Abstract
Fusarium culmorum is a major pathogen of grain crops. Infected plants accumulate deoxynivalenol (DON), 3-acetyl-deoxynivalenol (3-ADON), or nivalenol (NIV), which are mycotoxins of the trichothecene B group. These toxins are also produced by F. graminearum species complex. New trichothecenes structurally similar to trichothecenes B but lacking the carbonyl group on C-8, designated NX toxins, were recently discovered in atypical isolates of F. graminearum from North America. Only these isolates and a few strains of a yet to be characterized Fusarium species from South Africa are known to produce NX-2 and other NX toxins. Here, we report that among 20 F. culmorum strains isolated from maize, wheat, and oat in Europe and Asia over a period of 70 years, 18 strains produced NX-2 simultaneously with 3-ADON and DON or NIV. Rice cultures of strains producing 3-ADON accumulated NX-2 in amounts corresponding to 2–8% of 3-ADON (1.2–36 mg/kg). A strain producing NIV accumulated NX-2 and NIV at comparable amounts (13.6 and 10.3 mg/kg, respectively). In F. graminearum, producers of NX-2 possess a special variant of cytochrome P450 monooxygenase encoded by TRI1 that is unable to oxidize C-8. In F. culmorum, producers and nonproducers of NX-2 possess identical TRI1; the reason for the production of NX-2 is unknown. Our results indicate that the production of NX-2 simultaneously with trichothecenes B is a common feature of F. culmorum.
Keywords:
NX-2; NX toxins; Fusarium culmorum; 3-actyl-deoxynivalenol; nivalenol; trichothecenes; chemotype Key Contribution:
Isolates of Fusarium culmorum obtained from different hosts and locations over several decades produce NX-2, which is a type A trichothecene structurally related to 3-ADON.
1. Introduction
Fusarium head blight (FHB) is a cosmopolitan disease of small-grain cereals caused by several Fusarium species with a high economic impact [1]. Fusarium graminearum and F. culmorum belong to the predominant agents of FHB [2]. Infection of grain crops with Fusarium spp. causes yield losses and contamination of grains with toxic metabolites (mycotoxins), impairing food safety [3,4].
Fusarium culmorum belongs to the F. sambucinum species complex [5]. It infects a wide range of grain crops such as wheat [2,6], maize [7,8,9], barley [10,11], oat [10], triticale [11], and rye [11]. Fusarium culmorum typically co-occurs with other Fusarium species. In most reports, the incidence of F. culmorum in crops was second to F. graminearum, but in some crops, growing regions, and years, F. culmorum dominated [2,6,10]. Infected plant material often contains trichothecene mycotoxins deoxynivalenol (DON) and its derivatives 3-acetyl-deoxynivalenol (3-ADON) and 15-acetyl-deoxynivalenol (15-ADON), nivalenol (NIV) and its derivative fusarenon X (4-acetyl-nivalenol), and resorcylic acid lactone zearalenone (ZEN) [4,12].
Biosynthesis of trichothecenes in Fusarium spp. is encoded by TRI genes, comprising a core TRI cluster and one or two additional loci, depending on the species [13]. According to the dominant trichothecene produced, Fusarium strains producing trichothecenes B are partitioned into the 3-ADON chemotype, the 15-ADON chemotype, and the NIV chemotype [13,14].
The distribution of chemotypes among wheat-producing regions exhibits a strong geographic pattern, with most areas dominated by either the 3-ADON or 15-ADON chemotype (reviewed in [15]; a succinct overview can also be found in the introduction of [16]). The 3-ADON chemotype in the USA was assumed to be introduced from Europe [17]. Some studies have not found any relationship between the chemotype and aggressiveness of F. graminearum [18,19], but other studies found the 3-ADON chemotype was more aggressive than the 15-ADON chemotype [20,21,22,23]. As several authors suggested [20,21], the discrepancy between earlier and later studies may be accounted for by the use of different inoculation methods because DON is not required for initial infection, but it facilitates the spread of the pathogen along the spike. The greater aggressiveness of the 3-ADON chemotype as compared to the 15-ADON chemotype is in line with a shift in F. graminearum populations from 15-DON to 3-ADON producers in North America over the last two decades [16,21]. In several studies on the population structure of F. graminearum, a fitness advantage of 3-ADON producers due to their higher aggressiveness was postulated and correlations with growth rates and spore production were determined. The reason for the greater aggressiveness of 3-ADON producers as compared to 15-ADON producers has rarely been addressed. A plausible hypothesis was coined by Poppenberger et al. [24] based on their finding that 3-ADON was protected against glucosylation by UDP-glucosyltransferase from Arabidopsis thaliana (see also Section 3.5).
The trichothecene chemotype has been monitored extensively in F. graminearum (e.g., [9,16,18,19,20,21,22,23,25]; reviewed in [15]). Fewer studies monitored the chemotype in F. culmorum, which only comprises the 3-ADON and NIV types [9,26,27,28]. Few studies in F. culmorum have monitored fusarenon X in pure cultures; cultures of NIV producers typically accumulated fusarenon X, some at higher and some at lower concentrations than NIV. In the past, chemotypes were often assigned according to polymorphisms in TRI genes [25], but frequently reported discrepancies between the chemotype prediction by PCR-RFLP and the results of chemical analysis have questioned this approach [14,16,29].
During an investigation of F. graminearum strains colonizing wheat heads in North America, a new type A trichothecene mycotoxin, the NX-2, was discovered [30]. The structure of NX-2 is similar to 3-ADON, but it lacks a carbonyl group at C-8. A deacetylated derivative of NX-2, called NX-3, has also been reported. Very recently, these metabolites have also been found in a yet to be characterized species of the Fusarium sambucinum species complex from South Africa [31]. The first strains of F. graminearum from North America producing NX toxins did not produce any trichothecene type B under laboratory conditions. The lack of known trichothecenes actually motivated the investigation that led to the discovery of NX toxins [30]. Strains producing NX-2 were originally found at a very low frequency, but a recent study by Lofgren et al. [32] estimated that 20% of the F. graminearum population in the USA produces NX-2. An even more recent study of F. graminearum strains in Canada [16] identified producers of NX-2 (which they designated 3ANX for consistency with the nomenclature of ADONs) among strains of F. graminearum assigned to the 15-ADON chemotype. Contrary to the reports that NX-2-producing strains from the USA have not produced DON, NIV, or their acetylated derivatives [30,33], most strains in their work [16] produced NX-2 simultaneously with 15-ADON.
Here, we report that most isolates of F. culmorum collected in Europe and Asia produce NX-2 toxin simultaneously with DON and 3-ADON or NIV.
