Highlights
What are the main findings?
- Monocular depth estimation based on the Depth Anything v2 method applied for segmentation and calculation of vegetation indices for trees in an olive grove proved to be superior to other methods.
- The method used, the time of year when trees are photographed, and the resolution used significantly affect the final results of multispectral measurements.
What is the implication of the main finding?
- When determining vegetation indices in an olive grove, high-resolution sampling (drone) and the application of segmentation methods based on depth estimation are recommended.
- The seasonality and dependence of the calculated vegetation indices must be taken into account when drawing conclusions about the condition of trees. It is recommended to propose a new index that would take into account all these parameters and be simpler for interpretation by the user.
Abstract
This study investigates the application of unmanned aerial vehicles with multispectral cameras for monitoring the condition of olive groves with high accuracy. Multispectral images of olive groves provided detailed insight into the spectral data required for the analysis of vegetation indices. Using deep learning-based object detection, individual olive trees were identified within the images, which allowed the extraction of parts corresponding to each tree. To separate the background from the canopy, segmentation based on the monocular depth estimation algorithm, Depth Anything, was applied. In this way, elements that are not part of the tree’s crown were removed for more accurate analysis and calculation of the NDVI (Normalized Difference Vegetation Index) and NDRE (Normalized Difference Red Edge Index) indices. The obtained results were compared with the results obtained for unsegmented patches, threshold-based patches, and manually segmented patches. The comparison and analysis carried out shows that the proposed segmentation approach improved the accuracy of NDVI and NDRE by focusing exclusively on the crowns of the observed trees, excluding the noise of the surrounding vegetation and soil. In addition, measurements were carried out on three observed olive groves at different parts of the vegetation cycle, and the values of the vegetation indices were compared. This integrated method combining drone-based multispectral imaging, deep learning object detection, and advanced segmentation techniques highlights a robust approach to olive tree health monitoring and provides insight into seasonal vegetation dynamics, for winter and spring, to capture differences in vegetative activity.
1. Introduction
Olive trees (Olea europaea L.) are one of the most historically significant and economically valuable crops in the Mediterranean region. In the production year 2023/2024, the provisional total production of olive oil in the world was 2564 thousand tons, while the provisional total production of table olives was 2825.5 thousand tons. Almost 60% of the total production of olive oil is in the EU, while the percentage of production of table olives in the EU is lower and accounts for about 22% of world production [1]. It is worth noting that the stated percentages of production share vary considerably from year to year.
Olive oil, known for its high nutritional value, continues to grow in demand globally. The price of extra virgin olive oil recorded a positive trend since 2020, both in the EU member states and in the world (in the EU, the price was between EUR 8 and 9/kg) [1,2]. However, the sustainability and productivity of olive groves are increasingly threatened by various factors such as pests, diseases, climate-related stress (high and low temperatures, lack of water, etc.), and nutrient imbalances. For example, in the Republic of Croatia in 2023, high spring temperatures and, in some places, frequent precipitation during the flowering period had a negative effect on fruit set, and a long-term drought with high temperatures during the summer months on shallow soil in olive groves that are not largely irrigated had negative effects and consequences on the production of olive fruits and on the amount of olive oil produced. Therefore, compared to the year 2022, a significant reduction in olive fruit production by 25.6% and olive oil production by 30.6% was reported [3].
These challenges necessitate continuous and precise monitoring of olive tree health to optimize yield and minimize losses. Traditional monitoring methods, such as manual field inspections, are time-consuming and often inadequate for large-scale plantations. Satellite-based remote sensing technologies, while useful, depend on weather conditions (cloudiness) and often lack the necessary resolution for detecting subtle changes in individual trees. This gap has led to the increasing adoption of unmanned aerial vehicles (UAVs), or drones, equipped with advanced sensors, which provide high-resolution, real-time data over large areas, thereby enabling more precise and efficient monitoring [4]. UAVs outfitted with multispectral sensors are capable of capturing reflectance data across multiple wavelengths, including the visible, near-infrared (NIR), and red-edge bands of the electromagnetic spectrum.
These measurements allow for the calculation of widely used vegetation indices such as the Normalized Difference Vegetation Index (NDVI) and the Normalized Difference Red Edge Index (NDRE) [5]. NDVI is a well-established index used to assess plant vigor and photosynthetic activity by comparing the reflectance of red and NIR light. Higher NDVI values indicate healthy, actively photosynthesizing plants, while lower values suggest stress or disease. NDRE, which focuses on the red-edge portion of the spectrum, is more sensitive to changes in chlorophyll content, making it particularly effective for detecting early-stage stress or physiological changes in plants that may not yet be visible to the naked eye. Together, these indices provide powerful tools for assessing plant health and informing timely interventions in olive groves. Despite the utility of NDVI and NDRE, accurate application of these indices at the individual plant level can be challenging, particularly in heterogeneous agricultural environments.
First of all, if satellite images are used, for example, from ESA Sentinel-2A, the spatial resolution (1 pixel corresponds to an area of 10 m × 10 m) does not allow accurate determination of the vegetation index of an individual plant. Furthermore, the information about the state of the plant obtained by the measured vegetation index is not actually complete, since the computed values must be viewed in the context of seasonality. Namely, the values of the plant vegetation indices, including the NDVI and NDRE indices, vary significantly throughout the year [6]. In addition, multi-year change trends have been observed due to climate change [7,8]. When calculating the NDVI and NDRE index values, errors can occur due to inaccurate radiometric calibration [9]. This error can in some cases be greater than 10%, so, when comparing measurements in different weather conditions and days, it is definitely necessary to consider the need for sensor calibration [10].
