Abstract
The prediction of growing stock volume is one of the commonest applications of remote sensing to support the sustainable management of forest ecosystems. In this study, we used data from the 4th Spanish National Forest Inventory (SNFI-4) and from the 1st nationwide Airborne Laser Scanning (ALS) survey to develop predictive yield models for the three major commercial tree forest species (Eucalyptus globulus, Pinus pinaster and Pinus radiata) grown in north-western Spain. Integration of both types of data required prior harmonization because of differences in timing of data acquisition and difficulties in accurately geolocating the SNFI plots. The harmonised data from 477 E. globulus, 760 P. pinaster and 191 P. radiata plots were used to develop predictive models for total over bark volume, mean volume increment and total aboveground biomass by relating SNFI stand variables to metrics derived from the ALS data. The multiple linear regression methods and several machine learning techniques (k-nearest neighbour, random trees, random forest and the ensemble method) were compared. The study findings confirmed that multiple linear regression is outperformed by machine learning techniques. More specifically, the findings suggest that the random forest and the ensemble method slightly outperform the other techniques. The resulting stand level relative RMSEs for predicting total over bark volume, annual increase in total volume and total aboveground biomass ranged from 30.8–38.3%, 34.2–41.9% and 31.7–38.3% respectively. Although the predictions can be considered accurate, more precise geolocation of the SNFI plots and coincide temporarily with the ALS data would have enabled use of a much larger and robust field database to improve the overall accuracy of estimation.
1. Introduction
Information about timber resources and how these vary over time is both economically and environmentally important and is essential for landowners, enterprises, forest managers and researchers. Moreover, local, regional or global estimates of volume and biomass are fundamental for analyzing forest productivity, estimating biomass and carbon stocks and evaluating the ecosystem response to climatic change and anthropogenic influences [1].
Forest field inventory based on measuring diameter at breast height (dbh) and total height (h) of trees in numerous sample plots remains the most commonly used method of estimating timber and forest biomass stocks for forest management [2]. The individual tree model predictions are often aggregated at the plot level and used as training and/or accuracy assessment data for remote sensing-based applications or aggregated at the plot level and then added or averaged over plots to produce large area estimates [3]. Although this method is accurate, it is very expensive and often faces serious operational difficulties [4]. Some available public databases such as the National Forest Inventories do not always provided information about the plots at the level of detail required for forest management purposes. However, in recent decades, the combined use of these databases and the semi-automatic capture of forest state variables by various remote sensing techniques has substantially increased the amount of data available for stand level prediction purposes [5]. Remote sensing systems have proved to be an effective option for overcoming the above-mentioned limitations and enabling forest data to be obtained in large areas with a more reasonable effort [6] and even in areas not previously sampled [1]. Among these systems, Airborne Laser Scanning (ALS), an active remote sensing methodology which transmits pulses of laser light towards the ground, is recognized to be an accurate, efficient and cost-effective approach to predicting forest variables such as stand height, volume and biomass [4,7,8]. In a practical applications of ALS data, georeferenced plots are first used to develop empirical models of the relationships between field measurements and ALS-derived metrics, and the models are then applied to the entire area of interest, thus predicting the forest attributes on the basis of ALS metrics alone [1]. Although positioning accuracy may not be critical in some forest studies, it is of key importance when developing predictive models from very high-resolution remotely sensed data [9]. Accurate geographical co-registration of ALS data and field plots is necessary for the accurate prediction of stand properties, otherwise the laser-derived metrics will be subject to errors [10].
Since the early studies by Maclean and Krabill [11] and Nelson et al. [12], which first used ALS data for volume and biomass estimation [13], many efforts have been made to use this technology for predicting forest yields [14,15,16]. Multiple Linear Regression (MLR) has frequently been used to estimate the empirical model parameters (e.g., [17,18]). The simplicity and clarity of the resulting model are the main advantages of the technique, while the probability of selecting highly correlated predictors with little physical justification and the non-fulfilment of the assumptions of normality, homoscedasticity, independence and linearity are the main drawbacks [19]. In order to overcome the limitations of MLR, much attention has been given in recent years to non-parametric machine learning techniques [19,20,21]. The power of machine learning techniques is based on the fact that they do not depend on any a priori assumption about the data; however, they yield models that are usually complex and the role of the variables selected from the models may be difficult to understand [22].
Models developed using ALS data can be trained with data from existing NFI plots, allowing the development of accurate empirical yield models for predicting variables such as volume or biomass with errors lower than 25% and 27% respectively, as observed in Nordic countries [2]. So far, this methodology has been used in very few countries: Austria [23], Denmark [24] and Sweden [2]. In Spain, two public organizations have collected both ALS and NFI data and made them available to be downloaded free of charge by any interested party. Thus, the Spanish National Forest Inventory (SNFI), carried out by the Ministerio de Agricultura, Pesca, Alimentación y Medio Ambiente (MAPAMA), is the most important public database providing information about forest use, structure and yield of all forests around the country. Moreover, ALS data, which cover the whole country, have been compiled by the PNOA-LiDAR project of the Instituto Geográfico Nacional (IGN) since 2008 with multipurpose objectives, e.g., obtaining digital elevation, surface or hydrographic models, and automatic detection of terrain modifications. Despite the availability of this very valuable information, very few studies have used both types of information together (e.g., [4,25,26]), mainly because of the following important challenges: (i) differences in time of acquisition of both types of data in most regions, and (ii) problems associated with the precise geolocation of SNFI plots.
Northwestern Spain (encompassing the regions of Galicia, Asturias and Cantabria) is one of the most productive forest areas in Europe, and the major commercial forest species, grown in intensive forest plantations to produce panelboard, sawlog and pulpwood, are Eucalyptus globulus Labill, Pinus pinaster Ait. and Pinus radiata D. Don [27]. Together the three regions of NW Spain contribute about 58% of the timber harvesting (42% of coniferous and 79% of hardwoods) carried out annually in Spain [28]. Due to the economic importance of the timber, methods of quantifying the wood/biomass have become important for all agents involved in forest management and conservation. Therefore, the main objective of this study was to generate a high-resolution raster database with information about key forest yield variables such as total over bark volume (m3/ha) and total aboveground biomass (t/ha). Secondary objectives -necessary to achieve the first objective- include the following: (i) development of a procedure to harmonize the SNFI and the ALS data; (ii) selection of the best empirical models of relationships between field measures and ALS-derived metrics, by comparing a parametric technique (MLR) and several well-known non-parametric machine learning regression techniques; and (iii) to generate spatially-continuous maps of yield variables.
2. Materials and Methods
2.1. Study Area
Three of the most productive regions in Spain (Galicia, Asturias and Cantabria), covering a total area 45,499 km2, were chosen for this study. Galicia is divided into four provinces (A Coruña, Lugo, Ourense and Pontevedra), while Asturias and Cantabria are both single-province regions (Figure 1). This is important as province is the basic unit used in the SNFI to elaborate and present the data. The study area forms part of the European Atlantic Bio-geographical Region, except for south-eastern Galicia, which belongs to the Mediterranean Bio-geographical Region [29]. Forests occupy an area of 21,190 km2 [28] which represents 46.5% of the total surface area of the study area. Considering the area occupied, Eucalyptus globulus is the dominant forest species (22.5%) followed by Pinus pinaster (20.2%), Quercus robur (15.5%), Quercus pyrenaica (8%), Castanea sativa (8%), Pinus radiata (7.5%) and Fagus sylvatica (5.7%) [30].