2. Results
2.1. Mycotoxin Production in Rice Cultures
Our set of 20 F. culmorum strains consisted of 14 strains isolated from maize, wheat, and oat in Germany and 6 strains obtained from other laboratories and culture collections, which were isolated in different countries in a time span of 70 years. The analysis of rice culture extracts revealed an accumulation of NX-2 at concentrations larger than 1 mg/kg in 18 cultures (Table 1). All strains except one were primarily 3-ADON producers; one isolate was assigned to the NIV chemotype. The culture of this strain also accumulated fusarenon X, which is common in NIV producers. The concentration of 3-ADON in rice cultures of DON/ADON producers was high, exceeding 100 mg/kg in the cultures of 12 out of 20 strains. The concentrations of NX-2 and 3-ADON in the cultures of the strains of the 3-ADON chemotype were tightly correlated (Figure 1). NX-2 in these cultures accumulated to levels corresponding to 2–8% of 3-ADON. In the culture of the only isolate of the NIV chemotype (isolate 59.6st), the concentrations of NIV and NX-2 were comparable (Table 1).

Table 1.
Accumulation of trichothecenes in rice cultures of F. culmorum strains.
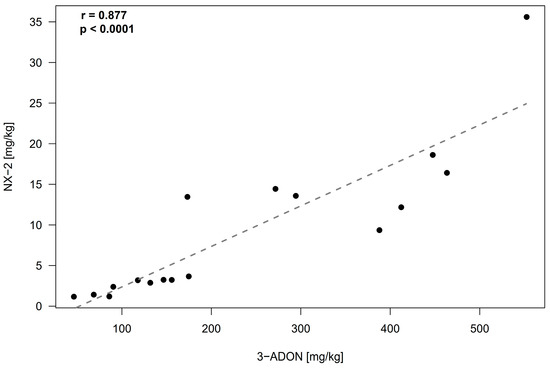
Figure 1.
Relationship (Pearson correlation) between NX-2 and 3-ADON concentrations in rice cultures of F. culmorum. Each culture (Table 1) is represented by a single data point (black dots). Isolate 59.6st, which was assigned to the NIV chemotype, and isolates 227.2 and IPP0999, in which the concentrations of NX-2 were below the limit of detection, were excluded.
2.2. Confirmation of the Structure of NX-2
Putative NX-2 accumulating in rice cultures of F. culmorum was originally identified by comparing its retention time in HPLC and MS/MS fragmentation with data obtained with purified NX-2 from Prof. Franz Berthiller (BOKU, Vienna, Austria). Because the production of NX-2 by F. culmorum has not been reported before and an isomer of NX-2 could possess the same retention time and generate product ions with the same m/z values, we purified putative NX-2 from rice cultures of F. culmorum 240.2sp (Figure 2) to verify its structure. Approximately 5 mg of pure metabolite was obtained from 480 g of dry rice culture (see Section 5.4 for details).
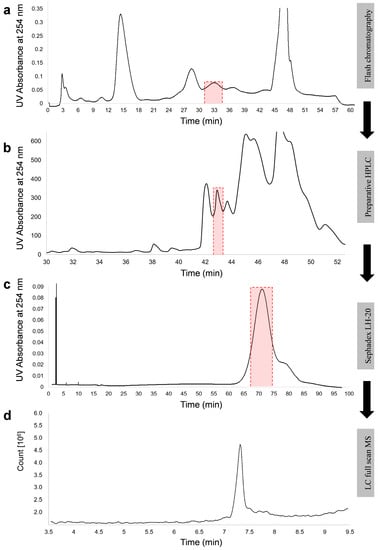
Figure 2.
Purification of NX-2 toxin from rice culture of F. culmorum 240.2sp. Three-week-old rice cultures were extracted with methanol/water/acetic acid and putative NX-2 was enriched by chromatography on a C18 cartridge (a), polar-modified C18 column (b), and Sephadex LH-20 (c). The purity of the metabolite was established by HPLC-MS in a full-scan mode (d). Red boxes mark fractions collected for purification.
One- (1H and 13C) and two-dimensional (1H,13C-HSQC, 1H,13C-HMBC, 1H,1H-COSY) NMR spectroscopic analysis was performed on the purified metabolite (Supplementary Figures S1–S5). The spectroscopic data were in accordance with published data for NX-2 (Supplementary Table S1). The structure of NX-2 is shown in Figure 3.
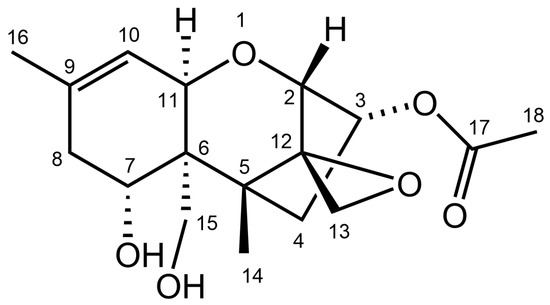
Figure 3.
Structure of NX-2 toxin.
2.3. Species Assignment and Investigation of Polymorphisms in TRI1
The examination of conidia assigned all isolates analyzed for trichothecene production (Table 1) to Fusarium culmorum. Morphological characterization was complemented by the analysis of the melting curves of amplicons of taxonomically informative genes encoding translation elongation factor 1α (TEF-1α) and the second largest component of the RNA polymerases II (RPB2) [34]. Species identification was further strengthened by the analysis of the full-length sequence (1753 nt) of the TRI1 gene, which encodes cytochrome P450 monooxygenase catalyzing oxygenation of calonectrin on C-7 and C-8. The sequences of TRI1 obtained from isolates of F. culmorum used in this study (GeneBank accession numbers OM144918 to OM144937) were aligned with a set of reference sequences (Supplementary Table S3) and used for phylogenetic analysis using the maximum-likelihood method. Separation of F. culmorum from other Fusarium species was highly supported, indicating that TRI1 is taxonomically informative in Fusarium species producing trichothecenes.
The reason for selecting the TRI1 gene for the analysis was that the product of TRI1 catalyzes biosynthetic steps distinguishing NX toxins from trichothecenes B, and that polymorphisms in TRI1 differentiating F. graminearum strains producing NX toxins from nonproducers were identified [30]. As expected, all F. graminearum strains not producing NX toxins were separated from all strains producing NX toxins. In contrast, no polymorphisms separating F. culmorum strains producing NX toxins from nonproducers were found in the TRI1 gene (Figure 4).
The amino acid sequences of the translation products of the TRI1 gene, designated Tri1, were identical for all isolates of F. culmorum in this study. Isolates of F. graminearum producing NX toxins differed from nonproducers in 14 amino acid residues within the heme-binding motif [35] (Table 2). In all F. culmorum strains used in this study, comprising 18 producers and 2 nonproducers of NX-2, these amino acid residues were identical, and they matched the corresponding residues in the strains of F. graminearum that did not produce NX toxins. Thus, the reason for the production of NX-2 by F. culmorum is not an NX-specific form of TRI1 found in NX-2 producers of F. graminearum.

