopsy Profiling. Cancers 2020, 12, 1588. [Google Scholar] [CrossRef]
- van der Leest, P.; Boonstra, P.A.; ter Elst, A.; van Kempen, L.C.; Tibbesma, M.; Koopmans, J.; Miedema, A.; Tamminga, M.; Groen, H.J.M.; Reyners, A.K.L.; et al. Comparison of Circulating Cell-Free DNA Extraction Methods for Downstream Analysis in Cancer Patients. Cancers 2020, 12, 1222. [Google Scholar] [CrossRef]
- Deans, Z.C.; Butler, R.; Cheetham, M.; Dequeker, E.M.C.; Fairley, J.A.; Fenizia, F.; Hall, J.A.; Keppens, C.; Normanno, N.; Schuuring, E.; et al. IQN path ASBL report from the first European cfDNA consensus meeting: Expert opinion on the minimal requirements for clinical ctDNA testing. Virchows Arch. 2019, 474, 681–689. [Google Scholar] [CrossRef]
- Keppens, C.; Palma, J.F.; Das, P.M.; Scudder, S.; Wen, W.; Normanno, N.; van Krieken, J.H.; Sacco, A.; Fenizia, F.; Gonzalez de Castro, D.; et al. Detection of EGFR Variants in Plasma: A Multilaboratory Comparison of a Real-Time PCR EGFR Mutation Test in Europe. J. Mol. Diagn. 2018, 20, 483–494. [Google Scholar] [CrossRef]
- Dearden, S.; Brown, H.; Jenkins, S.; Thress, K.S.; Cantarini, M.; Cole, R.; Ranson, M.; Jänne, P.A. EGFR T790M mutation testing within the osimertinib AURA Phase I study. Lung Cancer 2017, 109, 9–13. [Google Scholar] [CrossRef]
- Odogwu, L.; Mathieu, L.; Goldberg, K.B.; Blumenthal, G.M.; Larkins, E.; Fiero, M.H.; Rodriguez, L.; Bijwaard, K.; Lee, E.Y.; Philip, R.; et al. FDA Benefit-Risk Assessment of Osimertinib for the Treatment of Metastatic Non-Small Cell Lung Cancer Harboring Epidermal Growth Factor Receptor T790M Mutation. Oncologist 2018, 23, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.-K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef]
- Keppens, C.; Dequeker, E.M.C.; Patton, S.J.; Normanno, N.; Fenizia, F.; Butler, R.; Cheetham, M.; Fairley, J.A.; Williams, H.; Hall, J.A.; et al. International pilot external quality assessment scheme for analysis and reporting of circulating tumour DNA. BMC Cancer 2018, 18, 804. [Google Scholar] [CrossRef]
- List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools). Available online: https://www.fda.gov/medical-devices/vitro-diagnostics/list-cleared-or-approved-companion-diagnostic-devices-vitro-and-imaging-tools (accessed on 20 September 2020).
- Mosko, M.J.; Nakorchevsky, A.A.; Flores, E.; Metzler, H.; Ehrich, M.; van den Boom, D.J.; Sherwood, J.L.; Nygren, A.O.H. Ultrasensitive Detection of Multiplexed Somatic Mutations Using MALDI-TOF Mass Spectrometry. J. Mol. Diagn. 2016, 18, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ossandon, M.R.; Agrawal, L.; Bernhard, E.J.; Conley, B.A.; Dey, S.M.; Divi, R.L.; Guan, P.; Lively, T.G.; McKee, T.C.; Sorg, B.S.; et al. Circulating tumor DNA assays in clinical cancer research. J. Natl. Cancer Inst. 2018, 110, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Paweletz, C.P.; Kuang, Y.; Mach, S.L.; O’Connell, A.; Messineo, M.M.; Luke, J.J.; Butaney, M.; Kirschmeier, P.; Jackman, D.M.; et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 2014, 20, 1698–1705. [Google Scholar] [CrossRef] [PubMed]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Crucitta, S.; Gianfilippo, G.; Passaro, A.; Petrini, I.; Restante, G.; Michelucci, A.; Fogli, S.; de Marinis, F.; Porta, C.; et al. Understanding the Mechanisms of Resistance in EGFR-Positive NSCLC: From Tissue to Liquid Biopsy to Guide Treatment Strategy. Int. J. Mol. Sci. 2019, 20, 3951. [Google Scholar] [CrossRef]
- Gerber, T.; Taschner-Mandl, S.; Saloberger-Sindhöringer, L.; Popitsch, N.; Heitzer, E.; Witt, V.; Geyeregger, R.; Hutter, C.; Schwentner, R.; Ambros, I.M.; et al. Assessment of Pre-Analytical Sample Handling Conditions for Comprehensive Liquid Biopsy Analysis. J. Mol. Diagn. 2020, 22, 1070–1086. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- van der Leest, P.; Schuuring, E. The potential of combined mutation sequencing of plasma circulating cell-free DNA and matched white blood cells for treatment response prediction. Mol. Oncol. 2020, 14, 487–489. [Google Scholar] [CrossRef]