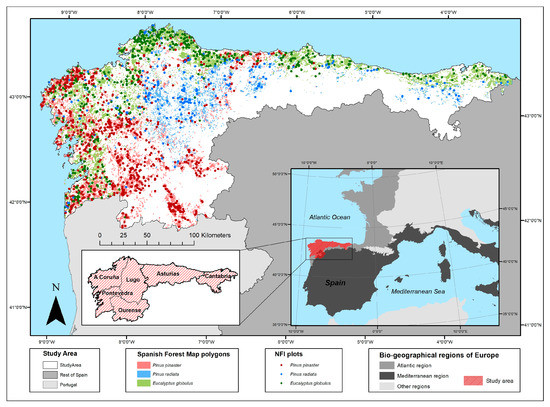
Figure 1.
Location of the study area and National Forest Inventory plots and Spanish Forest Map polygons dominated by the three major commercial tree forest species (Eucalyptus globulus, Pinus pinaster and Pinus radiata) in in north-western Spain.
2.2. Data Collection
Two different sources of data were used in this study: national forest inventory and nationwide airborne laser scanning data, both of which were obtained free of charge from different Spanish Governmental organizations.
2.2.1. SNFI Data
The field data used in this study were obtained from the Fourth Spanish National Forest Inventory (SNFI-4), in Galicia [31], Asturias [32] and Cantabria [33] in different surveys carried out between 2008 and 2012. The SNFI operates on a ten-year cycle and the sampling plots are established at the intersections of a 1×1 km UTM grid. Different field plots of variable radius, depending on the dbh of the trees, were sampled: a radius of 25 m for trees with dbh ≥ 42.5 cm, a radius of 15 m for trees with dbh ≥ 22.5 cm, a radius of 10 m for trees with dbh ≥ 12.5 cm and a radius of 5 m for trees with a dbh ≥ 7.5 cm. Trees with 2.5 ≤ dbh ≤ 7.5 cm were counted but not measured. For more details about the SNFI data, see Alberdi et al. [34].
Plots were selected according to the species composition, including pure stands (basal area ≥ 80% of the total basal area within the plot) of the three major commercial tree forest species grown in the area. Following this criterion, a total of 1428 field plots were used in this study (Figure 1). In total, 760 plots were dominated by P. pinaster, 191 by P. radiata and 477 by E. globulus. Stand-related variables such as the number of stems per hectare, basal area, dominant height (mean height of the 100 thickest trees per hectare), mean stand height, quadratic mean diameter, dominant diameter, total over bark volume and annual increase volume were calculated from tree measurements and by using appropriate expansion factors. An expansion factor can be defined as the relationship between the reference area (1 ha) and the area of subplots by adjusting the values of the number of sampled trees to a per hectare value [5].
The stand level species-specific allometric models developed for the same ecoregion by Castaño-Santamaría et al. [35] were used to estimate total aboveground biomass per plot.
Table 1 includes the summary statistics of the stand-related yield variables considered for the three forest species in the study area.

Table 1.
Descriptive statistics of total volume (TV), annual increase in total volume (AITV) and total aboveground biomass (AGB), extracted from the SNFI-4 plots with the dominant species equal to or greater than 80% of the total basal area of the final plots used after the harmonization procedure.
The distribution of the species under study and the classification of vegetation types were determined using the Spanish Forest Map (Figure 1) (scale 1:25,000, minimum mapping unit of 1 ha), developed in coordination with the SNFI-4.
2.2.2. ALS Data
The ALS data used in the study area were collected during different flights between 2009 and 2012 for the PNOA-LiDAR project. The point cloud was captured with up to four returns measured per pulse and a mean density of 0.5 points/m2 and vertical RMSE ≤ 0.20 m. Square ALS tiles of area 2 × 2 km in LASer (LAS) binary files were obtained from the National Geographic Information Centre (CNIG) (data available at http://centrodedescargas.cnig.es/CentroDescargas/buscadorCatalogo.do?codFamilia=LIDAR#, last accessed on 4 October 2018). A total of 13,396 LAS files were required in order to cover the study area. The ALS data were extracted using several processing programmes implemented in the FUSION/LDV software [36]. After separating the first and last returns and eliminating the possible outliers from the point cloud, a Digital Terrain Model (DTM) of cell size 5 m was generated. The “GridMetrics” program was used to compute metrics for ALS returns within 25 m grid cells, defining a grid cell size related to the size of the ground plot according to Canadian specifications [37]. The ALS metrics were computed by considering the first returns and all returns independently. A predefined threshold of between 2 and 50 m above ground level was used in order to compute canopy cover metrics. In total, 36 ALS metrics widely used as effective variables for volume or biomass estimations (e.g., [38,39,40]) were computed as independent variables for developing the empirical yield models (Table 2). Note that all these metrics were subsequently calculated for the SNFI plots using the “ClipData” and “CloudMetrics” programs; the normalized ALS point cloud was clipped with the limits of each field plot (25 m radius) thus creating an independent file per plot.

Table 2.
Summary of Airborne Laser Scanning (ALS) metrics extracted for each plot.
2.2.3. Harmonization of SNFI and ALS Data
The plot positioning procedures used in the SNFI-4 were applied by using handheld GPS equipment, which has an expected average accuracy of approximately 3–5 m [25]. In this respect, Gobakken and Næsset [10] observed that larger plot sizes (300–400 m2) compensate for errors in locating plots sampled for estimation of biophysical properties from ALS data. Thus, as the plot size used in the SNFI is around 1964 m2, for positioning errors of 5 and 10 m (much larger than the theoretical error), the areas overlapping a plot in a correct position and a plot located in an altered position are 84.3% and 74.7% respectively [25]. Although this may suggest that SNFI plot positioning is not difficult, practical errors will actually be much higher than theoretical errors. Thus, for example, Murgaš et al. [41] reported that up to 10% of the plots had positioning errors of 20 m or more within inventories of selected management units in Slovakia. As revisiting SNFI plots to capture more accurate coordinates is very costly and time-consuming, Spanish authorities are planning to capture new coordinates with errors less than 1 m in the next re-measurement of SNFI plots [26,42], thus making it easier to combine the field plot data with the information provided by remote sensing systems [34].
Another important problem related to the combined use of SNFI and ALS data is the difference in the time of data acquisition in some provinces (Table 3). This generates a new problem as some plots may be disturbed (cutting operations, wind damage, etc.) during the time between the laser scanning and the field inventory. Thus, prior to using both sources of data together, we developed a three-step process aimed at harmonizing the different types of data.

Table 3.
Dates of SNFI-4 and ALS surveys carried out in the different provinces in the study area.
We first carried out a forward or backward projection of the stand yield variables obtained from the SNFI to the same date as the ALS data, in order to correct the effect of the different years of ALS and SNFI surveys. Thus, stand total volume was projected by using the tree volume increment values available from the SNFI. For updating total aboveground biomass, we relied on the strong well-known relationship between stand volume and stand biomass (e.g., [43,44]) and we thus developed total stand volume-to-total stand aboveground biomass models to estimate biomass from the projected stand volume.