Table 2.
Amino acid residues specific for the production of NX-2 in the translation product of TRI1 of F. graminearum and the corresponding residues in F. culmorum. Species-specific positions are highlighted in purple for F. culmorum and blue for F. graminearum. Positions reported to distinguish NX-2-producing strains of F. graminearum [35] are marked red.
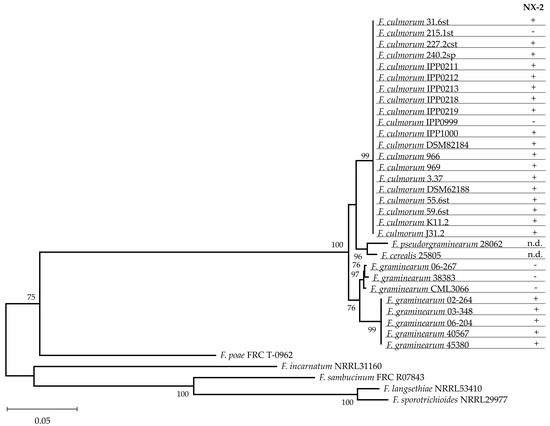
Figure 4.
Maximum likelihood estimation of the phylogenetic relationships among the TRI1 genes in Fusarium spp. Complete sequences of the TRI1 genes of the investigated isolates of F. culmorum (see Table 3) and reference sequences were subjected to maximum likelihood analysis [36] assuming the Tamura−Nei model [37]. Bootstrap values (1000 replications) are shown next to the nodes. The production of NX-2 was determined by HPLC-MS/MS; n.d. stands for no data available. Nucleotide sequences were deposited at NCBI with the accession nos. listed in Supplementary Table S3.
3. Discussion
3.1. Production of NX-2 Is a Characteristic Feature of F. culmorum
So far, production of NX-2 has exclusively been reported in F. graminearum, which shares a high level of genomic similarity with F. culmorum, produces the same mycotoxins, and has the same host range [6,38]. The production of NX-2 was discovered in an atypical population of F. graminearum from the Midwest of the USA, named the Northland population, which did not produce any known trichothecene [33]. Producers of NX-2 were later found in this population [30], but only a small fraction of F. graminearum strains collected in the area produced NX toxins. A later study on F. graminearum in wheat in Ontario (Canada), however, reported that 80% of the investigated strains produced NX toxins [16]. A key finding of the current study is that NX-2 production is not limited to F. graminearum, and, contrary to the previous assumption [35], it is not endemic to northern USA and southern Canada.
The ability to produce NX-2 appears to be a common feature of F. culmorum. Among the 20 strains of F. culmorum isolated in Europe and Asia over a period of 70 years, 18 strains produced NX-2 (Table 1). We suppose that the production of NX-2 toxins in cultures of F. culmorum remained unnoticed because NX toxins were not monitored in routine surveys. No commercial standards for NX-2 were available at the time of writing. Because infection of grain crops with F. culmorum is widespread, we assume that contamination of grains and grain products with NX-2 might be common.
3.2. Both Chemotypes of F. culmorum Produce NX-2
Isolates of F. culmorum belong to the 3-ADON and NIV chemotypes; the 15-ADON chemotype is absent [26,27,28,39,40]. Most F. culmorum isolates studied in this work belonged to the 3-ADON chemotype and produced NX-2 toxin. The only strain of the NIV chemotype produced NX-2 toxin, too (Table 1). Investigation of further isolates has yet to confirm that this generally holds for strains of the NIV chemotype. In F. graminearum, all producers of NX toxins identified in the first report from the USA belonged to the 3-ADON chemotype [30]. Thus, production of NX-2 by a strain of the NIV chemotype is another unique feature distinguishing NX-2 production in F. culmorum and F. graminearum. Another group studying F. graminearum strains from Canada confirmed this finding: NX-2 producers were rare, and all of them (including five isolates from Ontario, see below) were assigned to the 3-ADON chemotype [41]. The most recent study on F. graminearum from Ontario reported contradictory results, assigning most NX-2 producers to the 15-ADON chemotype [16]. Production of NX-2 was verified by chemical analysis in both studies. In the study of Kelly et al. [41], the 3-ADON/15-ADON chemotype was assessed by PCR-RFLP while Crippin at al. [16] used both PCR-RFLP and chemical analysis. Incongruencies between the chemotype prediction by PCR-RFLP and the results of chemical analysis, reported by several studies [14,16,29,42], cannot explain the contradictory chemotype assignments mentioned above because both studies used the same PCR assay (TRI3/TRI12), yet Crippin et al. [16] reported that all their NX-2 producers belonged to the 15-ADON chemotype while Kelly at al. [41] assigned all their NX-2 producers to the 3-ADON chemotype. Crippin at al. [16] also analyzed trichothecenes in cultures on three growth media by HPLC-MS/MS, using a special elution gradient for the separation of 3-ADON and 15-ADON (these mycotoxins often co-elute, and they cannot be reliably distinguished by MS fragmentation). They reported the concentrations of 15-ADON, but unfortunately not the concentrations of 3-ADON and/or DON.
The acetylation of C-3-OH of NX-2 (Figure 3) is reminiscent of 3-ADON. Varga et al. [30] showed that when part of the translation product of TRI1 in a strain of the 15-ADON chemotype was replaced with a TRI1 segment specific for the production of NX-2, the recombinant strain accumulated NX-4 (a derivative acetylated on C-15-OH, reminiscent of 15-ADON) rather than NX-2. The replacement of NX-specific polypeptide in an NX-2 producer with a protein segment from a 15-ADON producer converted the NX-2-producing strain into a 3-ADON producer. Both results strongly support the hypothesis that NX-2 producers in F. graminearum developed from strains of the 3-ADON chemotype. The controversy between the assignment of chemotypes to NX-2 producers from Ontario in [41] and [16] remains unresolved.
3.3. Fusarium culmorum Produces NX-2 Simultaneously with DON and 3-ADON or NIV
A search for new trichothecenes in F. graminearum that facilitated the discovery of NX toxins was motivated by a failure to detect any trichothecene in a group of atypical strains of F. graminearum from the USA [30,33]. These strains produced NX toxins but no trichothecenes B. The recent study from Ontario cited above [16] reported that cultures of F. graminearum accumulating NX-2 simultaneously accumulated 15-ADON.
Cultures of NX-2-producing F. culmorum strains in our study accumulated 3-ADON (most strains) or NIV (a single strain). In cultures of the 3-ADON chemotype, NX-2 accumulated at amounts corresponding to 2–8% of 3-ADON, and the content of NX-2 and trichothecenes B was tightly correlated (Figure 1). In contrast, the concentrations of NX-2 to 15-ADON in F. graminearum cultures in the study of Crippin et al. [16] were not correlated; the ratio NX-2/15-ADON varied from 0.003 to 33.