In order to address issues with low-resolution images, as well as the need to separate the pixels that represent the crown of trees (in our case olive trees) from the background (branches, grass, soil), it is necessary to (1) use higher resolution images (collected by a UAV) and (2) apply some of the advanced procedures for object detection and segmentation of digital images. In this research, we propose a complete procedure that includes the application of deep neural networks for the detection of individual trees [11,12] in images collected by an unmanned aerial vehicle with a multispectral camera. Furthermore, the images of the detected olive trees are segmented using Depth Anything v2 [13] in order to discard the background from the olive tree crown. Depth Anything v2 is an advanced and robust monocular depth estimation model that aims to predict accurate depth maps from a single 2D image without using stereo depth sensors.
In this study, a comprehensive procedure is proposed for collecting RGB and multispectral images by an unmanned aerial vehicle, detecting objects of interest (olive trees), and extracting only those parts of the detected patches that truly represent the tree crown (separation from surrounding vegetation, discarding pixels belonging to branches and soil). This improves the accuracy of calculating NDVI and NDRE values, allowing for more accurate monitoring of plant health. In addition, measurements were conducted on three olive groves during two distinct seasons (spring and winter) allowing for an analysis of how NDVI and NDRE values fluctuate throughout the growing cycle. These seasonal comparisons provide valuable insights into the temporal dynamics of olive tree health, highlighting periods of increased stress or growth that may require targeted management practices. Development of a more accurate and automated method for monitoring olive groves ultimately improves the efficiency of agricultural practices and contributes to the long-term sustainability of olive production.
A recent review article on the use of remote sensing imagery applications for precision management in olive growing highlighted the significant increase in published articles in this area over the past few years [14]. A trend of increasing use of UAVs was observed, as well as the integration of UAVs equipped with multispectral sensors. Satellites [15,16] and high-altitude flying airplanes [17] provide extensive coverage, but have limitations primarily regarding spatial resolution and sensitivity to cloud cover. Due to this fact, UAVs find extensive use in olive orchards for tree counting, yield estimation, biophysical parameter estimation [18], and disease detection and monitoring [19,20].
Multispectral sensors on UAVs allow for the collection of high-resolution data across critical spectral bands [21], including near-infrared (NIR) and red-edge, essential for monitoring plant health and detecting early signs of stress. Vegetation indices (VIs) that are obtained by a multispectral sensor on a UAV were applied to quantify the density of vegetation ground cover in olive groves [22]. The most popular index, NDVI, provides insight into the general lushness of vegetation, while NDRE, due to its sensitivity to chlorophyll levels, has proven very successful in detecting irrigation irregularities [23] and early stress symptoms, particularly those related to water deficiency and nutrient imbalance [24].
Advanced methods [25,26,27] such as RetinaNet and DeepForest have emerged as essential tools for isolating individual olive trees in dense orchards [28], a critical step to enhance NDVI and NDRE accuracy. RetinaNet, recognized for its effectiveness in small object detection, applies a focal loss function that emphasizes tree-specific features, helping to distinguish individual trees even in complex vegetation settings. Complementing this, DeepForest further refines tree canopy identification by leveraging its specialization in forest-type canopy detection, which aligns closely with the structural characteristics of olive groves [26,29].
Once tree crowns are located, it is necessary to separate the crown pixels from the background because the detected locations are rectangles surrounding a single tree. One of the approaches is the implementation of Otsu’s based thresholding [30]. Another solution is based on delineation of olive tree crowns using a height threshold method applied to the calculated digital elevation model [31].
A segmentation method that does not require prior definition of the number of clusters or additional information about the heights of the recorded terrain is the mean shift method [32]. Mean shift, as a non-parametric clustering algorithm, groups pixels based on spectral and spatial features, enabling precise canopy segmentation. This methodology has shown significant accuracy improvements in NDVI and NDRE readings [33]. However, if the soil is more similar in characteristics to the olive tree crown, the segmentation may be unreliable. Therefore, a new approach is proposed that is based on depth-based segmentation, i.e., separating the parts of the tree that belong to the crown from the background (soil).
Additionally, researchers have analyzed the seasonality of vegetation indices, highlighting its value in identifying critical growth phases and guiding interventions adapted to seasonal changes [34,35].
After this introduction with the current state of the art, the rest of the paper is organized as follows: Materials and methodology are described in Section 2. Section 3 presents the results of the analysis of the observed olive groves. The discussion of the results obtained and the final conclusions are given in Section 4 and Section 5.
2. Materials and Methods
2.1. Study Area
The study was conducted in three olive groves located in the Mediterranean region of Croatia [36], each characterized by specific environmental conditions that are representative of the broader olive cultivation landscape in the region. The first study location was in Mravinci, situated north of Split, Croatia (43°31′40.7″N, 16°30′57.8″E). This olive grove has 38 trees that were observed. The second (17 olive trees) and third (58 olive trees) study locations were in Tinj, a rural area located south-west of Benkovac in Zadar County, Croatia. The coordinates for these locations are 44°00′49″N, 15°28′13.2″E and 44°01′10.6″N, 15°28′19.5″E (Figure 1).
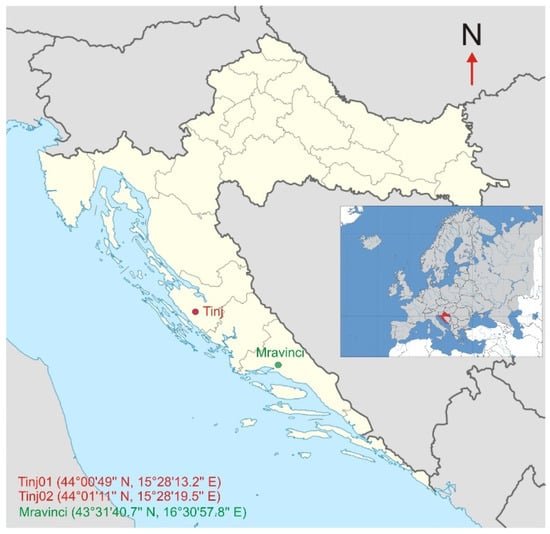
Figure 1.
Geographical locations of the study sites in Croatia. The map highlights Mravinci near Split and two sites in Tinj near Benkovac, Zadar County.