Secondly, with the aim of using dominant height to eliminate incorrectly georeferenced or disturbed plots, when necessary, we projected the values of this variable obtained from the SNFI plots forwards or backwards (between 1 and 3 years) to the same date when the ALS data were acquired. Site quality equations are essential to enable this important step to be carried out, and information about stand age and site index are also required. Stand age is required to enable the growth stage of the plot to be determined, and stand age and site index are used in site quality equations to determine the height growth rate. Site quality equations have already been published for the species in the region [45,46,47]. However, obtaining stand age and site index proved very challenging because the SNFI does not collect such data. To resolve this problem, we developed specific models for each species in order to predict stand age for each SNFI plot, and we used the spatially continuous site index maps recently developed for the three species in the ecoregion [48,49,50] to estimate the site index values. In order to project dominant height of SNFI plots to the date of the ALS survey, the following three steps were therefore undertaken for each SNFI plot: (i) estimation of site index, (ii) prediction of stand age and (iii) calculation of the dominant height growth ratio.
Finally, we compared the dominant height of the SNFI-4 plots projected to the ALS date and the 95th height percentile (h95) of the point cloud obtained using the laser scanning survey. Following the procedure used by Nilsson et al. [2] in Sweden, plots with differences in the upper 3 m of the dominant height were removed. Once this step was carried out, we relied on effective elimination of plots that were poorly geolocated or disturbed in the period between the laser scanning and the field work. As a result of this refinement process, 477 E. globulus plots (22.0% of total available plots), 760 P. pinaster plots (37.4%) and 191 P. radiata plots (30.8%) were used to develop predictive models of total over bark volume, mean volume increment and total aboveground biomass relating SNFI stand variables to metrics derived from the ALS data.
2.3. Data Analysis
2.3.1. Regression Techniques
We compared the performance of several regression techniques, including novel machine learning methods, for estimating stand level forest yield variables: (i) the parametric Multiple Linear Regression technique (MLR), and the non-parametric techniques (ii) k-Nearest Neighbour (kNN), (iii) Regression Trees (RT) with M5P trees used as the basic regression technique for developing this ensemble (base level algorithm), (iv) Random Forest (RF), and (v) the Ensemble Method (EM) with the metaclassifier Stacking Multiple Classifiers.
The parametric MLR technique is the most commonly used approach in this type of study [51]. Moreover, the model produced is easy to understand and is widely used in most scientific disciplines. However, unlike non-parametric approaches, MLR relies on certain assumptions such as the fundamental least squares assumption of independence and equal distribution of errors with zero mean and constant variance, which can be violated by various factors, including non-normality of variables, multicollinearity of variables and heteroscedasticity of error variance.
Nearest Neighbour (NN), a well-known machine learning technique used in remote sensing [52], makes predictions by using the information about the neighbours of the instance to be regressed [53]. The NN depends on a parameter, usually called k, which determines the number of neighbours used by the algorithm. The technique is therefore usually called kNN when more than one neighbour is used. Although the idea behind this type of technique is quite intuitive, the resulting model is not easy to interpret because the results depend on a training set.
Regression Trees (RT) using M5P as the basic regression technique are ordinary decision trees with linear regression models at the leaves that predict the value of observations that reach the leaves [54,55]. The nodes of the tree represent variables and branches represent split values. Model tree induction algorithms are derived from the divide-and-conquer decision tree methodology. Unlike classification trees, which choose the attribute and its splitting value for each node to maximize the information gain, model trees choose these to minimize the intra-subset variation in the class values down each branch and maximize the expected error reduction (standard deviation reduction). As the tree structure divides the sample space into regions and a linear regression model is found for each, the tree is somewhat open to interpretation.
Random Forest (RF), first proposed by Breiman [56], is a widely used non-parametric classification and regression approach consisting of an ensemble of decision trees. The success of this technique is based on the use of numerous trees developed with different independent variables that are randomly selected from the complete original set of variables. The randomized sampling leads to greater stability and better accuracy than a single decision tree approach [57]. The final regression estimate for each sample is obtained as a weighted mean value of the estimates of a large number of individual trees [56]. The number of predictors included in the trees and the number of trees are established by the user. This non-parametric approach is relatively insensitive to the number of input data and the multicollinearity of the data [58].
Stacking (sometimes called stacked generalization) is an Ensemble Method (EM) that allows several different types of prediction algorithms to be combined in a single algorithm [59]. This EM involves training a learning algorithm to combine the predictions of several other learning algorithms. In addition to selecting multiple sub-models, stacking enables specification of another model (meta-classifier) to learn how to best combine the predictions from the sub-models (base classifiers). Because a meta model is used to combine the predictions of sub-models, this technique is sometimes called blending, as in blending predictions together. Stacking typically performs better than any single one of the trained models [60].
WEKA open source software [61] was used to implement all of the techniques compared in this study. Thus, linear regression was used for MLR, IBk for kNN and M5P for RT, while RF was used to construct a forest of random trees and Stacking was used to develop the EM.
2.3.2. Feature Selection and Parametrization
A feature set describing a data instance might range in size from two to several hundred features. The representative quality of a feature set greatly influences the effectiveness of ML algorithms. Feature selection has three benefits: (i) learning and classification times are reduced by decreasing the number of features, (ii) the accuracy of classification is improved by removal of irrelevant or redundant features, (iii) the degree of overfitting in the training dataset is reduced (i.e., Valbuena et al., [62]). The number and type of features used to train the ML algorithm should therefore be carefully selected, in a process known as feature selection. Feature selection algorithms are broadly categorized in the filter or wrapper model.
In this study we used the search algorithms included in Wrapper methods to select the subsample of variables as this usually produces the best results [63]. This feature selection process selects the subsample of variables using a learning algorithm as part of the evaluation function. In the present study, the goodness of the fit was evaluated by the root mean square error (RMSE) obtained with a training set.
The different optimal parameters for each regression technique were configured using the CVParameterSelection method implemented in the WEKA software [64], and parameter selection was performed by cross-validation for the selected classifiers: (i) LinearRegression was used to fit the MLR model with no attribute selection; (ii) IBk was used to fit the kNN model by setting the number of neighbours to between 1 and 50; (iii) M5P was used to fit the RT by setting the minimum number of instances to allow at a leaf node between 0 to 50 and maximum depth of unlimited tree; (iv) RandomForest was used to fit the RF model by setting the number of trees to 1500, the number of features selected in each split to between 0 and 10 and the maximum depth of unlimited tree; and (v) Stacking was used to develop the EM to demonstrate the ability to improve predictive performance by combining four of the above base classifiers (MLR, kNN, RT and RF) by setting the meta-classifier as the MLR method for learning how to best combine the predictions.
