Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases
Abstract
1. Introduction
2. ART Structures
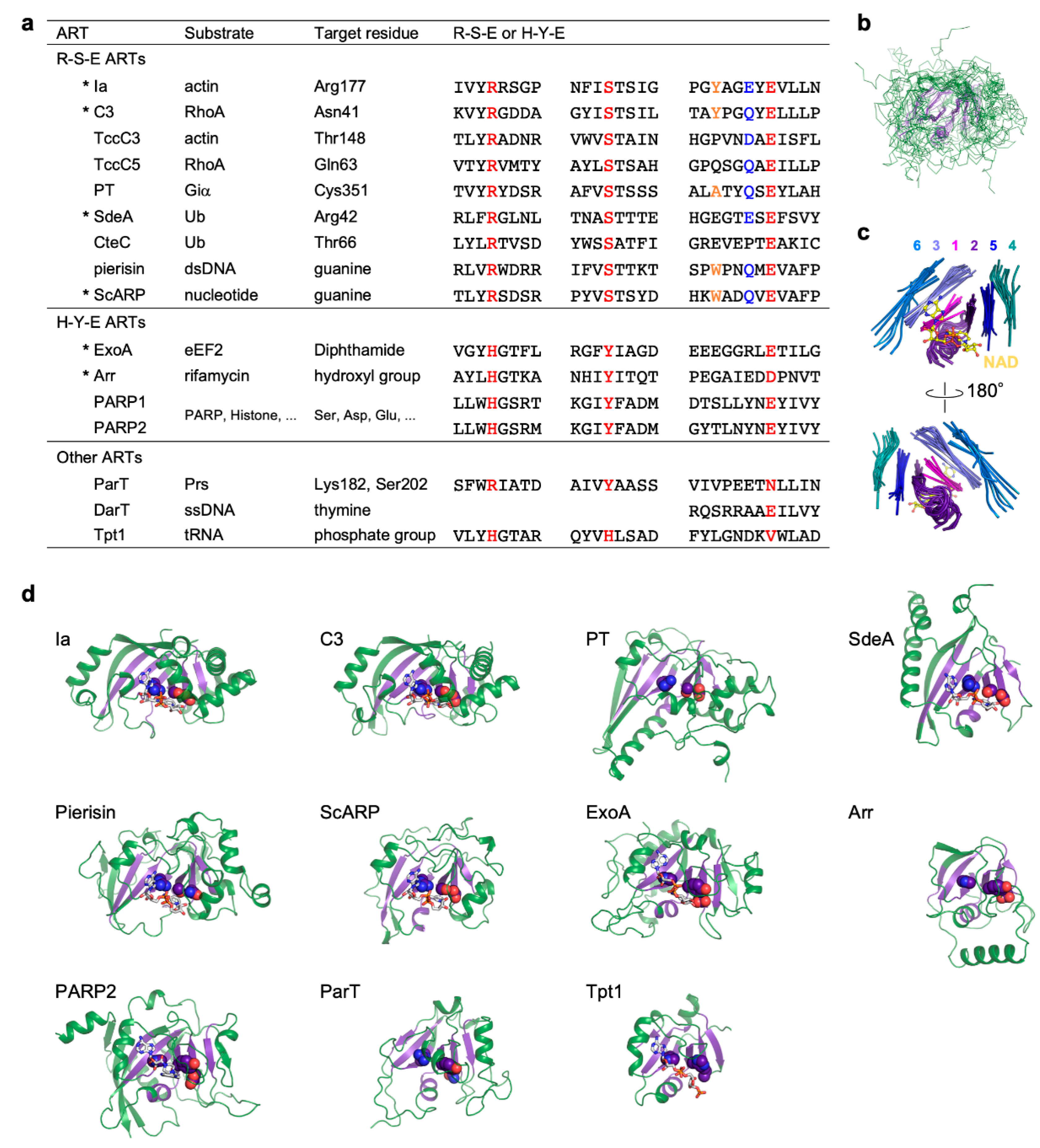
3. Substrates and Target Residues of R-S-E Class ARTs
4. Substrates and Target Residues of H-Y-E Class ARTs
5. Other ARTs
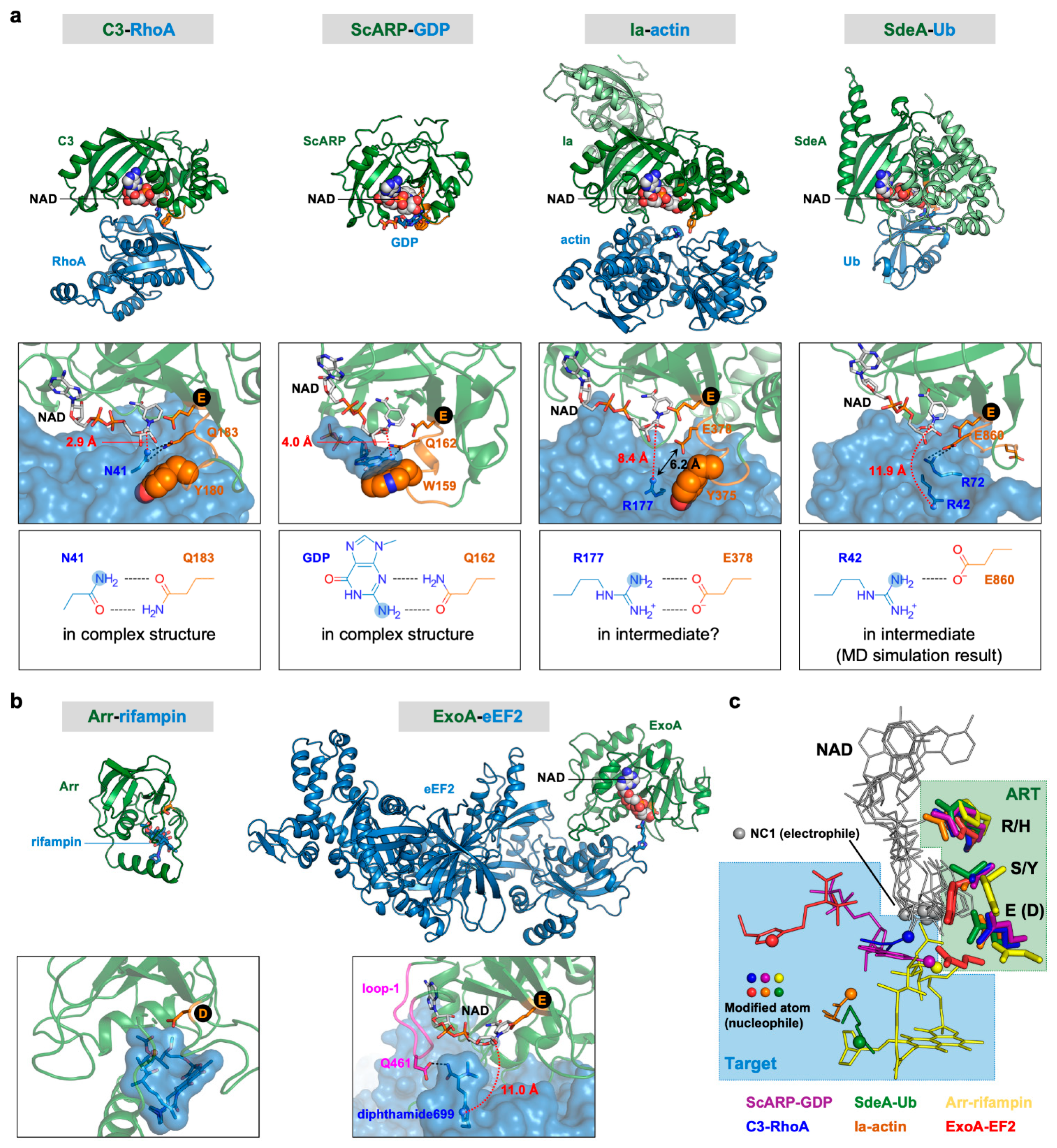
6. Substrate and Target Residue Recognition Mechanisms of R-S-E Class ARTs
7. Substrate and Target Residue Recognition Mechanisms of H-Y-E Class ARTs
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aktories, K. Bacterial Protein Toxins That Modify Host Regulatory GTPases. Nat. Rev. Microbiol. 2011, 9, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.C.; Aktories, K.; Barbieri, J.T. Novel Bacterial ADP-Ribosylating Toxins: Structure and Function. Nat. Rev. Microbiol. 2014, 12, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Unger, M.; Koch-Nolte, F.; Haag, F. ADP-Ribosylation of Arginine. Amino Acids 2011, 41, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, L.; Mikoč, A.; Ahel, I. ADP-Ribosylation: New Facets of an Ancient Modification. FEBS J. 2017, 284, 2932–2946. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Zhang, D.; de Souza, R.F.; Anand, S.; Iyer, L.M. The Natural History of ADP-Ribosyltransferases and the ADP-Ribosylation System. Curr. Top. Microbiol. Immunol. 2015, 384, 3–32. [Google Scholar] [CrossRef]
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a Unified Nomenclature for Mammalian ADP-Ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef]
- Sung, V.M.-H. Mechanistic Overview of ADP-Ribosylation Reactions. Biochimie 2015, 113, 35–46. [Google Scholar] [CrossRef]
- Margarit, S.M.; Davidson, W.; Frego, L.; Stebbins, C.E. A Steric Antagonism of Actin Polymerization by a Salmonella Virulence Protein. Struct. Lond. Engl. 1993 2006, 14, 1219–1229. [Google Scholar] [CrossRef]
- Aktories, K.; Bärmann, M.; Ohishi, I.; Tsuyama, S.; Jakobs, K.H.; Habermann, E. Botulinum C2 Toxin ADP-Ribosylates Actin. Nature 1986, 322, 390–392. [Google Scholar] [CrossRef]
- Perelle, S.; Gibert, M.; Bourlioux, P.; Corthier, G.; Popoff, M.R. Production of a Complete Binary Toxin (Actin-Specific ADP-Ribosyltransferase) by Clostridium Difficile CD196. Infect. Immun. 1997, 65, 1402–1407. [Google Scholar] [CrossRef]