The only strain in our work producing NX-2 at an amount comparable to trichothecenes B was a strain of the NIV chemotype (Table 1). In rice cultures of this strain, NX-2 reached 75% of the concentration of NIV and 66% of total trichothecenes B. Future studies must clarify whether the relatively high production of NX-2 is a typical feature of strains of F. culmorum with the NIV chemotype. In F. graminearum, no strain of the NIV chemotype has so far been reported to produce NX-2.
3.4. NX-2 Production by F. culmorum Is not Caused by a Variant of TRI1
The reason for the production of NX toxins by certain strains of F. graminearum is that these strains harbor a special variant of the TRI1 gene [30]. The gene, which is only present in genomes of trichothecene-producing Fusarium species [43], encodes cytochrome P450 monooxygenase (calonectrin C7/C8 hydroxylase) Tri1, which catalyzes oxidation of C-7 and C-8 [44,45]. The difference between TRI1 of NX-2 producers and nonproducers allowed a PCR-RFLP assay for NX-2 producers [46]. We have not found such a polymorphism in the TRI1 gene of F. culmorum.
In F. graminearum, changes in Tri1 specific for NX toxins occurred in the heme-binding motif close to the C-terminus. Furthermore, Ramdass et al. [47] found a new potential glycosylation site in the enzyme of NX-2 producers. It was located at a large distance from the heme-binding site, but the authors suggested that glycosylation may modulate enzyme activity via protein folding. None of these changes occurred in the TRI1 gene of NX-2-producing F. culmorum. Some residues in the heme-binding segment differed from corresponding residues in F. graminearum (purple in Table 2), but these residues occurred in NX-2 producers and nonproducers.
Factors other than the amino acid sequence of Tri1 must suppress its activity towards C-8 of calonectrin in F. culmorum strains producing NX-2. The enzyme is located inside the endoplasmic reticulum, with two hydrophobic segments close to the ends of the protein crossing the membrane [47]. Other proteins and especially other cytochromes P450, which compete with Tri1 for the same NADPH-cytochrome P450 reductase, share this location. Cytochromes P450 are known to interact with each other and form heterooligomers, which modifies their activity [48]. The interaction of Tri1 with other proteins anchored in the membrane of endoplasmic reticulum may suppresses the oxidation of C-8 in NX-2-producing strains of F. culmorum.
3.5. NX-2 and the Aggressiveness of F. graminearum and F. culmorum
In F. graminearum, a shift from the 15-ADON to the 3-ADON chemotype observed in the last decades supports the hypothesis that the 3-ADON chemotype is more aggressive. For instance, an increase in the 3-ADON chemotype at locations where 15-ADON producers used to be predominant was reported in a study from North America [49]. Many studies of the population structure in F. graminearum elucidated the relationship between genotype, chemotype, and aggressiveness, aiming to explain how selection and gene flow shaped the populations (e.g., [17,20,35,41,46,49,50,51,52]). Field studies documented the success of the 3-ADON chemotype of F. graminearum, but they could not address its cause, which requires a biochemical approach. According to a hypothesis from the lab of Gerhard Adam [24], acetylation of C-3-OH prevents detoxification of DON, which is a virulence factor, by plant UDP-glycosyltransferases. In NX-2, the hydroxyl on C-3 is also protected by acetylation, and Varga at al. [30] suggested that the production of NX-2 may benefit F. graminearum during the infection in the same way. Glucosylation of DON takes place in the cytoplasm while the target of DON is protein synthesis in rough endoplasmic reticulum, where the acetyl group would have to be removed. This is plausible because the endoplasmic reticulum is rich in hydrolases [53]. Varga et al. [30] also speculated that the lack of carbonyl on C-8 circumvents detoxification by glutathionylation. This hypothesis holds for F. cumorum, too.
Can the effect of NX-2 on the aggressiveness of F. graminearum or F. culmorum be proved? Field trials with natural isolates are unlikely to generate a conclusive proof. The same situation exists for the claim that the 3-ADON chemotype is more aggressive than the 15-ADON chemotype, which was supported by some studies [20,21,22,23] yet rejected by others [18,19,52]. Field isolates differ in many properties modulating aggressiveness. Some of them are likely linked to the chemotype. Isogenic strains differing only in the gene in question are required. Regarding the relative aggressiveness of the chemotypes 3-ADON and 15-ADON, strains with swapped TRI8 genes or TRI8 chimeras, constructed by Alexander et al. [54], could be used. Similarly, infection experiments with F. graminearum strains harboring TRI1 with swapped domains controlling the NX-2 production, constructed by Varga at al. [30], would reveal the effect of NX-2 production on aggressiveness.
3.6. Do further Fusarium Species Produce NX Toxins?
Comparison of the amino acid residues in the Tri1 sequence distinguishing NX-2-producing isolates from nonproducers in F. graminearum (Table 2) showed that certain residues specific for the production of NX-2 are also present in Tri1 of other Fusarium species. For instance, Tri1 in a particular F. sambucinum strain shared three residues with NX-2-producing strains of F. graminearum, and it differed from Tri1 of strains that did not produce NX-2 in another three positions. We suggest that this species should be examined for NX-2 production. Fusarium cerealis and F. pseudograminearum harbor TRI1 genes very similar to TRI1 of F. culmorum (Figure 4), and the amino acid sequence in the heme-binding segment of their Tri1 protein is identical with the corresponding sequence in F. culmorum (Table 2). Examination of these species for the production of NX toxins, too, appears worthwhile. A universal PCR-based assay for NX-2 production does not seem feasible.
3.7. Can F. culmorum Produce DON and NIV Simultaneously?
It is generally assumed that strains of F. culmorum and F. graminearum produce either NIV or DON but not both. We believe that this view is biased by the low sensitivity of analytical methods in the past, which only detected trichothecenes at high concentrations (e.g., [27]). Reports of the production of both trichothecenes by a single strain were largely overlooked. For instance, Foroud et al. [3] write in their recent review “NIV chemotypes do not produce DON”. Experimental studies such as [20,55,56], the first two of which are cited in [3], clearly showed simultaneous production of DON and NIV by single strains of F. culmorum and F. graminearum.
DON and NIV have a common precursor. In NIV producers, the precursor is hydroxylated at C-4 by Tri13 [13]. We suggest that before the entire pool is hydroxylated, some precursor enters the path leading to DON, and thus all cultures producing NIV accumulate small amounts of DON and/or its acetylated derivatives. The presence of NIV in cultures of DON producers can be explained by a residual activity of TRI13 or the activities of hydroxylases with a relaxed substrate specificity. In line with this reasoning, cultures of the NIV-producing strain 59.6st in our work accumulated relatively large amounts of DON while cultures of most DON and ADON producers contained small amounts of NIV (Table 1). Similar results were reported in F. graminearum (Table 6 in [56]).