2.3.3. Performance Evaluation
Several approaches can be used to train models and test data sets for validation of supervised learning algorithms. We used the common method of k-fold cross validation. In this process, the data set is divided into k subsets. Each time, one of the k subsets is used as the test set and the other k-1 subsets form the training set. Error statistics are calculated across all k trials. This provides a good indication of how well the classifier will perform with unseen data. We considered a set of a 10-fold cross-validation (i.e., models were fitting using 90% of the data for training and the remaining 10% for model evaluation) and computed several standard performance metrics to compare the regression techniques. Thus, we calculated three goodness-of-fit statistics: the pseudo-coefficient of determination (R2), the bias, the absolute values of mean error (MAE) and the relative root mean squared error (rRMSE). We also used the paired t-test (corrected), based on Student’s t-criterion, to detect any significant differences in the results caused by the five approaches considered (α = 0.05). All of the values represent the mean and the standard deviation of 100 model runs (i.e., 10-fold cross-validation repeated 10 times using the training datasets).
Selected fitted models were also compared for each dependent variable on the basic of graphical analysis of observed against predicted values of the dependent variable.
3. Results
As a result of the feature selection process, an optimal subset size of between 9 and 17 (of the 36) variables was selected by the Wrapper method (Figure 2). The results indicated that the features for estimating TV, AITV and AGB by the different regression techniques evaluated can be classified into two different groups. In order of decreasing importance, the first group comprises a combination of height metrics (height percentiles and central trend metrics) and the second group comprises the canopy cover metrics, with an average relative importance of variables of >38% and <5% respectively.
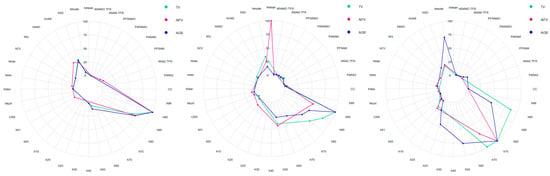
Figure 2.
Variables included in the models (VIM), for the best techniques, including type and variable importance for E. globulus (column 1), P. pinaster (column 2) and P. radiata (column 3). To ensure that values of variable importance are expressed on comparable scales for each of the response variable, the scores of all the predictors selected were normalized so that they added up to a unitary value (normalized importance), or they were expressed as relative values: Relative importance = (VIM−VIMmin)/(VIMmax−VIMmin).
The goodness-of-fit statistics yielded by the different regression methods used to model total volume, TV (m3/ha), annual increase in total volume, AITV (m3/ha year), and total aboveground biomass, AGB (t/ha), for the major commercial tree species in north-western Spain are shown in Table 4.

Table 4.
Summary of the goodness-of-fit statistics yielded by regression methods for total volume, TV (m3/ha), annual increase in total volume, AITV (m3/ha year), and total aboveground biomass, AGB (t/ha), for the major commercial tree species in north-western Spain. Methods included Multiple Linear Regression (MLR), k-Nearest Neighbour (kNN), Regression Trees (RT), Random Forest (RF) and the Ensemble method (EM). All values represent the mean and standard deviation (std) of 100 model runs (i.e., 10 replicates, each with 10-fold cross validation). The performance of the regression methods was compared by using different statistics based on the model errors (mean field values were considered as true values): coefficient of determination (R2), bias (Bias), the relative root mean square error (rRMSE); and a paired t-test (corrected) based on Student’s t-criterion (α = 0.05); significantly differences are indicated in bold type.
The best results for the goodness-of-fit statistics are summarised as follows: (i) for TV and E. globulus the RF model yielded R2 = 0.83 and RMSE = 64.33 m3/ha; for P. pinaster the EM model yielded R2 = 0.74 and RMSE = 61.63 m3/ha; and for P. radiata the RF model yielded R2 = 0.79 and RMSE = 66.52 m3/ha. (ii) for AITV and E. globulus the EM model yielded R2 = 0.68 and RMSE = 5.77 m3/ha; for P. pinaster the EM model yielded R2 = 0.56 and RMSE = 3.86 m3/ha; and for P. radiata the RF model yielded R2 = 0.69 and RMSE = 4.35 m3/ha. (iii) for AGB and E. globulus the RF model yielded R2 = 0.82 and RMSE = 44.29 m3/ha; for P. pinaster the EM model yielded R2 = 0.74 and RMSE = 35.26 m3/ha, and for P. radiata the RF model yielded R2 = 0.80 and RMSE = 35.04 m3/ha. The goodness-of-fit statistics of the fitted models were highest for E. globulus with all the approaches used. Regarding the variables to be modelled, AITV always yielded the poorest goodness-of-fit statistics (Table 4).
The MAE values obtained were used for graphical analysis of performance between the regression techniques. The comparison was summarized in histograms showing the relative positions (percentile rank) for each technique (Figure 3). Qualitatively, the MLR and kNN technique produced the best and worst results respectively. Nevertheless, the EM and RF technique generally produced the best results.
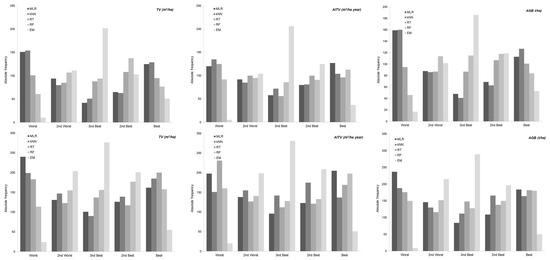
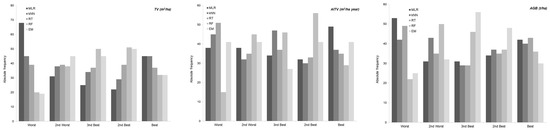
Figure 3.
Absolute frequency of relative position achieved (percentile rank) by each technique with the best parameterization for total volume (m3/ha), annual increase in total volume (m3/ha year) and total aboveground biomass (t/ha) in E. globulus (row 1), P. pinaster (row 2) and P. radiata (row 3).
The results of the paired t-test (corrected) based on Student’s t-criterion indicated significant differences in the results produced by the five approaches considered (α = 0.05) for the E. globulus and P. pinaster models, for which the best estimates were obtained with EM and RF, thus confirming the idea previously outlined in Figure 3. However, for P. radiata, there were no significant differences between the different approaches for any of the dependent variables (Table 4).
Graphical analysis of observed against predicted values of TV, AITV and AGB, estimated with each respective best technique, is shown in Figure 4. The linear model fitted to the scatter plot did not reveal any important problems related to heteroscedasticity or lack of normality, although there appeared to be a slight tendency towards underestimation of high values of TV, AITV and AGB (Figure 4).
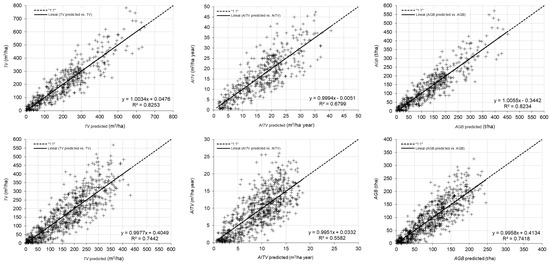
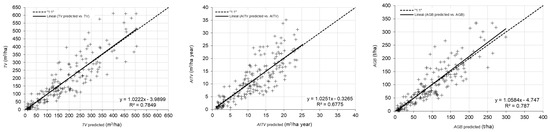
Figure 4.
Plot-level predicted against observed values for the best techniques for total volume (m3/ha), annual increase in total volume (m3/ha year) and total aboveground biomass (t/ha) in E. globulus (row 1), P. pinaster (row 2) and P. radiata (row 3). The solid line represents the linear model fitted to the scatter plot of data and the dashed line represents the line of slope equal to 1.