Both regions where the study was conducted experience a Mediterranean climate, which is characterized by dry summers and mild, wet winters. This climatic context is crucial for understanding the growth patterns and health of the olive trees in these areas. The olive orchard at the Mravinci location can be classified as extensive, and is characterized by irregular pruning and the non-uniform shaping of trees. Most of the trees are free vise-shaped, while a smaller number of trees have monoconical and globe-shaped plants. The trees are densely planted and, as a rule, there is no space between the crowns. Olive trees at the Tinj locations were vise-shaped and rather heavy pruned. In this location, there is a distance of one to three meters between the crowns of the trees. All observed sites have a Mediterranean climate characterized by dry summers and mild, wet winters.
2.2. Vegetation Index Maps
The ability of vegetation indices to provide insights into the health and vigor of olive trees [37] relies heavily on the principles of electromagnetic radiation, specifically how light at different wavelengths interacts with the leaves and canopy of olive trees. Light is a form of electromagnetic radiation that propagates in waves. The electromagnetic spectrum includes a broad range of wavelengths, from very short gamma rays to very long radio waves. However, in remote sensing and particularly in agricultural applications like olive farming, we focus on a narrower range of wavelengths, including visible light (400–700 nm), near-infrared (NIR, 700–1300 nm), and the red edge (690–730 nm).
Vegetation, particularly healthy olive leaves, reflects a significant portion of near-infrared (NIR, 700–1300 nm) light. This reflection is due to the internal structure of the leaves, which scatters NIR light. The NIR band is crucial for assessing plant biomass and overall health. Furthermore, the red edge (690–730 nm) is a narrow spectral region between the red and NIR bands. It is highly sensitive to changes in chlorophyll content, making it an important indicator for detecting early stress in vegetation, including olive trees.
When electromagnetic radiation strikes the surface of olive trees, it interacts in three primary ways: absorption, reflection, and transmission. These interactions are influenced by the biochemical and structural properties of the leaves and can be measured and analyzed using remote sensing techniques. Chlorophyll pigments in olive leaves absorb light primarily in the blue and red regions of the spectrum for photosynthesis. This absorption is critical for the plant’s energy production and results in lower reflectance values in these bands. Olive leaves reflect light differently depending on the wavelength. In the green band, some light is reflected, giving leaves their green appearance. However, in the NIR region, healthy leaves reflect a substantial amount of light, which is why NIR reflectance is a key component in vegetation indices like NDVI. Finally, some light passes through the leaf, particularly in the NIR region. This transmitted light may provide additional information about the leaf’s internal structure, water content, and health [37,38].
Vegetation indices are mathematical combinations of reflectance values at specific wavelengths, designed to highlight specific characteristics of vegetation. NDVI is calculated using the reflectance in the red and NIR bands. Healthy olive trees, which have high NIR reflectance and low red reflectance, will exhibit higher NDVI values, indicating good health and vigor.
NDRE is like NDVI but uses the red-edge band instead of the red band. NDRE is particularly useful in detecting early signs of stress in olive trees, such as nutrient deficiencies or water stress, which may not yet be apparent in visible light or NIR reflectance alone. NDRE is especially valuable in precision agriculture for managing nutrient applications and optimizing irrigation practices in olive orchards. In olive farming, understanding how light interacts with the trees enables the effective use of NDVI and NDRE to monitor and manage the orchard. These indices provide essential data that can guide decisions on irrigation, fertilization, and pest control, ultimately improving the quality and yield of the olive harvest.
The formula for NDVI calculation is as follows:
NDVI = (NIR − RED)/(NIR + RED)
Here, NIR represents the near-infrared light, which vegetation strongly reflects, and RED represents visible red light, which vegetation absorbs. The resulting value can range from −1 to 1, with higher values indicating healthier vegetation. This calculation provided a measure of the photosynthetic activity and overall health of the olive trees.
The formula for NDRE calculation is as follows:
where RE (red edge) represents a visible red light spectral band which is more sensitive to chlorophyll changes than the red band used in NDVI.
NDRE = (NIR − RE)/(NIR + RE)
Accordingly, the formed vegetation index maps provide insight into additional information about the observed plant than that obtained exclusively from the visible spectrum, i.e., a standard camera.
2.3. Procedure
This study utilizes a DJI (Da-Jiang Innovations, Shenzen, China) Phantom P4 drone to collect multispectral and RGB imagery from three olive groves. The primary objective was to obtain NDVI (Normalized Difference Vegetation Index) and NDRE (Normalized Difference Red Edge) values for each olive tree within the groves.
The collected RGB and multispectral images are the input for the proposed deep learning-based object detection system. The system allows for the extraction of individual image patches corresponding to each olive tree. These patches are further analyzed by estimating the depth for each pixel. This information, together with the pixel color, is used to extract the olive crown pixels from the other pixels in the observed patch.
Finally, the NDVI and NDRE values of the original patches and values from the segmented parts of the patches were compared to evaluate the effectiveness of the segmentation process. This comprehensive approach, combining multispectral imaging, deep learning, and advanced segmentation techniques, aims to enhance the precision and efficiency of olive grove monitoring and management in the region. A complete and more detailed overview of the procedure is shown in Figure 2.
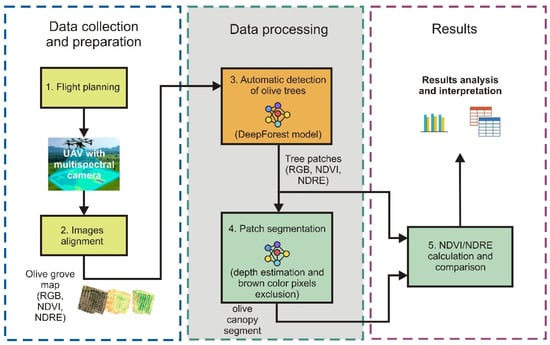
Figure 2.
Workflow of NDVI/NDRE analysis for olive tree monitoring.