We anticipate that with the widespread use of sensitive analytical methods, small amounts of NIV will often be found in cultures of Fusarium strains producing DON, and substantial amounts of DON will be found in cultures of all strains producing NIV.
4. Conclusions
Most strains of Fusarium culmorum produce NX-2 toxin simultaneously with deoxynivalenol, 3-acetyldeoxynivalenol, or nivalenol. Strains producing NX-2 do not possess a specific variant of the TRI1 gene known from NX-2-producing strains of F. graminearum.
5. Materials and Methods
5.1. Fungal Strains
The strains of F. culmorum are listed in Table 3. Strains isolated in the course of this study were obtained from maize grains, rachis, stalks, and oat grains according to Leslie and Summerell [57]. Briefly, samples were surface sterilized for 10 min with 0.1% silver nitrate or 3% sodium hypochlorite, rinsed, and placed on potato dextrose agar (PDA). Isolates were purified via single-spore cultures and grown on PDA for colony characteristics and on low-nutrient agar (SNA, [58]) under long-wave UV light for morphological characterization of spores. For long-term storage, fungal cultures were freeze-dried.

Table 3.
Strains of F. culmorum used in this study.
Table 3.
Strains of F. culmorum used in this study.
| Species | Origin | Year | Isolate | Host |
|---|---|---|---|---|
| F. culmorum | Germany | 2017 | 31.6st | Maize stalk |
| F. culmorum | Germany | 2018 | 215.1st | Maize stalk |
| F. culmorum | Germany | 2018 | 227.2cst | Maize stalk |
| F. culmorum | Germany | 2018 | 240.2sp | Maize rachis |
| F. culmorum | - | <1984 a | IPP0211 b | Wheat |
| F. culmorum | Italy | <1993 a | IPP0212 b | Wheat |
| F. culmorum | - | 1993 | IPP0213 b | Barley |
| F. culmorum | Hungary | 1991 | IPP0618 b | Wheat |
| F. culmorum | Germany | 1991 | IPP0619 b | Wheat |
| F. culmorum | Germany | 2010 | IPP0999 b | Wheat |
| F. culmorum | Germany | 2010 | IPP1000 b | Wheat |
| F. culmorum | Germany | 1952 | DSM62184 c | Maize grain |
| F. culmorum | Syria | 2009–2010 | 966 d | Wheat |
| F. culmorum | Syria | 2009–2010 | 969 d | Wheat |
| F. culmorum | Germany | 2004 | 3.37 b | Wheat |
| F. culmorum | Germany | 1990 | DSM62188 c | Maize stalk |
| F. culmorum | Germany | 2017 | 55.6st | Maize stalk |
| F. culmorum | Germany | 2017 | 59.6st | Maize stalk |
| F. culmorum | Germany | 2021 | K11.2 | Oat |
| F. culmorum | Germany | 2021 | J31.2 | Oat |
a Isolates were collected before the specified year. b Isolates of the Plant Phytopathology and Crop Protection Section at the University of Göttingen, Göttingen, Germany. c Strains were obtained from the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. d The isolates were described by Alkadri et al. [40].
5.2. DNA Methods
Fungal DNA was extracted using a CTAB-based protocol [59] from 10 mg of lyophilized mycelium and dissolved in 50 µL of TE (10 mM Tris, 1 mM EDTA, pH 8.0). Segments of marker genes TEF-1α and RPB2 were amplified, and the PCR products subjected to high-resolution melting curve (HRM) analysis as described previously [34]. The TRI1 gene was amplified for sequencing as four overlapping fragments (Table 4), which were sequenced by the Sanger method and assembled to full-length gene sequences. PCR was carried out in a peqSTAR 96 thermocycler (PEQLAB, Erlangen, Germany) in a total reaction volume of 25 µL. Reaction mixtures were composed of a buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.3 at 25 °C; New England Biolabs, Beverly, MA, USA) adjusted to a 2 mM final MgCl2 concentration containing 100 µM of each deoxyribonucleoside triphosphate (Bioline, Luckenwalde, Germany), 0.3 µM of each primer, 0.62 U HotStart-polymerase (New England Biolabs, Beverly, MA, USA), and 1 µL of template DNA solution diluted 100 times. The thermocycling conditions are specified in Supplementary Table S2. PCR products were precipitated with isopropanol, washed with 80% ethanol, and sequenced at the facilities of Macrogen Europe (Macrogen Europe, Amsterdam, The Netherlands).

Table 4.
Oligonucleotides used for the sequence analysis of the gene TRI1.
The sequences were quality-trimmed with Chromas version 2.6.6 (Technelysium Pty. Ltd., South Brisbane, Australia) and assembled to full-length gene sequences; the accession numbers are listed in Supplementary Table S3. Multiple sequence alignments were performed with ClustalW [60] in MEGA version 10.1.8 [36]. Phylogenetic relationships among TRI1 genes in Fusarium spp. were investigated using maximum-likelihood analysis using MEGA X under the assumption of Tamura’s and Nei’s substitution model [37].
5.3. Mycotoxin Extraction and HPLC-MS/MS
Rice cultures were prepared in 50-mL Falcon tubes (Sarstedt, Nümbrecht, Germany) by autoclaving 3 g of dry polished rice with 5 mL of tap water. The tubes were inoculated with plugs of PDA (0.5 cm diameter) overgrown with 5-day-old mycelium. Rice medium incubated with agar plugs without mycelium served as a control. The cultures were incubated for 21 days at 25 °C in the dark. Fungal metabolites were extracted by shaking with 30 mL acetonitrile/water/acetic acid (84:15:1 (v/v/v)) overnight. Extracts were dried in a vacuum concentrator (Martin Christ, Osterode am Harz, Germany) and residues re-dissolved in methanol/water (20:80 (v/v)) as described previously [61]. Mycotoxin analysis was carried out using a 1290 Infinity II HPLC system (Agilent Technologies, Waldbronn, Germany) coupled with a 6460 triple quadrupole detector (Agilent Technologies, Waldbronn, Germany). The separation was performed on a Zorbax Eclipse Plus C18 column, 50 × 2.1 mm with 1.8 µm particle size (Agilent Technologies, Waldbronn, Germany). The column oven temperature was 40 °C. Mobile phase A was water with 0.1% formic acid (v/v), and phase B was methanol with 0.1% formic acid (v/v). The gradient was as follows: 0 to 0.2 min, 5% B; 0.2 to 8 min, 5% to 35% B; 8 to 8.5 min, 35% to 98% B; 8.5 to 12 min, 98% B; 12 to 12.5 min, 98% to 5% B; 12.5 to 16 min, 5% B. The calibration curve included 11 concentrations from 0.48 to 500 μg/L. A blank was analyzed after every 7th sample and a quality control standard after every 15th sample. The metabolites were detected in a multiple reaction monitoring (MRM) mode. The acquisition parameters and the limits of detection (LOD) and quantification (LOQ) are listed in Table 5.