The spatial distribution of the total aboveground biomass resulting from the application of the best modelling technique for the three major commercial tree species (E. globulus, P. pinaster and P. radiata) grown in forests across north-western Spain is presented in Figure 5 The high spatial resolution of the maps reveals how the forest develops into fragmented environments, as a consequence of the smallholding structure. We also observed that E. globulus is mainly concentrated on the coast and P. radiata appears mainly in the interior areas of most provinces, but is almost non-existent in those located to the south (Pontevedra and Ourense); P. pinaster is distributed indistinctly both on the coast and in interior areas.
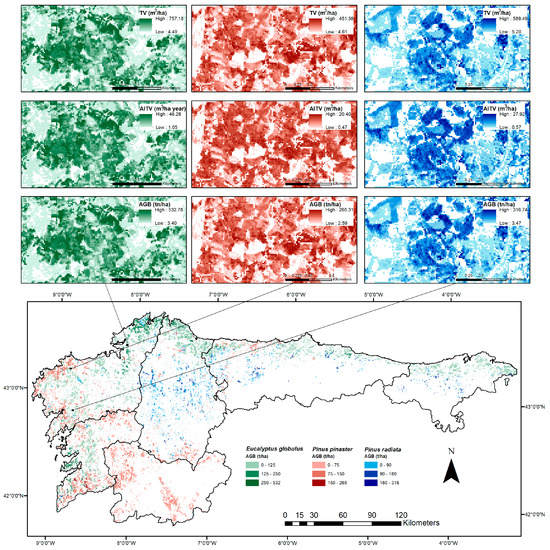
Figure 5.
Spatial distribution of the total aboveground biomass (t/ha) in north-western Spain (bottom). Top: Map details (25-m spatial resolution) for TV (m3/ha), AITV (m3/ha year) and AGB (t/ha) in E. globulus (row 1), P. pinaster (row 2) and P. radiata (row 3).
Finally, average values per hectare and total ALS-based wall-to-wall predictions for TV, AITV and AGB for the three species occupying areas greater or equal to 70% according Spanish Forest Map were generated per province by applying the best model developed in this study (Figure 6). Both volume and biomass productivity were higher in the coastal provinces (Asturias, Cantabria, A Coruña and Pontevedra) than in the interior areas (Lugo and Ourense) (Figure 6). Regarding the timber and forest biomass stocks for the different species, for E. globulus and P. pinaster, forest production was higher on the north central coast and on the west coast (Asturias, Pontevedra and A Coruña) and lower in the interior regions (Ourense and Lugo) and Cantabria. However, P. radiata forest production is higher on the northeast coast (Cantabria) of the study area than in the other provinces, where there are no significant differences. In global terms, the timber and forest biomass stocks of E. globulus were highest in the province of A Coruña (NW).
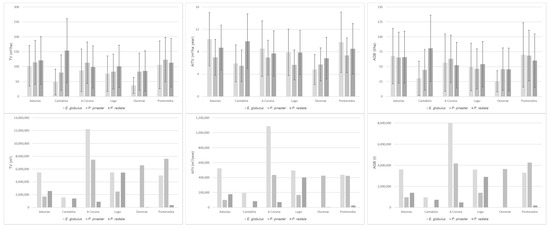
Figure 6.
Graphs showing the average and standard deviation (error bars) values per hectare (top) and total (bottom) of the ALS-based wall-to-wall predictions per province for total volume (TV), annual increase in total volume (AITV) and total aboveground biomass (AGB), for the three forest species studied. Results were obtained by applying the best model obtained in this study and the species distribution provided by the Spanish Forest Map.
4. Discussion
The study findings show that total over bark volume and total aboveground biomass yields for the three major commercial tree species (E. globulus, P. pinaster and P. radiata) grown in north-western Spain can be modelling using previously harmonized (by the proposed procedure) SNFI and ALS data, with a precision comparable to that obtained in other studies published in the relevant international literature.
Several non-parametric methods were tested: the k-Nearest Neighbour method [65]; Regression Trees [54], with M5P trees as the base level algorithm; the Random Forest method [56]; and the Ensemble Method (EM), with a metaclassifier stacking multiple classifiers [59]. Multiple Linear Regression [51] was also carried out to compare how it performs relative to non-parametric methods for estimating forest yield variables at stand level. There were statistically significant differences for validation in performance between the non-parametric methods and Multiple Linear Regression based on the results of the paired t-test (corrected) with Student’s t-criterion (α = 0.05). On the other hand, there were no differences between the use of R2 and RMSE for validation. There is some variation in the magnitude of the errors (slight differences in terms of MAE and RMSE for validation) in the models analyzed, but very large errors are unlikely to have occurred. The results obtained for the averaged models show that RF and EM are the most robust techniques in the case of E. globulus, while EM is most robust for P. pinaster and RF for P. radiata. Both of these methods are well known and frequently used in the field of remote sensing, especially in forestry applications [2,6,66,67].
The R2 yielded by the TV, AITV and AGB models for the three forest species considered and the different regression techniques ranged from 0.71 to 0.83, from 0.51 to 0.69, and from 0.71 to 0.82 respectively. The rRMSE values ranged from 30.71% to 40.81%, from 34.24% to 44.09%, and from 31.59% to 42.28% respectively (Table 4).
The study findings showed that the most important features for estimating TV, AITV and AGB by the different modelling techniques evaluated correspond to a combination of height metrics (height percentiles and central trend metrics), which are more sensitive to changes in both the vertical arrangement of canopy elements and the degree of canopy openness [68,69]. This pattern is consistent with those observed in other studies in relation to the prevalence of height variables (especially high percentiles) as key elements for extracting information from LiDAR data [19,38,70]. Analysis of the relative importance of the variables indicated a normal distribution skewed towards the top of the tree canopy profile of E. globulus and P. pinaster plantations (h95 and h80 percentiles), together with the symmetrical normal distribution of canopy elements in P. radiata plantations (h75 and h50 percentiles); these results are consistent with the observations of Teobaldelli et al. [8]. According to the results obtained for other pure coniferous forests [38,71,72,73], the set of models confirmed that the combination of height and canopy cover metrics represents a sufficient and concise quantitative description of a homogeneous vertical structure of the species analyzed.
In this study considerably better goodness-of-fit statistics were obtained than in previous studies carried out with the same type of data in Spain (Spanish NFI and laser pulse density of 0.5 points/m2). For example, the AGB models used by Jiménez et al. [4] in Galicia yielded R2 values of 0.55, 0.60–0.65 and 0.62, and rRMSE values of 64.52%, 46.92–57.71% and 48.31%, for Eucalyptus spp., P. pinaster and P. radiata respectively. The AGB model used by Lekuona-Zuazo et al. [74] in Bizcaia yielded R2 = 0.67 and rRMSE = 32.66% in P. radiata forests. The TV and IATV models used by Tomé Morán et al. [75] in Murcia yielded rRMSE values of respectively 47.2% and 44.4% for P. halepensis forests and rRMSE values of respectively 45.8% and 43.9% for mixed stands of pines (P. nigra, P. halepensis and P. pinaster). We only found one study [26], carried out in La Rioja, which reported values in the same range as those obtained in the present study (R2 = 0.75 and rRMSE = 26.1–32.3 for P. sylvestris).