2.3.1. Data Collection and Preparation
RGB and multispectral images were collected using the camera on the DJI Phantom 4 Multispectral drone The recording schedule is shown in Table 1. This UAV is equipped with a camera with six 1/2.9″ CMOS (complementary metal-oxide semiconductor) image sensors. One CMOS sensor is the RGB sensor for visible light imaging, while the other five sensors are narrow-band sensors used for multispectral imaging. Each sensor has 2.08 megapixels (MP). The spectral bands and their respective central wavelengths are as follows: Blue: 450 nm ± 16 nm; Green: 560 nm ± 16 nm; Red: 650 nm ± 16 nm; Red Edge: 730 nm ± 16 nm; Near-Infrared (NIR): 840 nm ± 26 nm. The DJI Phantom 4 multispectral camera calibrates its bands relative to the near-infrared (NIR) band. To obtain true reflectance factors, a radiometric calibration target is necessary.

Table 1.
Shooting dates in test olive groves.
These sensors allow the capture of reflectance data across different wavelengths, enabling the calculation of vegetation indices such as the NDVI and NDRE [37,39]. The DJI P4 multispectral camera is integrated with a stabilized 3-axis gimbal, ensuring that the sensors remain steady during flight, thereby reducing motion blur and ensuring high-quality data capture. The camera system is also synchronized with the UAV’s GPS, which allows for accurate georeferencing of the images, crucial for creating reliable and repeatable datasets for analysis. Data was collected under consistent flight conditions to maintain uniformity across different study sites. Flights were conducted at an altitude of 35 m, with a flight speed of 5 m/s. The forward and sideway image overlaps were set at 75%, ensuring comprehensive coverage of the areas surveyed. The resulting ground sampling distance (GSD) was 2 cm, providing high-resolution imagery suitable for detailed analysis of the olive groves and providing a spatial resolution sufficient to identify and analyze individual olive trees.
DJI Terra v4.0.0 software was utilized as the primary flight planning software for this study [40]. In addition to flight planning, DJI Terra facilitated the stitching of collected RGB images (images alignment module in Figure 2), enabling the creation of high-resolution 2D terrain maps of the monitored olive orchards. During reconstruction with P4 Multispectral photos, DJI Terra obtains the reflectivity based on the camera’s DN value (reflection intensity) and the incident intensity obtained by the spectral sunlight sensor, which will then provide the reflectivity so as to calculate the NDVI and other indices.
2.3.2. Data Processing
The next phase of the procedure is the automatic detection of olive trees (Module 3 in Figure 2). The proposed approach is based on the integration of the RetinaNet deep learning model and the DeepForest Python package, designed to accurately identify and delineate individual olive trees within dense agricultural environments [11,24]. The standard, pretrained DeepForest model, developed primarily on images featuring a variety of general tree types, did not include olive trees in its original training set. Therefore, in [11], authors applied transfer learning for adapting RetinaNet to the local olive tree dataset. The described procedure is carried out on RGB maps of olive trees. Detection results of individual olive trees on maps Tinj1, Tinj2, and Mravinci are given in Figure 3. Patches are stored from each map of olive groves, which represent the rectangles that enclose the trees (Figure 4).

Figure 3.
UAV imagery with identified olive trees in three study locations: (a) Tinj1 (May, 2021), (b) Tinj2 (May, 2021), and (c) Mravinci (May, 2021).

Figure 4.
Example of individual olive tree patches extracted from the Tinj1 site using object detection algorithm.
As presented in Figure 2, DJI Terra software package, in addition to producing the RGB map of the monitored olive grove, produced corresponding NDVI and NDRE maps as well (Figure 5).
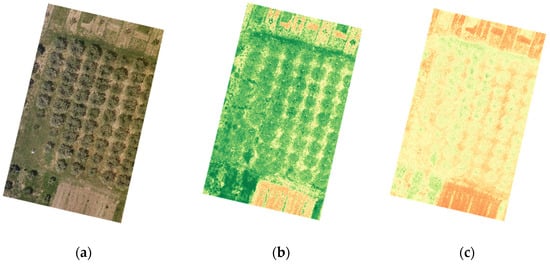
Figure 5.
Multispectral and vegetation index analysis of the olive grove Tinj 2 May: (a) original image, (b) NDVI map, and (c) NDRE map.
Patch coordinates for each map were stored in an XML file, and a Python 3.10 program was written to extract NDVI and NDRE patches at the same coordinates. So, now, three patches are stored for each tree (patches with RGB, NDVI, and NDRE values).
The patch segmentation procedure (Module 4 in Figure 2) written in Python 3.10 consists of several steps that are performed for every patch of olive grove map:
- Step 1
- Read location of current patch on original map, patch width and height;
- Step 2
- Store new RGB patch with same location but double width and height;
- Step 3
- Calculate depth map for the new RGB patch (Depth_map);
- Step 4
- ndvi channel = extracted ndvi values from NDVI patch;
- Step 5
- ndre channel = extracted ndre values from NDRE patch;
- Step 6
- Make combined NDVI mask for the original patch using pixels from (Depth_map, (depth value > depth threshold) AND ndvi channel (ndvi value > ndvi threshold));
- Step 7
- Make combined NDRE mask for the original patch using pixels from (Depth_map (depth value > depth threshold) AND ndre channel (ndre value > ndre threshold));
- Step 8
- Make color mask (excluding brown pixels);
- Step 9
- Make final NDVI mask (combine NDVI mask and color mask);
- Step 10
- Store segmented NDVI patch (original NDVI AND final NDVI mask);
- Step 11
- Make final NDRE mask (combine NDRE mask and color mask);
- Step 12
- Store segmented NDRE patch (original NDRE AND final NDRE mask).
Some additional explanation of the steps presented is needed. In Step 2, a new patch is created that, in addition to the pixels of the original patch, also includes surrounding pixels from the original map. This enlarged patch is used to better estimate depth in the next step (Step 3), when Depth Anything v2 model is applied, because the original patch may have insufficient background pixels for reliable depth estimation.