Table 5.
Parameters for HPLC-MS/MS analysis of trichothecenes.
5.4. Purification of NX-2
F. culmorum isolate 240.2sp, which produced the largest amounts of NX-2, was cultivated on rice media. The cultures were prepared in 4 l flat penicillin flasks with 120 g of organic polished rice and 270 mL of tap water, which were autoclaved and inoculated with 12 agar-plugs (0.6 cm diameter) from 5-day-old fungal cultures. Rice cultures were incubated at 25 °C for 3 weeks in the dark. NX-2 was extracted with a 5-time excess of methanol/water/acetic acid (90:9:1) by shaking overnight. The extract was concentrated to approximately 15% of its original volume using a rotary evaporator R-100 (Buchi, Flawil, Switzerland). The concentrated extract was partitioned with the same volume of ethyl acetate (EtOAc) three times. Combined EtOAc fractions were dried in a rotary evaporator. The residue was dissolved in methanol/water (30:70) and cleared by centrifugation. The supernatant was subjected to flash chromatography (Sepacore Flash system X10/X50, Buchi, Flawil, Switzerland) equipped with a binary pump (modules C-601/C-605) and a UV detector (module C-635). Metabolites were separated on a reverse-phase column (Chromoband Flash cartridge RP C18 ec 40–63 µm, 262 × 37 mm, Machery-Nagel, Düren, Germany). Mobile phase A was water with 0.2% acetic acid; phase B was methanol with 0.2% acetic acid. The gradient elution program was as follows: 0–2 min, 5% B; 2–42 min, from 5% to 50% B; 42–45 min, from 50% to 98% B; 45–55 min, 98% B; 55–58 min, from 98% to 5% B; 58–66 min, 5% B. The flow rate was 50 mL min−1. The separation was monitored at 254 nm. Fractions containing NX-2 were collected at 33 min and the solvent was evaporated in a rotary evaporator. The residue was re-dissolved in methanol/water (30:70) and subjected to a preparative-HPLC (PU-2086 plus, Jasco, Gross-Umstadt, Germany) equipped with a preparative reverse-phase column (Nucleodur C18 pyramid, 5 µm, 250 × 21mm, Macherey-Nagel, Düren, Germany) and coupled to a UV/VIS detector (UV-970, Jasco, Gross-Umstadt, Germany). NX-2 was eluted by a gradient of water with 0.2% acetic acid (v/v) (solvent A) and methanol with 0.2% acetic acid (solvent B) as follows: 0–5 min, 5% B; 5–50 min, from 5% to 50% B; 50–55 min, from 50% to 98% B; 55–70 min, 98% B; 70–75 min, from 98% to 5% B; 75–90 min, 5% B. The injection volume was 3 mL, and the flow rate was 14 mL min−1. The separation was monitored at 254 nm. NX-2 was collected at 43 min and solvent was evaporated in a vacuum concentrator (Christ, Osterode am Harz, Germany). NX-2 was further polished on Sephadex LH-20 (Sigma-Aldrich, Darmstadt, Germany) in a 460 × 15 mm column eluted isocratically with methanol at a flow rate of 1 mL min−1. The eluent was monitored at 254 nm.
5.5. Nuclear Magnetic Resonance (NMR) Spectroscopy
1D (1H and 13C) NMR spectra were obtained from Bruker Avance 500 Hz and 2D (1H,13C-HSQC, 1H,13C-HMBC, 1H,1H-COSY) NMR spectra were obtained from an Agilent DD2 400 MHz system. The spectra were recorded at 500/400 MHz (1H) and 126/101 MHz (13C). Chemical shifts were referenced to internal TMS (δH 0, 1H) or MeOH-d4 (δc 49.0, 13C). The data are provided as Supplementary Figures S1–S5.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/toxins14070456/s1, Figure S1: 1H NMR spectrum of NX-2; Figure S2: 13C NMR spectrum of NX-2; Figure S3: 1H, 13C HSQC NMR spectrum of NX-2; Figure S4: 1H, 13C HMBC spectrum of NX-2; Figure S5: 1H, 1H COSY spectrum of NX-2. TableS1: Comparison of NMR spectra of NX-2 with metabolite purified from F. culmorum 240.2. Table S2: Conditions used for the amplification of TRI1 gene. Table S3: Acc. nos. of sequences of the TRI1 gene.
Author Contributions
Conceptualization, P.K. and A.v.T.; methodology, S.S., M.A., C.R., L.B., T.B. and P.K.; validation, S.S., M.A., C.R. and T.B.; formal analysis, S.S., M.A., L.B. and P.K.; investigation, S.S., M.A., C.R. and T.B.; resources, P.K. and A.v.T.; data curation, S.S. and M.A.; manuscript writing, all authors; visualization, S.S., M.A., C.R., T.B. and L.B.; supervision, P.K. and A.v.T.; funding acquisition, P.K. and A.v.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Federal Office for Agriculture and Food (BLE), grant numbers 2818208315 and 28AIN02B20.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary materials. DNA sequences were desposited in NCBI; their accession nos. are listted in Supplementary Table S3.