- Simpson, L.L.; Stiles, B.G.; Zepeda, H.; Wilkins, T.D. Production by Clostridium Spiroforme of an Iotalike Toxin That Possesses Mono(ADP-Ribosyl)Transferase Activity: Identification of a Novel Class of ADP-Ribosyltransferases. Infect. Immun. 1989, 57, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Craig, J.A.; Putnam, C.D.; Carozzi, N.B.; Tainer, J.A. Evolution and Mechanism from Structures of an ADP-Ribosylating Toxin and NAD Complex. Nat. Struct. Biol. 1999, 6, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Irikura, D.; Monma, C.; Suzuki, Y.; Nakama, A.; Kai, A.; Fukui-Miyazaki, A.; Horiguchi, Y.; Yoshinari, T.; Sugita-Konishi, Y.; Kamata, Y. Identification and Characterization of a New Enterotoxin Produced by Clostridium Perfringens Isolated from Food Poisoning Outbreaks. PLoS ONE 2015, 10, e0138183. [Google Scholar] [CrossRef] [PubMed]
- Yonogi, S.; Matsuda, S.; Kawai, T.; Yoda, T.; Harada, T.; Kumeda, Y.; Gotoh, K.; Hiyoshi, H.; Nakamura, S.; Kodama, T.; et al. BEC, a Novel Enterotoxin of Clostridium Perfringens Found in Human Clinical Isolates from Acute Gastroenteritis Outbreaks. Infect. Immun. 2014, 82, 2390–2399. [Google Scholar] [CrossRef]
- Aktories, K.; Lang, A.E.; Schwan, C.; Mannherz, H.G. Actin as Target for Modification by Bacterial Protein Toxins. FEBS J. 2011, 278, 4526–4543. [Google Scholar] [CrossRef]
- Aktories, K.; Braun, U.; Rösener, S.; Just, I.; Hall, A. The Rho Gene Product Expressed in E. Coli Is a Substrate of Botulinum ADP-Ribosyltransferase C3. Biochem. Biophys. Res. Commun. 1989, 158, 209–213. [Google Scholar] [CrossRef]
- Genth, H.; Schmidt, M.; Gerhard, R.; Aktories, K.; Just, I. Activation of Phospholipase D1 by ADP-Ribosylated RhoA. Biochem. Biophys. Res. Commun. 2003, 302, 127–132. [Google Scholar] [CrossRef]
- Genth, H.; Gerhard, R.; Maeda, A.; Amano, M.; Kaibuchi, K.; Aktories, K.; Just, I. Entrapment of Rho ADP-Ribosylated by Clostridium Botulinum C3 Exoenzyme in the Rho-Guanine Nucleotide Dissociation Inhibitor-1 Complex. J. Biol. Chem. 2003, 278, 28523–28527. [Google Scholar] [CrossRef]
- Just, I.; Mohr, C.; Schallehn, G.; Menard, L.; Didsbury, J.R.; Vandekerckhove, J.; van Damme, J.; Aktories, K. Purification and Characterization of an ADP-Ribosyltransferase Produced by Clostridium Limosum. J. Biol. Chem. 1992, 267, 10274–10280. [Google Scholar] [CrossRef]