In the fourth and fifth step of this procedure, NDVI and NDRE values are extracted from the stored NDVI and NDRE patches. Based on these values and the depth mask obtained by applying the Depth Anything v2 model, appropriate filtering masks are created, taking into account the set acceptance threshold values that are defined in the initialization section for all patches (Steps 6 and 7).
In Step 8, an additional color mask is generated. The original RGB patch is converted to the HSV color model, and pixels with a color within a defined range of brown shades are excluded from the new matrix–mask. By combining the color mask and the masks generated in Steps 6 and 7, final filtering masks are created, which are applied to extract only the desired parts of the original patches that represent parts of the olive tree crown (Steps 9 to 12).
An illustration of the application of the Depth Anything v2 model to one of the original patches is shown in Figure 6.
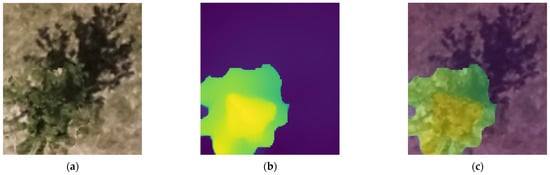
Figure 6.
Olive tree crown extraction process using monocular depth estimation: (a) original RGB patch (step 1), (b) depth map obtained by Depth Anything v2 (step 3), and (c) depth overlay on original patch.
By comparing the NDVI/NDRE values derived from masked and unmasked images, the effectiveness of the proposed segmentation procedure in enhancing index accuracy can be quantitatively evaluated. Typically, masked calculations reduce the noise introduced by non-vegetative elements, providing a clearer and more accurate measure of the olive tree’s condition.
3. Results
The following parameter values were used for the proposed procedure: depth_threshold = 0.5 (threshold is set to 50% of maximum depth); NDVI_threshold = 0.4; and NDRE_threshold = 0.3. Hue-saturation-value (HSV) range for brown color exclusion was set from (13,70,30) to (18,100,150).
In order to compare the obtained results, in addition to whole (non-segmented) patches, NDVI and NDRI values were also calculated for patches where all pixels whose NDVI or NDRE channel value was above the set threshold (0.4 or 0.3, respectively) were discarded. Also, all individual patches are manually labeled in order to single out only the part of the crown with leaves of each tree. The generated masks were used to calculate “Manually labeled” patches and the corresponding NDVI and NDRE values that included only the pixels of the mask (Figure 7).
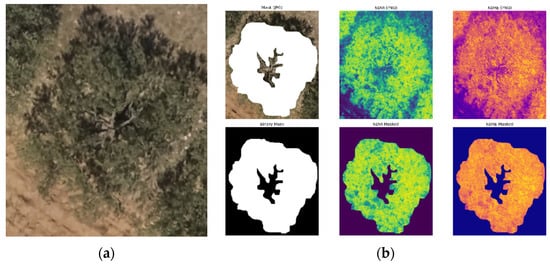
Figure 7.
Example of manually labeled patch. Tinj 1 (May, 2021), tree #13: (a) original patch and (b) mask applied to NDVI and NDRE channels.
Furthermore, to compare the calculated values of the observed indices, it was necessary to pair the detections obtained for different seasons. Although this procedure could be solved programmatically (by comparing the x, y coordinates of the patches on the maps), in this case, the alignment of the tree numbering was performed manually. It should be noted that, due to the extremely dense distribution of trees at the Mravinci location, the DeepForest detection algorithm dropped several trees, and only those pairs of trees that were correctly detected in both seasonal surveys were considered for this location in the calculation. It should also be noted that even a human observer could not reliably determine the boundaries of the trees in these cases.
3.1. NDVI Results
The NDVI analysis for olive groves in different locations (Tinj 1, Tinj 2, and Mravinci) across two time periods (May and December/January) is presented in Table 2.

Table 2.
Comparison of mean NDVI Values by segmentation and season for three monitored olive grove locations. Non-segmented: all patch pixels are used for the calculation of the index; Depth Seg: index values calculated only for pixels remained after patch segmentation procedure; NDVI 0.4: only pixels with NDVI > 0.4 included in calculation; Manually segmented: only pixels included with manually segmented mask used for calculation.
Seasonal comparison for non-segmented and segmented image processing methods is given in Figure 8. The results indicate significant differences in NDVI values between the segmented and non-segmented approaches for the Tinj 1 location in May (0.68 vs. 0.77), and also for the Tinj 2 location (0.69 vs. 0.79 in May and 0.73 vs. 0.80 for winter measurement). For the olive grove at the Mravinci location, the differences in unsegmented and segmented values are small. This can be explained by the small number of pixels representing the background in the patch and the dense arrangement of trees and touching crowns.
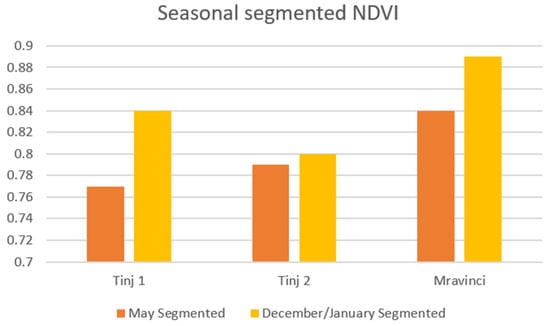
Figure 8.
Seasonal comparison of segmented NDVI values in olive groves.
Seasonal variations are noticeable for all three locations—the calculated NDVI values are higher in the winter measurement compared to May.
Assuming that the manually segmented set is “Ground truth”, f-tests and t-tests [41] were performed for the pairs of presented methods (1–4, 2–4, 3–4). The analysis of the obtained p-values for the Depth Seg method in most cases showed a value > 0.05, while the other methods had worse values (Table 3).

Table 3.
Comparison of t-test NDVI results between four methods. p-values > 0.05 are in bold.