Considering the study area, although similar studies have been conducted in the region of Galicia, all these were carried out using research plots rather than SNFI. Thus, for pure P. radiata forests, the TV models used by González-Ferreiro et al. [76] yielded R2 = 0.69 and rRMSE = 30% for 0.5 points/m2, and R2 = 0.79 and rRMSE = 25% for 8 points/m2; the AGB models used by the same authors yielded R2 = 0.75 and rRMSE = 26.8% for 0.5 points/m2, and R2 = 0.80 and rRMSE = 23.7% for 8 points/m2. In a later study of pure E. globulus stands, the AGB model used by González-Ferreiro et al. [77] yielded R2 = 0.63–0.83 and rRMSE = 28.3–19.2% for 0.5 points/m2, and R2 = 0.76–0.86 and rRMSE = 22.8–17.6% for 4 points/m2. The TV model used by Gonçalves-Seco et al. [78] for E. globulus forests yielded R2 = 0.81 for a density of 4 points/m2. García-Gutiérrez et al. [79] reported R2 values of 0.66–0.70 and 0.64–0.79 for TV models fitted to LiDAR data sets of respectively 0.5 and 8 points/m2, and for AGB models, R2 values of 0.64–0.74 for 0.5 points/m2, 0.63–0.84 for 0.5 and 4 points/m2, and 0.61–0.77 for a laser pulse density of 8 points/m2, in P. radiata and E. globulus.
Local studies carried out in other regions of Spain produced slightly higher results to those outlined above, but most of the studies were carried out with higher pulse density and research plots. For example, the TV model used by Navarro et al. [80] in Madrid yielded R2 = 0.79 and rRMSE = 25.6% for a laser pulse density of 2.96 points/m2 in research plots of P. pinaster forests. Domingo et al. [81] in Aragon reported that the AGB model yielded R2 values of 0.78–0.87 for 1.5 points/m2 for research plots of Pinus halepensis. The results obtained in Extremadura by Guerra-Hernández et al. [38] for AGB model yielded R2 = 0.57–0.74 and rRMSE = 25.9–33.1% in pure Pinus pinea forests for a laser pulse density of 0.5 points/m2 and plots belonging to the Extremadura Forest Service, and Hernando et al. [82] estimated the AGB yield R2 = 0.64 and rRMSE = 16.72% for 1.15 pulses m2 in research plots of Pinus sylvestris-dominated forest located in Spain.
The present study findings were also similar to those obtained in Northern European countries, such as Finland, where the TV model used by Järnstedt et al. [83] yielded rRMSE = 31.3% for a laser pulse density of 0.5–2 points/m2 in specifically measured field data of Finnish forests and Kotivuori et al. [84] yielded rRMSE = 27.8% for nationwide and ranged from 22.9% to 31.8% for regional TV models for (0.5–1 points/m2) in nine Finnish Forest Centre inventory projects situated in various parts of Finland; or in Denmark, where the AGB model used by Nord-Larsen & Schumacher [24] yielded R2 = 0.78 and rRMSE = 33.1% and their TV model yielded R2 = 0.83 and rRMSE = 38.5% for a laser pulse density of 0.5 points/m2 in NFI plots of coniferous forests. However, the rRMSE values obtained are slightly lower than those reported by Nilsson et al. [2] in Sweden for the TV model (rRMSE = 18.9–22.5%) and NFI plots of Picea abies, P. sylvestris and Betula spp. with a laser pulse density of 0.5–1 points/m2.
Slightly higher values were also obtained in other countries outside of Europe, probably as a consequence of the use of higher density LiDAR data and research plots, which provide greater precision in the geolocation of plots and coincide temporarily with the ALS data. Thus, Stephens et al. [85] reported that their AGB model yielded R2 = 0.81 and rRMSE = 22% for a laser pulse density of 3 points/m2 in P. radiata, Pseudotsuga menziesii and Eucalyptus spp. forests in New Zealand. The AGB model used by Gleason & Im [21] yielded an rRMSE = 18.1–32.4% for a laser pulse density of 12.7 points/m2 in coniferous and deciduous forests in New York state (USA); and the TV model used by Görgens et al. [39] yielded R2 values of 0.88–0.90 and rRMSE values of 12.6% to 26.9% for a laser pulse density of 5 points/m2 in forests of Eucalyptus grandis and Eucalyptus urophylla in Sao Paulo (Brazil).
After comparing our results for the study area with those obtained in the previously mentioned studies, we can conclude the following: (i) our results produced similar results than those obtained studies carried out in large scale with data obtained from SFNI plots and ALS data with the same pulse density, and (ii) the accuracy of our results was slightly lower to those of studies developed in smalls scale using research plots data and a higher pulse density. Although our predictions can be considered accurate, more precise geolocation of the SNFI plots and coincide temporarily with the ALS data would have enabled use of a much larger and robust field database to improve the overall accuracy of estimation.
5. Conclusions
This paper presents a procedure to harmonize the 4th Spanish National Forest Inventory (SNFI-4) and the 1st nationwide Airborne Laser Scanning (ALS) data and a comparison between common regression techniques in machine learning and the MLR-based methods for estimating forest variables to predict yield estimations for E. globulus, P. pinaster and P. radiata in north-western Spain. The results showed that Random Forest and Ensemble Method statistically out-performed the other techniques. Nevertheless, the results confirmed recently reported findings, as machine learning techniques produced better results than those produced by the parametric MLR technique.
Accurate positioning of field plots and coincident timing of NFI and ALS data are key points when developing predictive models for stand properties from remote sensing data, and therefore both conditions are desirable. Nevertheless, ideal conditions are often not met and approaches that integrate both data sources are required. Despite some difficulties, we have demonstrated that is possible to integrate both types of publicly available data in Spain and to combine them in order to generate an accurate raster database of yield predictions for Spanish forests. These predictions can, of course, be improved by increasing the accuracy of SNFI plot positioning as is being accomplished by the Spanish Government. This will be a key element in using the SNFI database together with the large amount of remote sensing data available nowadays.
Author Contributions
A.N.-F., M.B.-A. and C.A.L.-S. conceived, designed and performed the experiments and wrote the manuscript. C.R. and A.C.-O. reviewed drafts of the manuscript.
Funding
The research was partially supported by the Hunosa Chair at the University of Oviedo (Project Reference SV-17-HUNOSA-1).