Acknowledgments
The authors would like to thank Luciana Macis and Nahid Najafi Hajiwar for technical support and Franz Berthiller (BOKU, Wien, Austria) for providing us with NX toxins. We acknowledge support by the Open Access Publication Funds of the University of Goettingen.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Birr, T.; Hasler, M.; Verreet, J.-A.; Klink, H. Composition and predominance of Fusarium species causing fusarium head blight in winter wheat grain depending on cultivar susceptibility and meteorological factors. Microorganisms 2020, 8, 617. [Google Scholar] [CrossRef] [PubMed]
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains—An update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef] [PubMed]
- Polak-Śliwińska, M.; Paszczyk, B. Trichothecenes in food and feed, relevance to human and animal health and methods of detection: A systematic review. Molecules 2021, 26, 454. [Google Scholar] [CrossRef]
- O’Donnell, K.; Rooney, A.P.; Proctor, R.H.; Brown, D.W.; McCormick, S.P.; Ward, T.J.; Frandsen, R.J.N.; Lysøe, E.; Rehner, S.A.; Aoki, T.; et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important Fusaria. Fungal Genet. Biol. 2013, 52, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Logrieco, A.; Mulè, G.; Moretti, A.; Bottalico, A. Toxigenic Fusarium species and mycotoxins associated with maize ear rot in Europe. Eur. J. Plant Pathol. 2002, 108, 597–609. [Google Scholar] [CrossRef]
- Pfordt, A.; Ramos Romero, L.; Schiwek, S.; Karlovsky, P.; von Tiedemann, A. Impact of environmental conditions and agronomic practices on the prevalence of Fusarium species associated with ear- and stalk rot in maize. Pathogens 2020, 9, 236. [Google Scholar] [CrossRef]
- Basler, R. Diversity of Fusarium species isolated from UK forage maize and the population structure of F. graminearum from maize and wheat. PeerJ 2016, 4, 2143. [Google Scholar] [CrossRef][Green Version]
- Sundheim, L.; Brodal, G.; Hofgaard, I.S.; Rafoss, T. Temporal variation of mycotoxin producing fungi in Norwegian cereals. Microorganisms 2013, 1, 188–198. [Google Scholar] [CrossRef]
- Nielsen, L.K.; Jensen, J.D.; Nielsen, G.C.; Jensen, J.E.; Spliid, N.H.; Thomsen, I.K.; Justesen, A.F.; Collinge, D.B.; Jørgensen, L.N. Fusarium head blight of cereals in Denmark: Species complex and related mycotoxins. Phytopathology 2011, 101, 960–969. [Google Scholar] [CrossRef]
- Ropejko, K.; Twarużek, M. Zearalenone and its metabolites—General overview, occurrence, and toxicity. Toxins 2021, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: From simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Villafana, R.T.; Ramdass, A.C.; Rampersad, S.N. TRI genotyping and chemotyping: A balance of power. Toxins 2020, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- van der Lee, T.; Zhang, H.; van Diepeningen, A.; Waalwijk, C. Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. 2015, 32, 453–460. [Google Scholar] [CrossRef]
- Crippin, T.; Renaud, J.B.; Sumarah, M.W.; Miller, J.D. Comparing genotype and chemotype of Fusarium graminearum from cereals in Ontario, Canada. PLoS ONE 2019, 14, e0216735. [Google Scholar] [CrossRef]
- Gale, L.R.; Ward, T.J.; Balmas, V.; Kistler, H.C. Population subdivision of Fusarium graminearum sensu stricto in the upper midwestern United States. Phytopathology 2007, 97, 1434–1439. [Google Scholar] [CrossRef]
- Gilbert, J.; Clear, R.M.; Ward, T.J.; Gaba, D.; Tekauz, A.; Turkington, T.K.; Woods, S.M.; Nowicki, T.; O’Donnell, K. Relative aggressiveness and production of 3- or 15-acetyl deoxynivalenol and deoxynivalenol by Fusarium graminearum in spring wheat. Can. J. Plant Pathol. 2010, 32, 146–152. [Google Scholar] [CrossRef]
- von der Ohe, C.; Gauthier, V.; Tamburic-Ilincic, L.; Brule-Babel, A.; Fernando, W.G.D.; Clear, R.; Ward, T.J.; Miedaner, T. A comparison of aggressiveness and deoxynivalenol production between Canadian Fusarium graminearum isolates with 3-acetyl and 15-acetyldeoxynivalenol chemotypes in field-grown spring wheat. Eur. J. Plant Pathol. 2010, 127, 407–417. [Google Scholar] [CrossRef]
- Puri, K.D.; Zhong, S. The 3ADON population of Fusarium graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology 2010, 100, 1007–1014. [Google Scholar] [CrossRef]
- Malihipour, A.; Gilbert, J.; Piercey-Normore, M.; Cloutier, S. Molecular phylogenetic analysis, trichothecene chemotype patterns, and variation in aggressiveness of Fusarium isolates causing head blight in wheat. Plant Dis. 2012, 96, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Sun, H.Y.; Li, W.; Xia, Y.L.; Deng, Y.Y.; Zhang, A.X.; Chen, H.G. Fitness of three chemotypes of Fusarium graminearum species complex in major winter wheat-producing areas of China. PLoS ONE 2017, 12, e0174040. [Google Scholar] [CrossRef] [PubMed]
- Amarasinghe, C.; Sharanowski, B.; Dilantha Fernando, W.G. Molecular phylogenetic relationships, trichothecene chemotype diversity and aggressiveness of strains in a global collection of Fusarium graminearum species. Toxins 2019, 11, 263. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, M.; Beyer, M.; Logrieco, A.; Audenaert, K.; Balmas, V.; Basler, R.; Boutigny, A.L.; Chrpová, J.; Czembor, E.; Gagkaeva, T.; et al. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Front. Microbiol. 2016, 7, 406. [Google Scholar] [CrossRef]
- Jennings, P.; Coates, M.E.; Turner, J.A.; Chandler, E.A.; Nicholson, P. Determination of deoxynivalenol and nivalenol chemotypes of Fusarium culmorum isolates from England and Wales by PCR assay. Plant Pathol. 2004, 53, 182–190. [Google Scholar] [CrossRef]
- Quarta, A.; Mita, G.; Haidukowski, M.; Santino, A.; Mulè, G.; Visconti, A. Assessment of trichothecene chemotypes of Fusarium culmorum occurring in Europe. Food Addit. Contam. 2005, 22, 309–315. [Google Scholar] [CrossRef]
- Matny, O.N.; Bates, S.T.; Song, Z. Geographic distribution of Fusarium culmorum chemotypes associated with wheat crown rot in Iraq. J. Plant. Prot. Res. 2017, 57, 43–49. [Google Scholar] [CrossRef]
- Kulik, T.; Buśko, M.; Bilska, K.; Ostrowska-Kołodziejczak, A.; Van Diepeningen, A.D.; Perkowski, J.; Stenglein, S. Depicting the discrepancy between TRI genotype and chemotype on the basis of strain CBS 139514 from a field population of F. graminearum sensu stricto from Argentina. Toxins 2016, 8, 330. [Google Scholar] [CrossRef]
- Varga, E.; Wiesenberger, G.; Hametner, C.; Ward, T.J.; Dong, Y.; Schöfbeck, D.; Mccormick, S.; Broz, K.; Stückler, R.; Schuhmacher, R.; et al. New tricks of an old enemy: Isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ. Microbiol. 2015, 17, 2588–2600. [Google Scholar] [CrossRef]