Table 4 shows the sum of absolute differences in NDVI values calculated for each tree between individual methods and normalized with total number of trees in an olive grove, and these values clearly show that the smallest difference is between depth segmentation and manual segmentation.

Table 4.
Comparison of absolute differences (per each tree) between four methods. The best results are in bold.
The absolute difference (AD) between the obtained vegetation index values using different methods was obtained using the following formula:
where
N is the total number of trees in an olive grove;
NDVIA,i is the NDVI value for the i-th tree calculated by method A;
NDVIB,i is the NDVI value for the i-th tree calculated by method B.
To illustrate the differences between individual methods for each individual tree, Table 5 shows the NDVI values at the location Tinj 1 for the measurements in May (N = 17).

Table 5.
NDVI values for each tree at location Tinj 1 (May).
Although only the overall results at the level of the entire olive grove are presented here, the procedure itself provides insight into individual index values for each observed tree. It should be noted that no significant deviations in the values of individual trees were observed, which is an indicator that there were no trees under any kind of stress (biotic or abiotic).
It can be noted that the observed differences between NDVI values measured in May and in December and January are in line with measurements carried out in olive groves by Vanderlinden et al. [42].
3.2. NDRE Results
Again, the NDRE analysis for the olive groves at three locations (Tinj 1, Tinj 2, and Mravinci) in May and December/January demonstrates the effectiveness of image segmentation in enhancing NDRE values by focusing on the tree crowns (Table 6). In this case too, larger deviations of unsegmented and segmented values were registered for the Tinj 1 location in May (0.53 vs. 0.58) and the Tinj 2 location (0.45 vs. 0.50 in May and 0.56 vs. 0.61 in winter measurements). For the olive grove at the Mravinci location, the differences in unsegmented and segmented values are small. As with the NDVI values, for the olive grove at the Mravinci location, the differences between the unsegmented and segmented mean NDRE index values are small. The reason is the same: very little of the ground is visible in the images because the trees are densely spaced and their crowns touch.

Table 6.
Comparison of NDRE values by segmentation and season for three monitored olive grove locations. Non-segmented: all patch pixels are used for the calculation of the index; Depth Seg: index values calculated only for pixels remained after patch segmentation procedure; NDRE 0.3: only pixels with NDRE > 0.3 included in calculation; Manually segmented: only pixels included with manually segmented mask used for calculation.
Seasonal NDRE variations are noted (Figure 9), which reflect changes in chlorophyll content associated with vegetative cycles. The difference in the increase in seasonal values of this index is more significant than in the NDVI index–maximum for the location Tinj 2 location (0.50 vs. 0.61) and the location Mravinci (0.49 vs. 0.60).
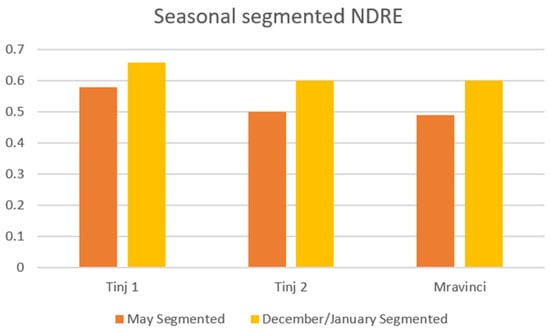
Figure 9.
Seasonal comparison of segmented NDRE values in olive groves.
4. Discussion
In many cases, depth segmentation gives higher NDVI and NDRE index values than manual segmentation (although the differences are not large). This indicates that, although we took manual segmentation as a kind of ground truth, depth segmentation is actually better due to the fact that it is carried out at the pixel level, while manual segmentation is related to larger areas.
It can be noted that, for orchards where only one tree species is present, the first step involving the implementation of a DeepForest model that is pre-trained to detect (in this case) only olive trees could be omitted. In that case, only the crown extraction phase based on monocular depth estimation could be used to detect individual trees from the input orchard maps.
Also, it is necessary to point out that the incomplete system calibration procedure used for this work may cause errors in the input values of the vegetation indices. This deficiency is also present in the seasonal results presented in this paper. Therefore, for a more reliable comparison, it would be necessary to include a complete radiometric calibration procedure during each survey. For farmers themselves, this process can be complex, so the realistic scenario in which such calibration would be included in seasonal measurements and monitoring of annual trends in orchards can be discussed.
5. Conclusions
A new segmentation approach based on the application of deep neural networks in two steps was proposed, first for the detection of individual trees and then for the monocular depth estimation, which is essential for distinguishing the canopy from the ground. The results of this study demonstrate the effectiveness of using the proposed procedure.
The application of the proposed segmentation procedure significantly enhanced the reliability and accuracy of NDVI and NDRE measurements, providing a clearer picture of vegetation health across the observed olive orchards. These findings have important implications for remote sensing applications in vegetation monitoring, demonstrating the potential for advanced image processing algorithms to improve the accuracy of vegetation health assessments in dynamic and complex environments.
Also, the observed differences in the calculated values of the vegetation indices depending on the applied processing method, as well as seasonal changes, lead to the conclusion that it would be interesting for the end user to enable a more reliable measure of the condition of the plant. That new measure should include information on the measured value of the vegetation index and the part of the season when it was measured. It would be useful to think about the range and number of possible values it could have, because the current ranges and quite unclear boundaries between the descriptions that are associated with certain amounts of vegetation indices are often not informative enough for the farmers.
In conclusion, a proposed state-of-the-art approach, combining UAV-enabled multispectral imaging, deep learning-based object detection, and advanced segmentation techniques, represents a cutting-edge framework for monitoring olive groves. Such methodologies provide detailed, high-resolution data that enhance sustainable agriculture practices, improve yield optimization, and foster overall olive grove health.