- Wilde, C.; Vogelsgesang, M.; Aktories, K. Rho-Specific Bacillus Cereus ADP-Ribosyltransferase C3cer Cloning and Characterization. Biochemistry 2003, 42, 9694–9702. [Google Scholar] [CrossRef]
- Wilde, C.; Chhatwal, G.S.; Schmalzing, G.; Aktories, K.; Just, I. A Novel C3-like ADP-Ribosyltransferase from Staphylococcus Aureus Modifying RhoE and Rnd3. J. Biol. Chem. 2001, 276, 9537–9542. [Google Scholar] [CrossRef]
- Turner, M.; Tremblay, O.; Heney, K.A.; Lugo, M.R.; Ebeling, J.; Genersch, E.; Merrill, A.R. Characterization of C3larvinA, a Novel RhoA-Targeting ADP-Ribosyltransferase Toxin Produced by the Honey Bee Pathogen, Paenibacillus Larvae. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, J.; Fünfhaus, A.; Knispel, H.; Krska, D.; Ravulapalli, R.; Heney, K.A.; Lugo, M.R.; Merrill, A.R.; Genersch, E. Characterization of the Toxin Plx2A, a RhoA-Targeting ADP-Ribosyltransferase Produced by the Honey Bee Pathogen Paenibacillus Larvae. Environ. Microbiol. 2017, 19, 5100–5116. [Google Scholar] [CrossRef] [PubMed]
- Lang, A.E.; Schmidt, G.; Schlosser, A.; Hey, T.D.; Larrinua, I.M.; Sheets, J.J.; Mannherz, H.G.; Aktories, K. Photorhabdus Luminescens Toxins ADP-Ribosylate Actin and RhoA to Force Actin Clustering. Science 2010, 327, 1139–1142. [Google Scholar] [CrossRef]
- Ting, S.-Y.; Bosch, D.E.; Mangiameli, S.M.; Radey, M.C.; Huang, S.; Park, Y.-J.; Kelly, K.A.; Filip, S.K.; Goo, Y.A.; Eng, J.K.; et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 2018, 175, 1380–1392.e14. [Google Scholar] [CrossRef] [PubMed]
- Katada, T. The Inhibitory G Protein G(i) Identified as Pertussis Toxin-Catalyzed ADP-Ribosylation. Biol. Pharm. Bull. 2012, 35, 2103–2111. [Google Scholar] [CrossRef]
- Littler, D.R.; Ang, S.Y.; Moriel, D.G.; Kocan, M.; Kleifeld, O.; Johnson, M.D.; Tran, M.T.; Paton, A.W.; Paton, J.C.; Summers, R.J.; et al. Structure-Function Analyses of a Pertussis-like Toxin from Pathogenic Escherichia Coli Reveal a Distinct Mechanism of Inhibition of Trimeric G-Proteins. J. Biol. Chem. 2017, 292, 15143–15158. [Google Scholar] [CrossRef]