- Laraba, I.; McCormick, S.P.; Vaughan, M.M.; Geiser, D.M.; O’Donnell, K. Phylogenetic diversity, trichothecene potential, and pathogenicity within Fusarium sambucinum species complex. PLoS ONE 2021, 16, e0250812. [Google Scholar] [CrossRef] [PubMed]
- Lofgren, L.; Riddle, J.; Dong, Y.; Kuhnem, P.R.; Cummings, J.A.; Del Ponte, E.M.; Bergstrom, G.C.; Kistler, H.C. A high proportion of NX-2 genotype strains are found among Fusarium graminearum isolates from northeastern New York State. Eur. J. Plant Pathol. 2018, 150, 791–796. [Google Scholar] [CrossRef]
- Gale, L.R.; Ward, T.J.; Kistler, H.C. A subset of the newly discovered Northland population of Fusarium graminearum from the U.S. does not produce the B-type trichothecenes DON, 15ADON, 3ADON or NIV. In Proceedings of the National Fusarium Head Blight Forum, Milwaukee, WI, USA, 7–9 December 2010; pp. 48–49. [Google Scholar]
- Schiwek, S.; Beule, L.; Vinas, M.; Von Tiedemann, A.; Karlovsky, P. High-resolution melting (HRM) curve assay for the identification of eight Fusarium species causing ear rot in maize. Pathogens 2020, 9, 270. [Google Scholar] [CrossRef]
- Kelly, A.; Proctor, R.H.; Belzile, F.; Chulze, S.N.; Clear, R.M.; Cowger, C.; Elmer, W.; Lee, T.; Obanor, F.; Waalwijk, C.; et al. The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet. Biol. 2016, 95, 39–48. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Goswami, R.S.; Kistler, H.C. Heading for Disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef]
- Oufensou, S.; Balmas, V.; Scherm, B.; Rau, D.; Camilio, S.; Prota, V.A.; Ben-Attia, M.; Gargouri, S.; Pasquali, M.; El-Bok, S. Genetic variability, chemotype distribution, and aggressiveness of Fusarium culmorum on durum wheat in Tunisia. Phytopathol. Mediterr. 2019, 58, 103–113. [Google Scholar] [CrossRef]
- Alkadri, D.; Nipoti, P.; Döll, K.; Karlovsky, P.; Prodi, A.; Pisi, A. Study of fungal colonization of wheat kernels in Syria with a focus on Fusarium species. Int. J. Mol. Sci. 2013, 14, 5938–5951. [Google Scholar] [CrossRef]
- Kelly, A.C.; Clear, R.M.; O’Donnell, K.; McCormick, S.; Turkington, T.K.; Tekauz, A.; Gilbert, J.; Kistler, H.C.; Busman, M.; Ward, T.J. Diversity of Fusarium head blight populations and trichothecene toxin types reveals regional differences in pathogen composition and temporal dynamics. Fungal Genet. Biol. 2015, 82, 22–31. [Google Scholar] [CrossRef]
- Pasquali, M.; Beyer, M.; Bohn, T.; Hoffmann, L. Comparative analysis of genetic chemotyping methods for Fusarium: Tri13 polymorphism does not discriminate between 3- and 15-acetylated deoxynivalenol chemotypes in Fusarium graminearum. J. Phytopathol. 2011, 159, 700–704. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Kim, H.-S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Harris, L.J.; Alexander, N.J.; Ouellet, T.; Saparno, A.; Allard, S.; Desjardins, A.E. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl. Environ. Microbiol. 2004, 70, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 2007, 71, 2105–2123. [Google Scholar] [CrossRef]
- Liang, J.M.; Xayamongkhon, H.; Broz, K.; Dong, Y.; McCormick, S.P.; Abramova, S.; Ward, T.J.; Ma, Z.H.; Kistler, H.C. Temporal dynamics and population genetic structure of Fusarium graminearum in the upper Midwestern United States. Fungal Genet. Biol. 2014, 73, 83–92. [Google Scholar] [CrossRef]
- Ramdass, A.C.; Villafana, R.T.; Rampersad, S.N. Comparative Sequence Analysis of TRI1 of Fusarium. Toxins 2019, 11, 689. [Google Scholar] [CrossRef]
- Davydov, D.R.; Davydova, N.Y.; Sineva, E.V.; Halpert, J.R. Interactions among cytochromes P450 in microsomal membranes. J. Biol. Chem. 2015, 290, 3850–3864. [Google Scholar] [CrossRef]
- Ward, T.J.; Clear, R.M.; Rooney, A.P.; O’Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.E.; Gilbert, J.; Geiser, D.M.; Nowicki, T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 45, 473–484. [Google Scholar] [CrossRef]
- Castiblanco, V.; Castillo, H.E.; Miedaner, T. Candidate genes for aggressiveness in a natural Fusarium culmorum population greatly differ between wheat and rye head blight. J. Fungi 2018, 4, 14. [Google Scholar] [CrossRef]
- Miedaner, T.; Vasquez, A.; Castiblanco, V.; Castillo, H.E.; Foroud, N.; Würschum, T.; Leiser, W. Genome-wide association study for deoxynivalenol production and aggressiveness in wheat and rye head blight by resequencing 92 isolates of Fusarium culmorum. BMC Genom. 2021, 22, 1–16. [Google Scholar] [CrossRef]
- Mueller, B.; Groves, C.L.; Smith, D.L. Chemotype and aggressiveness evaluation of Fusarium graminearum and Fusarium culmorum isolates from wheat fields in Wisconsin. Plant Dis. 2021, 105, 3686–3693. [Google Scholar] [CrossRef] [PubMed]
- Hara-Nishimura, I.; Matsushima, R.; Shimada, T.; Nishimura, M. Diversity and formation of endoplasmic reticulum-derived compartments in plants. are these compartments specific to plant cells? Plant Physiol. 2004, 136, 3435–3439. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; McCormick, S.P.; Waalwijk, C.; van der Lee, T.; Proctor, R.H. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet. Biol. 2011, 48, 485–495. [Google Scholar] [CrossRef]
- Gang, G.; Miedaner, T.; Schuhmacher, U.; Schollenberger, M.; Geiger, H.H. Deoxynivalenol and nivalenol production by Fusarium culmorum isolates differing in aggressiveness toward winter rye. Phytopathology 1998, 88, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.J.; Mateo, R.; Hinojo, M.J.; Llorens, A.; Jiménez, M. Liquid chromatographic determination of toxigenic secondary metabolites produced by Fusarium strains. J. Chromatogr. A 2002, 955, 245–256. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Kansas State University: Manhattan, NY, USA, 2006; ISBN 9780813819198. [Google Scholar] [CrossRef]
- Nirenberg, H. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitt. Biol. Bundesanst. Land und Forstwirtsch. Berlin-Dahlem 1976, 169, 1–117. [Google Scholar]
- Brandfass, C.; Karlovsky, P. Upscaled CTAB-based DNA extraction and real-time PCR assays for Fusarium culmorum and F. graminearum DNA in plant material with reduced sampling error. Int. J. Mol. Sci. 2008, 9, 2306–2321. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Beule, L.; Lehtsaar, E.; Rathgeb, A.; Karlovsky, P. Crop diseases and mycotoxin accumulation in temperate agroforestry systems. Sustainability 2019, 11, 2925. [Google Scholar] [CrossRef]
- Wenzl, T.; Haedrich, J.; Schaechtele, A.; Robouch, P.; Stroka, J. Guidance Document on the Estimation of LOD and LOQ for Measurements in the Field of Contaminants in Feed and Food; EUR 28099; Publications Office of the European Union: Luxembourg, 2016; ISBN 978-92-79-61768-3. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).