Author Contributions
Conceptualization, V.P.; methodology, V.P. and N.B.; software, V.P., N.B. and. I.M.; validation, V.P., S.G. and J.G.; formal analysis, V.P. and N.B.; investigation, V.P.; resources, V.P. and J.G.; data curation, V.P.; writing—original draft preparation, V.P. and N.B.; writing—review and editing, V.P., J.G. and S.G.; visualization, N.B. and V.P.; supervision, V.P.; project administration, V.P.; funding acquisition, V.P. and S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Croatian Science Foundation under the project number HRZZ IP-2024-05-6393.
Data Availability Statement
Dataset and code available on request from the authors.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- International Olive Council (IOC). World Market of Olive Oil and Table Olives—Data from December 2024. Available online: https://www.internationaloliveoil.org/world-market-of-olive-oil-and-table-olives-data-from-december-2024/ (accessed on 16 July 2025).
- Mili, S.; Bouhaddane, M. Forecasting Global Developments and Challenges in Olive Oil Supply and Demand: A Delphi Survey from Spain. Agriculture 2021, 11, 191. [Google Scholar] [CrossRef]
- State Bureau of Statistics of the Republic of Croatia. Olive Oil Production in the Republic of Croatia. 2024. Available online: https://poljoprivreda.gov.hr/UserDocsImages//dokumenti/poljoprivredna_politika/zeleno_izvjesce//2024_08_21%20Zeleno%20izvješće%202023_3.pdf (accessed on 27 October 2024).
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
- Avola, G.; Di Gennaro, S.F.; Cantini, C.; Riggi, E.; Muratore, F.; Tornambè, C.; Matese, A. Remotely Sensed Vegetation Indices to Discriminate Field-Grown Olive Cultivars. Remote Sens. 2019, 11, 1242. [Google Scholar] [CrossRef]
- Yang, H.; Yang, X.; Heskel, M.; Sun, S.; Tang, J. Seasonal variations of leaf and canopy properties tracked by ground-based NDVI imagery in a temperate forest. Sci. Rep. 2017, 17, 267. [Google Scholar] [CrossRef]
- Klimavičius, L.; Rimkus, E.; Stonevičius, E.; Mačiulytė, V. Seasonality and long-term trends of NDVI values in different land use types in the eastern part of the Baltic Sea basin. Oceanologia 2023, 65, 171–181. [Google Scholar] [CrossRef]
- Eisfelder, C.; Asam, S.; Hirner, A.; Reiners, P.; Holzwarth, S.; Bachmann, M.; Gessner, U.; Dietz, A.; Huth, J.; Bachofer, F.; et al. Seasonal Vegetation Trends for Europe over 30 Years from a Novel Normalised Difference Vegetation Index (NDVI) Time-Series—The Timeline NDVI Product. Remote Sens. 2023, 15, 3616. [Google Scholar] [CrossRef]
- da Silva, T.v.d.W.; Gomes Pereira, L.; Oliveira, B.R.F. Assessing Geometric and Radiometric Accuracy of DJI P4 MS Imagery Processed with Agisoft Metashape for Shrubland Mapping. Remote Sens. 2024, 16, 4633. [Google Scholar] [CrossRef]
- Daniels, L.; Eeckhout, E.; Wieme, J.; Dejaegher, Y.; Audenaert, K.; Maes, W.H. Identifying the Optimal Radiometric Calibration Method for UAV-Based Multispectral Imaging. Remote Sens. 2023, 15, 2909. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Aubry-Kientz, M.; Vincent, G.; Senyondo, H.; White, E.P. Deepforest: A python package for RGB deep learning tree crown delineation. Methods Ecol. Evol. 2020, 11, 1743–1751. [Google Scholar] [CrossRef]
- Marin, I.; Gotovac, S.; Papić, V. Development and Analysis of Models for Detection of Olive Trees. Adv. Sci. Technol. Eng. Syst. J. 2023, 8, 87–96. [Google Scholar] [CrossRef]
- Yang, L.; Kang, B.; Huang, Z.; Zhao, Z.; Xu, X.; Feng, J.; Zhao, H. Depth Anything V2. arXiv 2024, arXiv:2406.09414. [Google Scholar] [CrossRef]
- Marques, P.; Pádua, L.; Sousa, J.J.; Fernandes-Silva, A. Advancements in Remote Sensing Imagery Applications for Precision Management in Olive Growing: A Systematic Review. Remote Sens. 2024, 16, 1324. [Google Scholar] [CrossRef]
- Pádua, L.; Vanko, J.; Hruška, J.; Adão, T.; Sousa, J.J.; Peres, E.; Morais, R. UAS, Sensors, and Data Processing in Agroforestry: A Review towards Practical Applications. Int. J. Remote Sens. 2017, 38, 2349–2391. [Google Scholar] [CrossRef]
- Yao, L.; Liu, T.; Qin, J.; Lu, N.; Zhou, C. Tree counting with high spatial-resolution satellite imagery based on deep neural networks. Ecol. Indic. 2021, 125, 107591. [Google Scholar] [CrossRef]
- Chemin, Y.H.; Beck, P.S.A. A Method to Count Olive Trees in Heterogenous Plantations from Aerial Photographs. Preprints 2017, 2017100170. [Google Scholar]
- Catania, P.; Roma, E.; Orlando, S.; Vallone, M. Evaluation of Multispectral Data Acquired from UAV Platform in Olive Orchard. Horticulturae 2023, 9, 133. [Google Scholar] [CrossRef]
- Di Nisio, A.; Adamo, F.; Acciani, G.; Attivissimo, F. Fast Detection of Olive Trees Affected by Xylella Fastidiosa from UAVs Using Multispectral Imaging. Sensors 2020, 20, 4915. [Google Scholar] [CrossRef]
- Blekos, K.; Tsakas, A.; Xouris, C.; Evdokidis, I.; Alexandropoulos, D.; Alexakos, C.; Katakis, S.; Makedonas, A.; Theoharatos, C.; Lalos, A. Analysis, Modeling and Multi-Spectral Sensing for the Predictive Management of Verticillium Wilt in Olive Groves. J. Sens. Actuator Netw. 2021, 10, 15. [Google Scholar] [CrossRef]