- Qiu, J.; Sheedlo, M.J.; Yu, K.; Tan, Y.; Nakayasu, E.S.; Das, C.; Liu, X.; Luo, Z.-Q. Ubiquitination Independent of E1 and E2 Enzymes by Bacterial Effectors. Nature 2016, 533, 120–124. [Google Scholar] [CrossRef]
- Bhogaraju, S.; Kalayil, S.; Liu, Y.; Bonn, F.; Colby, T.; Matic, I.; Dikic, I. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 2016, 167, 1636–1649.e13. [Google Scholar] [CrossRef]
- Yan, F.; Huang, C.; Wang, X.; Tan, J.; Cheng, S.; Wan, M.; Wang, Z.; Wang, S.; Luo, S.; Li, A.; et al. Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination. Mol. Cell 2020, 78, 641–652.e9. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kono, T.; Matsushima-Hibiya, Y.; Kanazawa, T.; Nishisaka, N.; Kishimoto, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Molecular Cloning of an Apoptosis-Inducing Protein, Pierisin, from Cabbage Butterfly: Possible Involvement of ADP-Ribosylation in Its Activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10608–10613. [Google Scholar] [CrossRef] [PubMed]
- Takamura-Enya, T.; Watanabe, M.; Totsuka, Y.; Kanazawa, T.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Mono(ADP-Ribosyl)Ation of 2’-Deoxyguanosine Residue in DNA by an Apoptosis-Inducing Protein, Pierisin-1, from Cabbage Butterfly. Proc. Natl. Acad. Sci. USA 2001, 98, 12414–12419. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kono, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Purification of Pierisin, an Inducer of Apoptosis in Human Gastric Carcinoma Cells, from Cabbage Butterfly, Pieris Rapae. Jpn. J. Cancer Res. 1998, 89, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Matsushima-Hibiya, Y.; Yamamoto, M.; Takahashi-Nakaguchi, A.; Fukuda, H.; Ono, M.; Takamura-Enya, T.; Kinashi, H.; Totsuka, Y. ADP-Ribosylation of Guanosine by SCO5461 Protein Secreted from Streptomyces Coelicolor. Toxicon Off. J. Int. Soc. Toxinol. 2013, 63, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Lyons, B.; Ravulapalli, R.; Lanoue, J.; Lugo, M.R.; Dutta, D.; Carlin, S.; Merrill, A.R. Scabin, a Novel DNA-Acting ADP-Ribosyltransferase from Streptomyces Scabies. J. Biol. Chem. 2016, 291, 11198–11215. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, J.; Wieden, H.-J.; Rodnina, M.V.; Aktories, K. Inactivation of the Elongation Factor Tu by Mosquitocidal Toxin-Catalyzed Mono-ADP-Ribosylation. Appl. Environ. Microbiol. 2002, 68, 4894–4899. [Google Scholar] [CrossRef]