Acknowledgments
Special thanks to Christine Francis for revising the English of the manuscript. We also thank the anonymous referees for their valuable and very constructive comments and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Moser, P.; Vibrans, A.C.; McRoberts, R.E.; Næsset, E.; Gobakken, T.; Chirici, G.; Mura, M.; Marchetti, M. Methods for variable selection in Lidar-assisted forest inventories. Forestry 2017, 90, 112–124. [Google Scholar] [CrossRef]
- Nilsson, M.; Nordkvist, K.; Jonzén, J.; Lindgren, N.; Axensten, P.; Wallerman, J.; Egberth, M.; Larsson, S.; Nilsson, L.; Eriksson, J.; et al. A nationwide forest attribute map of Sweden predicted using airborne laser scanning data and field data from the National Forest Inventory. Remote Sens. Environ. 2017, 194, 447–454. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Westfall, J.A. Effects of uncertainty in model predictions of individual tree volume on large area volume estimates. For. Sci. 2014, 60, 34–42. [Google Scholar] [CrossRef]
- Jiménez, E.; Vega, J.A.; Fernández-Alonso, J.M.; Vega-Nieva, D.; Ortiz, L.; López-Serrano, P.M.; López-Sánchez, C.A. Estimation of aboveground forest biomass in Galicia (NW Spain) by the combined use of LiDAR, LANDSAT ETM+ and National Forest Inventory data. IFOREST 2017, 10, 590–596. [Google Scholar] [CrossRef]
- Álvarez-González, J.G.; Cañellas, I.; Alberdi, I.; Gadow, K.V.; Ruiz-González, A.D. National Forest Inventory and forest observational studies in Spain: Applications to forest modelling. For. Ecol. Manag. 2014, 316, 54–64. [Google Scholar] [CrossRef]
- López-Serrano, P.M.; López-Sánchez, C.A.; Álvarez-González, J.G.; García-Gutiérrez, J. A Comparison of Machine Learning Techniques Applied to Landsat-5 TM Spectral Data for Biomass Estimation. Can. J. Remote Sens. 2016, 42, 690–705. [Google Scholar] [CrossRef]
- Corona, P.; Cartisano, R.; Salvati, R.; Chirici, G.; Floris, A.; Di Martino, P.; Marchetti, M.; Scrinzi, G.; Clementel, F.; Travaglini, D.; et al. Airborne laser scanning to support forest resource management under alpine, temperate and Mediterranean environments in Italy. Eur. J. Remote Sens. 2012, 45, 27–37. [Google Scholar] [CrossRef]
- Teobaldelli, M.; Cona, F.; Saulino, L.; Migliozzi, A.; Dùrso, G.; Langella, G.; Manna, P.; Saracino, A. Detection of diversity and stand parameters in Mediterranean forests using leaf-off discrete return LiDAR data. Remote Sens. Environ. 2017, 192, 126–138. [Google Scholar] [CrossRef]
- Mauro, F.; Valbuena, R.; Manzanera, J.A.; García-Abril, A. Influence of Global Navigation Satellite System errors in positioning inventory plots for tree-height distribution studies. Can. J. For. Res. 2011, 41, 11–23. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E. Assessing effects of positioning errors and sample plot size on biophysical stand properties derived from airborne laser scanning data. Can. J. For. Res. 2009, 39, 1036–1052. [Google Scholar] [CrossRef]
- Maclean, G.A.; Krabill, W.B. Gross-merchantable timber volume estimating using an airborne lidar systems. Can. J. Remote Sens. 1986, 12, 7–18. [Google Scholar] [CrossRef]
- Nelson, R.; Krabill, W.; Tonelli, J. Estimating forest and volume using airborne laser data. Remote Sens. Environ. 1988, 24, 247–267. [Google Scholar] [CrossRef]
- Nelson, R. How did we get here? An early history of forestry lidar. Can. J. Remote Sens. 2013, 39, S6–S17. [Google Scholar] [CrossRef]
- Means, J.E.; Acker, S.A.; Fitt, B.J.; Renslow, M.; Emerson, L.; Hendrix, C.J. Predicting forest stand characteristics with airborne scanning Lidar. Photogramm. Eng. Remote Sens. 2000, 66, 1367–1371. [Google Scholar]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Scrivani, J.A. Fusion of small-footprint lidar and multispectral data to estimate plot-level volume and biomass in deciduous and pine forests in Virginia. USA For. Sci. 2004, 50, 551–565. [Google Scholar]
- Tesfamichael, S.; Ahmed, F.; van Aardt, J. Investigating the impact of discrete return lidar point density on estimations of mean and dominant plot-level tree height in Eucalyptus grandis plantations. Int. J. Remote Sens. 2010, 31, 2925–2940. [Google Scholar] [CrossRef]
- Dalponte, M.; Martinez, C.; Rodeghiero, M.; Gianelle, D. The role of ground reference data collection in the prediction of stem volume with lidar data in mountain areas. ISPRS J. Photogramm. Remote Sens. 2011, 66, 787–797. [Google Scholar] [CrossRef]
- García-Gutiérrez, J.; Martínez-Álvarez, F.; Troncoso, A.; Riquelme, J.C. A comparison of machine learning regression techniques for LiDAR-derived estimation of forest variables. Neurocomputing 2015, 167, 24–31. [Google Scholar] [CrossRef]
- Chen, G.; Hay, G.J.; St-Onge, B. A GEOBIA framework to estimate forest parameters from lidar transects, Quickbird imagery and machine learning: A case study in Quebec, Canada. Int. J. Appl. Earth Obs. Geoinf. 2012, 15, 28–37. [Google Scholar] [CrossRef]
- Gleason, C.J.; Im, J. Forest biomass estimation from airborne LiDAR data using machine learning approaches. Remote Sens. Environ. 2012, 125, 80–91. [Google Scholar] [CrossRef]
- Lary, D.J.; Alavi, A.H.; Gandomi, A.H.; Walker, A.L. Machine learning in geosciences and remote sensing. Geosci. Front. 2016, 7, 3–10. [Google Scholar] [CrossRef]
- Hollaus, M.; Dorigo, W.; Wagner, W.; Schadauer, K.; Höfle, B.; Maier, B. Operational wide-area stem volume estimation based on airborne laser scanning and national forest inventory data. Int. J. Remote Sens 2009, 30, 5159–5175. [Google Scholar] [CrossRef]
- Nord-Larsen, T.; Schumacher, J. Estimation of forest resources from a country wide laser scanning survey and national forest inventory data. Remote Sens. Environ. 2012, 119, 148–157. [Google Scholar] [CrossRef]
- González-Ferreiro, E.; Arellano-Pérez, S.; Castedo-Dorado, F.; Hevia, A.; Vega, J.A.; Vega-Nieva, D.; Álvarez-González, J.G.; Ruiz-González, A.D. Modelling the vertical distribution of canopy fuel load using national forest inventory and low-density airborne laser scanning data. PLoS ONE 2017, 12, e0176114. [Google Scholar] [CrossRef]
- Fernández-Landa, A.; Fernández-Moya, J.; Tomé, J.L.; Algeet-Abarquero, N.; Guillén-Climent, M.L.; Vallejo, R.; Sandoval, V.; Marchamalo, M. High resolution forest inventory of pure and mixed stands at regional level combining National Forest Inventory field plots, Landsat, and low density lidar. Int. J. Remote Sens. 2018, 39, 14. [Google Scholar] [CrossRef]
- Merino, A.; Balboa, M.A.; Rodríguez-Soalleiro, R.; Alvarez-González, J.G. Nutrient exports under different harvesting regimes in fast growing forest plantations in southern Europe. For. Ecol. Manag. 2005, 207, 325–339. [Google Scholar] [CrossRef]
- MAPAMA. Anuario de Estadística. Avance 2017. Ministerio de Agricultura, Pesca y Alimentación: Madrid, 2018. Available online: https://www.mapa.gob.es/es/desarrollo-rural/estadisticas/avance_2017_web_tcm30-510675.pdf (accessed on 15 July 2019).