- Kim, S.; Le, T.-H.; Choi, Y.; Lee, H.; Heo, E.; Lee, U.; Kim, S.; Chae, S.; Kim, Y.A.; Yoon, H. Electrical monitoring of photoisomerization of block copolymers intercalated into graphene sheets. Nat. Commun. 2020, 11, 1324. [Google Scholar] [CrossRef]
- Jacot, W.; Dalenc, F.; Lopez-Crapez, E.; Chaltiel, L.; Durigova, A.; Gros, N.; Lozano, N.; Lacaze, J.-L.; Pouderoux, S.; Gladieff, L.; et al. PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res. Treat. 2019, 177, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Lamy, P.-J.; Zornback, K. Clinical implication of ctDNA analysis in advanced lung carcinoma patients using two different technologies: Realtime PCR and MALDI-TOF. Clin. Exp. Metastasis 2018, 35, 216. [Google Scholar]
- Demetrick, D.; Shell, S.; Perizzolo, M.; Stuart, A.; Santos, S.; Charlton, C.; Lozano, N.; Hummel, S.; Sartori, A.; Mosko, M.; et al. Establishing the Sensitivity, Specificity, Interlaboratory Reproducibility, and Analytical Limit of Detection of the UltraSEEK Liquid Biopsy Application Using Well-Defined Seraseq Reference Materials. In Proceedings of the AMP Annual Meeting 2019, Baltimore, MD, USA, 7–9 November 2019. Poster TT040. [Google Scholar]

| Tube Type | SNP Calls | Amplifiable Copies | WBC Contamination | Yield (ng/µL) | Qubit (ng/µL) | No Result * |
|---|---|---|---|---|---|---|
| Streck (n = 42) | 20 (16–21) | 179 (57–734) | 3% (0–28%) | 0.40 (0.13–1.6) | 0.62 (0.43–3.3) | 0 |
| EDTA (n = 42) | 20 (15–21) | 177 (66–1010) | 1% (0–35%) | 0.39 (0.15–2.3) | 0.63 (0.49–4.2) | 0 |
| Patient’s Cohort | SNP Calls | Amplifiable Copies | WBC Contamination | Yield (ng/µL) | Input (ng) | No Result * |
|---|---|---|---|---|---|---|
| MTP cohort (n = 100) | 20 (15–21) | 86 (30–891) | 0% (0–74%) | 0.19 (0.067–2.0) | 6.8 (2.7–25) | 24 |
| GRO cohort (n = 37) | 20 (15–21) | 257 (50–2136) | 1% (0–44%) | 0.57 (0.11–4.8) | 9.4 (2.3–10) | 1 |
| Input * | Cases | Concordant | Cobas Detected More Mutations | UltraSEEK Detected More Mutations | Grouped Concordance | Excluding Wildtype ‡ |
|---|---|---|---|---|---|---|
| Unevaluable † | 25 | 21 (84%) | 1 (4%) | 3 (12%) | 21 (84%) | 8/12 (67%) |
| 2–5 ng | 32 | 29 (91%) | 2 (6%) | 1 (3%) | 46 (79%) | 27/39 (69%) |
| 5–8 ng | 26 | 17 (65%) | 5 (19%) | 4 (15%) | ||
| 8–10 ng | 31 | 28 (90%) | 2 (6%) | 1 (3%) | 51 (94%) | 37/40 (93%) |
| >10 ng | 23 | 23 (100%) | 0 (0%) | 0 (0%) |
| Input * | Cases | Concordant | Cobas Detected More Mutations | UltraSEEK Detected More Mutations | Grouped Concordance | Excluding Wildtype ‡ |
|---|---|---|---|---|---|---|
| Unevaluable † | 15 | 11 (73%) | 1 (7%) | 3 (20%) | 11 (73%) | 0/4 (0%) |
| 2–5 ng | 15 | 13 (87%) | 0 (0%) | 2 (13%) | 19 (76%) | 7/13 (54%) |
| 5–8 ng | 10 | 6 (60%) | 0 (0%) | 4 (40%) | ||
| 8–10 ng | 7 | 7 (100%) | 0 (0%) | 0 (0%) | 23 (100%) | 16/16 (100%) |
| >10 ng | 16 | 16 (100%) | 0 (0%) | 0 (0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamy, P.-J.; van der Leest, P.; Lozano, N.; Becht, C.; Duboeuf, F.; Groen, H.J.M.; Hilgers, W.; Pourel, N.; Rifaela, N.; Schuuring, E.; et al. Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study. Cancers 2020, 12, 3002. https://doi.org/10.3390/cancers12103002
Lamy P-J, van der Leest P, Lozano N, Becht C, Duboeuf F, Groen HJM, Hilgers W, Pourel N, Rifaela N, Schuuring E, et al. Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study. Cancers. 2020; 12(10):3002. https://doi.org/10.3390/cancers12103002
Chicago/Turabian StyleLamy, Pierre-Jean, Paul van der Leest, Nicolas Lozano, Catherine Becht, Frédérique Duboeuf, Harry J. M. Groen, Werner Hilgers, Nicolas Pourel, Naomi Rifaela, Ed Schuuring, and et al. 2020. "Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study" Cancers 12, no. 10: 3002. https://doi.org/10.3390/cancers12103002
APA StyleLamy, P.-J., van der Leest, P., Lozano, N., Becht, C., Duboeuf, F., Groen, H. J. M., Hilgers, W., Pourel, N., Rifaela, N., Schuuring, E., & Alix-Panabières, C. (2020). Mass Spectrometry as a Highly Sensitive Method for Specific Circulating Tumor DNA Analysis in NSCLC: A Comparison Study. Cancers, 12(10), 3002. https://doi.org/10.3390/cancers12103002