- Wolf, P.; Elsässer-Beile, U. Pseudomonas Exotoxin A: From Virulence Factor to Anti-Cancer Agent. Int. J. Med. Microbiol. IJMM 2009, 299, 161–176. [Google Scholar] [CrossRef]
- Oppenheimer, N.J.; Bodley, J.W. Diphtheria Toxin. Site and Configuration of ADP-Ribosylation of Diphthamide in Elongation Factor 2. J. Biol. Chem. 1981, 256, 8579–8581. [Google Scholar] [CrossRef]
- Jørgensen, R.; Purdy, A.E.; Fieldhouse, R.J.; Kimber, M.S.; Bartlett, D.H.; Merrill, A.R. Cholix Toxin, a Novel ADP-Ribosylating Factor from Vibrio Cholerae. J. Biol. Chem. 2008, 283, 10671–10678. [Google Scholar] [CrossRef]
- Baysarowich, J.; Koteva, K.; Hughes, D.W.; Ejim, L.; Griffiths, E.; Zhang, K.; Junop, M.; Wright, G.D. Rifamycin Antibiotic Resistance by ADP-Ribosylation: Structure and Diversity of Arr. Proc. Natl. Acad. Sci. USA 2008, 105, 4886–4891. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Suskiewicz, M.J.; Palazzo, L.; Hughes, R.; Ahel, I. Progress and Outlook in Studying the Substrate Specificities of PARPs and Related Enzymes. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-Ribosyl)Hydrolases: Structure, Function, and Biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef]
- Piscotta, F.J.; Jeffrey, P.D.; Link, A.J. ParST Is a Widespread Toxin-Antitoxin Module That Targets Nucleotide Metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 826–834. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Lawarée, E.; Jankevicius, G.; Cooper, C.; Ahel, I.; Uphoff, S.; Tang, C.M. DNA ADP-Ribosylation Stalls Replication and Is Reversed by RecF-Mediated Homologous Recombination and Nucleotide Excision Repair. Cell Rep. 2020, 30, 1373–1384.e4. [Google Scholar] [CrossRef] [PubMed]
- Culver, G.M.; McCraith, S.M.; Zillmann, M.; Kierzek, R.; Michaud, N.; LaReau, R.D.; Turner, D.H.; Phizicky, E.M. An NAD Derivative Produced during Transfer RNA Splicing: ADP-Ribose 1”-2” Cyclic Phosphate. Science 1993, 261, 206–208. [Google Scholar] [CrossRef]
- Spinelli, S.L.; Kierzek, R.; Turner, D.H.; Phizicky, E.M. Transient ADP-Ribosylation of a 2’-Phosphate Implicated in Its Removal from Ligated TRNA during Splicing in Yeast. J. Biol. Chem. 1999, 274, 2637–2644. [Google Scholar] [CrossRef]