- Candiago, S.; Remondino, F.; De Giglio, M.; Dubbini, M.; Gattelli, M. Evaluating Multispectral Images and Vegetation Indices for Precision Farming Applications from UAV Images. Remote Sens. 2015, 7, 4026–4047. [Google Scholar] [CrossRef]
- Lima-Cueto, F.J.; Blanco-Sepúlveda, R.; Gómez-Moreno, M.L.; Galacho-Jiménez, F.B. Using Vegetation Indices and a UAV Imaging Platform to Quantify the Density of Vegetation Ground Cover in Olive Groves (Olea europaea L.) in Southern Spain. Remote Sens. 2019, 11, 2564. [Google Scholar] [CrossRef]
- Jorge, J.; Vallbé, M.; Soler, J.A. Detection of irrigation inhomogeneities in an olive grove using the NDRE vegetation index obtained from UAV images. Eur. J. Remote Sens. 2019, 52, 169–177. [Google Scholar] [CrossRef]
- Gao, L.; Wang, X.; Johnson, B.A.; Tian, Q.; Wang, Y.; Verrelst, J.; Mu, X.; Gu, X. Remote sensing algorithms for estimation of fractional vegetation cover using pure vegetation index values: A review. ISPRS J. Photogramm. Remote Sens. 2020, 159, 364–377. [Google Scholar] [CrossRef]
- Putra, Y.C.; Wijayanto, A.W. Automatic detection and counting of oil palm trees using remote sensing and object-based deep learning. Remote Sens. Appl. Soc. Environ. 2023, 29, 100914. [Google Scholar] [CrossRef]
- Marin, I.; Gotovac, S.; Papić, V. Individual Olive Tree Detection in RGB Images. In Proceedings of the 2022 International Conference on Software, Telecommunications and Computer Networks (SoftCOM), Split, Croatia, 22–24 September 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Dakov, V.; Petrova-Antonova, D. Urban Tree Detection from Remote Sensing Data based on DeepForest Model. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2024, X-4/W4-2024, 35–41. [Google Scholar] [CrossRef]
- Wijaya, D.; Irwansyah, E. Tree Counting with Deep Learning Algorithm and Carbon Evaluation using Pleiades Satellite Imagery Data in Kulon Progo, Yogyakarta, Indonesia. In Proceedings of the 2023 International Conference on Informatics, Multimedia, Cyber and Informations System (ICIMCIS), Jakarta Selatan, Indonesia, 7–8 November 2023; pp. 22–26. [Google Scholar] [CrossRef]
- Jemaa, H.; Bouachir, W.; Leblon, B.; Bouguila, N. Computer Vision System for Detecting Orchard Trees from UAV Images. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2022, XLIII-B4-2022, 661–668. [Google Scholar] [CrossRef]
- Khan, A.; Khan, U.; Waleed, M.; Khan, A.; Kamal, T.; Marwat, S.N.; Maqsood, M.; Aadil, F. Remote Sensing: An Automated Methodology for Olive Tree Detection and Counting in Satellite Images. IEEE Access 2018, 6, 77816–77828. [Google Scholar] [CrossRef]
- Komura, R.; Kubo, M.; Muramoto, K. Delineation of tree crown in high resolution satellite image using circle expression and watershed algorithm. In Proceedings of the IGARSS 2004. 2004 IEEE International Geoscience and Remote Sensing Symposium, Anchorage, AK, USA, 20–24 September 2004; Volume 3, pp. 1577–1580. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, J.; Wang, Q. Mean-shift-based color segmentation of images containing green vegetation. Comput. Electron. Agric. 2009, 65, 93–98. [Google Scholar] [CrossRef]
- YaKun, W.; Soh, Y.S.; Schultz, H. Individual tree crown segmentation in aerial forestry images by mean shift clustering and graph-based cluster merging. Int. J. Comput. Sci. Netw. Secur. 2006, 6, 40–45. [Google Scholar]
- Rehman, T.H.; Lundy, M.E.; Linquist, B.A. Comparative Sensitivity of Vegetation Indices Measured via Proximal and Aerial Sensors for Assessing N Status and Predicting Grain Yield in Rice Cropping Systems. Remote Sens. 2022, 14, 2770. [Google Scholar] [CrossRef]
- Raya-Sereno, M.D.; Ortiz-Monasterio, J.I.; Alonso-Ayuso, M.; Rodrigues, F.A., Jr.; Rodríguez, A.A.; González-Perez, L.; Quemada, M. High-Resolution Airborne Hyperspectral Imagery for Assessing Yield, Biomass, Grain N Concentration, and N Output in Spring Wheat. Remote Sens. 2021, 13, 1373. [Google Scholar] [CrossRef]
- Papić, V.; Bugarin, N.; Gugić, J. On Olive Groves Analysis using UAVs. In Proceedings of the 2021 International Conference on Software, Telecommunications and Computer Networks (SoftCOM), Split, Croatia, 23–25 September 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A.R. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Govander, M.; Chetty, K.; Bulcock, H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water SA 2007, 33, 145–151. [Google Scholar] [CrossRef]
- Osco, L.P.; dos Santos de Arruda, M.; Junior, J.M.; da Silva, N.B.; Marques Ramos, A.P.; Saito Moryia, E.A.; Imai, N.N.; Pereira, D.R.; Creste, J.E.; Matsubara, E.T.; et al. A convolutional neural network approach for counting and geolocating citrus trees in UAV multispectral imagery. ISPRS J. Photogramm. Remote Sens. 2020, 160, 97–106. [Google Scholar] [CrossRef]
- DJI Terra. Available online: https://www.dji.com/hr/downloads/products/dji-terra#other_software (accessed on 17 July 2025).
- Student. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Vanderlinden, K.; Martinez, G.; Ramos, M.; Mateos, L. Relevance of NDVI, soil apparent electrical conductivity and topography for variable rate irrigation zoning in an olive grove. Precis. Agric. 2024, 25, 3086–3108. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).