- Yang, C.-S.; Jividen, K.; Spencer, A.; Dworak, N.; Ni, L.; Oostdyk, L.T.; Chatterjee, M.; Kuśmider, B.; Reon, B.; Parlak, M.; et al. Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516.e5. [Google Scholar] [CrossRef]
- Chatrin, C.; Gabrielsen, M.; Buetow, L.; Nakasone, M.A.; Ahmed, S.F.; Sumpton, D.; Sibbet, G.J.; Smith, B.O.; Huang, D.T. Structural Insights into ADP-Ribosylation of Ubiquitin by Deltex Family E3 Ubiquitin Ligases. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Tainer, J.A. The ARTT Motif and a Unified Structural Understanding of Substrate Recognition in ADP-Ribosylating Bacterial Toxins and Eukaryotic ADP-Ribosyltransferases. Int. J. Med. Microbiol. IJMM 2002, 291, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Vogelsgesang, M.; Aktories, K. Exchange of Glutamine-217 to Glutamate of Clostridium Limosum Exoenzyme C3 Turns the Asparagine-Specific ADP-Ribosyltransferase into an Arginine-Modifying Enzyme. Biochemistry 2006, 45, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Toda, A.; Tsurumura, T.; Yoshida, T.; Tsumori, Y.; Tsuge, H. Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure. J. Biol. Chem. 2015, 290, 19423–19432. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Tsuge, H. Substrate N2 Atom Recognition Mechanism in Pierisin Family DNA-Targeting, Guanine-Specific ADP-Ribosyltransferase ScARP. J. Biol. Chem. 2018, 293, 13768–13774. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, H.; Nagahama, M.; Oda, M.; Iwamoto, S.; Utsunomiya, H.; Marquez, V.E.; Katunuma, N.; Nishizawa, M.; Sakurai, J. Structural Basis of Actin Recognition and Arginine ADP-Ribosylation by Clostridium Perfringens Iota-Toxin. Proc. Natl. Acad. Sci. USA 2008, 105, 7399–7404. [Google Scholar] [CrossRef] [PubMed]
- Tsurumura, T.; Tsumori, Y.; Qiu, H.; Oda, M.; Sakurai, J.; Nagahama, M.; Tsuge, H. Arginine ADP-Ribosylation Mechanism Based on Structural Snapshots of Iota-Toxin and Actin Complex. Proc. Natl. Acad. Sci. USA 2013, 110, 4267–4272. [Google Scholar] [CrossRef]
- Tsuge, H.; Nagahama, M.; Nishimura, H.; Hisatsune, J.; Sakaguchi, Y.; Itogawa, Y.; Katunuma, N.; Sakurai, J. Crystal Structure and Site-Directed Mutagenesis of Enzymatic Components from Clostridium Perfringens Iota-Toxin. J. Mol. Biol. 2003, 325, 471–483. [Google Scholar] [CrossRef]
- Dong, Y.; Mu, Y.; Xie, Y.; Zhang, Y.; Han, Y.; Zhou, Y.; Wang, W.; Liu, Z.; Wu, M.; Wang, H.; et al. Structural Basis of Ubiquitin Modification by the Legionella Effector SdeA. Nature 2018, 557, 674–678. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, M.; Feng, H.; Zhu, Y.; Liu, S.; Gao, A.; Gao, P. Structural Insights into Non-Canonical Ubiquitination Catalyzed by SidE. Cell 2018, 173, 1231–1243.e16. [Google Scholar] [CrossRef]
- Jørgensen, R.; Merrill, A.R.; Yates, S.P.; Marquez, V.E.; Schwan, A.L.; Boesen, T.; Andersen, G.R. Exotoxin A-EEF2 Complex Structure Indicates ADP Ribosylation by Ribosome Mimicry. Nature 2005, 436, 979–984. [Google Scholar] [CrossRef]
- Jørgensen, R.; Wang, Y.; Visschedyk, D.; Merrill, A.R. The Nature and Character of the Transition State for the ADP-Ribosyltransferase Reaction. EMBO Rep. 2008, 9, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Suskiewicz, M.J.; Zobel, F.; Ogden, T.E.H.; Fontana, P.; Ariza, A.; Yang, J.-C.; Zhu, K.; Bracken, L.; Hawthorne, W.J.; Ahel, D.; et al. HPF1 Completes the PARP Active Site for DNA Damage-Induced ADP-Ribosylation. Nature 2020, 579, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Bilokapic, S.; Suskiewicz, M.J.; Ahel, I.; Halic, M. Bridging of DNA Breaks Activates PARP2-HPF1 to Modify Chromatin. Nature 2020, 585, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-Ribosylation Depends on HPF1. Mol. Cell 2017, 65, 932–940.e6. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, T.; Tsuge, H. Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases. Toxins 2021, 13, 40. https://doi.org/10.3390/toxins13010040
Yoshida T, Tsuge H. Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases. Toxins. 2021; 13(1):40. https://doi.org/10.3390/toxins13010040
Chicago/Turabian StyleYoshida, Toru, and Hideaki Tsuge. 2021. "Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases" Toxins 13, no. 1: 40. https://doi.org/10.3390/toxins13010040
APA StyleYoshida, T., & Tsuge, H. (2021). Common Mechanism for Target Specificity of Protein- and DNA-Targeting ADP-Ribosyltransferases. Toxins, 13(1), 40. https://doi.org/10.3390/toxins13010040