- EEA. Biogeographical Regions. European Environment Agency: Copenhagen, Denmark, 2016. Available online: https://www.eea.europa.eu/data-and-maps/data/biogeographical-regions-europe-3 (accessed on 2 October 2018).
- MAPAMA. Mapa Forestal de España 1:25.000 (MFE25). 2012. Available online: https://www.miteco.gob.es/es/cartografia-y-sig/ide/descargas/biodiversidad/mfe.aspx (accessed on 2 October 2018).
- MARM. Cuarto Inventario Forestal Nacional; Comunidad Autónoma de Galicia, Dirección General del Medio Natural y Política Forestal: Madrid, Spain, 2011.
- MARM. Cuarto Inventario Forestal Nacional. Principado de Asturias; Ministerio de Medio Ambiente, y Medio Rural y Marino, Dirección General de Desarrollo Rural y Política Forestal: Madrid, Spain, 2012.
- MARM. Cuarto Inventario Forestal Nacional. Cantabria; Ministerio de Medio Ambiente, y Medio Rural y Marino, Dirección General de Desarrollo Rural y Política Forestal: Madrid, Spain, 2012.
- Alberdi, I.; Cañellas, I.; Vallejo, R. The Spanish National Forest Inventory: History, development, challenges and perspectives. Pesqui. Florest. Bras. 2017, 37, 361. [Google Scholar] [CrossRef]
- Castaño-Santamaría, J.; Barrio-Anta, M.; Álvarez-Álvarez, P. Potential above ground biomass production and total tree carbon sequestration in the major forest species in NW Spain. Int. For. Rev. 2013, 15, 273–289. [Google Scholar] [CrossRef]
- McGaughey, R.J. FUSION/LDV: Software for LIDAR Data Analysis and Visualization. In US Department of Agriculture, F.S.; Pacific Northwest Research Station: Seattle, WA, USA, 2014; p. 123. Available online: http://forsys.cfr.washington.edu/fusion/fusionlatest.html (accessed on 15 July 2019).
- White, J.; Tompalski, P.; Vastaranta, M.; Wulder, M.; Saarinen, N.; Stepper, C.; Coops, N. A Model Development and Application Guide for Generating an Enhanced Forest Inventory Using Airborne Laser Scanning Data and an Area-Based Approach; CWFC Information Report FI-X-018. Canadian Forest Service, Pacific Forestry Centre: Victoria, BC, Canada, 2017. [CrossRef]
- Guerra-Hernández, J.; Bastos Görgens, E.; García-Gutiérrez, J.; Estraviz Rodriguez, L.C.; Tomé, M.; González-Ferreiro, E. Comparison of ALS based models for estimating aboveground biomass in three types of Mediterranean forest. Eur. J. Remote Sens. 2016, 49, 185–204. [Google Scholar] [CrossRef]
- Görgens, E.B.; Montaghi, A.; Estraviz Rodriguez, L.C. A performance comparison of machine learning methods to estimate the fast-growing forest plantation yield based on laser scanning metrics. Comput. Electron. Agric. 2015, 116, 221–227. [Google Scholar] [CrossRef]
- Næsset, E.; Gobakken, T. Estimation of above-and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Murgaš, V.; Sačkov, I.; Sedliak, M.; Tunák, D.; Chudý, F. Assessing horizontal accuracy of inventory plots in forests with different mix of tree species composition and development stage. J. For. Sci. 2018, 64, 478–485. [Google Scholar] [CrossRef]
- Alberdi, I.; Sandoval, V.; Condés, S.; Cañellas, I.; Vallejo, R. El Inventario Forestal Nacional Español, una herramienta para el conocimiento, la gestión y la conservación de los ecosistemas forestales arbolados. Ecosistemas 2016, 25, 88–97. [Google Scholar] [CrossRef]
- Smith, J.E.; Heath, L.S.; Jenkins, J.C. Forest Volume-to-Biomass Models and Estimates of Mass for Live and Standing Dead Trees of U.S. Forests; Gen. Tech. Rep. NE-298. U.S. Department of Agriculture, Forest Service, Northeastern Research Station: Newtown Square, PA, USA, 2003.
- Boudewyn, P.A.; Song, X.; Magnussen, S.; Gillis, M.D. Model-Based, Volume-to-Biomass Conversion for Forested and Vegetated Land in Canada; Information Report BC-X-411. Natural Resources Canada, Canadian Forest Service, Pacific Forestry Centre: Victoria, BC, Canada, 2007.
- Arias-Rodil, M.; Barrio-Anta, M.; Diéguez-Aranda, U. Developing a dynamic growth model for maritime pine in Asturias (NW Spain): Comparison with nearby regions. Ann. For. Sci. 2016, 73, 297–320. [Google Scholar] [CrossRef]
- Diéguez-Aranda, U.; Burkhart, H.E.; Rodríguez-Soalleiro, R. Modelling dominant height growth of radiata pine (Pinus radiata D. Don) plantations in north-western Spain. For. Ecol. Manag. 2005, 215, 271–284. [Google Scholar] [CrossRef]
- García-Villabrille, D. Modelización del Crecimiento y la Producción de Plantaciones de Eucalyptus globulus Labill. en el noroeste de España. Ph.D. Thesie, Universidad de Santiago de Compostela, Higher Polytechnic Engineering School, Lugo, Spain, 2015; 181p. Available online: https://core.ac.uk/download/pdf/75994613.pdf (accessed on 2 October 2018).
- Barrio-Anta, M.; Cámara-Obregón, A.; Castedo-Dorado, F.; López-Sanchez, C.A. Modelling and mapping the current and future optimal habitat and productivity for maritime pine stands under climate change in Northwestern Spain. 2019; in preparation. [Google Scholar]
- López-Sánchez, C.A.; Cámara-Obregón, A.; Castedo-Dorado, F.; Barrio-Anta, M. Modelling and mapping current and future optimal distribution and site productivity for radiata pine stands in Northwestern Spain. 2019; in preparation. [Google Scholar]
- López-Sánchez, C.A.; Cámara Obregón, A.; Oliveros, A.; Barrio-Anta, M. Predicting and mapping Eucalyptus globulus productivity from biophysical variables in Northwestern Spain. 2019; in preparation. [Google Scholar]
- Fassnacht, F.E.; Hartig, F.; Latifi, H.; Berger, C.; Hernández, J.; Corvalán, V.; Koch, B. Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 2014, 154, 102–114. [Google Scholar] [CrossRef]
- Shataee, S. Forest attributes estimation using aerial laser scanner and TM Data. For. Syst. 2013, 22, 484–496. [Google Scholar]
- Cover, T.; Hart, P. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Quinlan, R.J. Learning with Continuous Classes. In Proceedings of the 5th Australian Joint Conference on Artificial Intelligence, Hobart, Tasmania, 16–18 November 1992; World Scientific: Singapore, 1992; pp. 343–348. [Google Scholar]
- Wang, Y.; Witten, I.H. Induction of model trees for predicting continuous classes. In Proceedings of the 9th European Conference on Machine Learning, Prague, Czech Republic, 23–25 April 1997. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Gislason, P.O.; Benediktsson, J.A.; Sveinsson, J.R. Random Forests for land cover classification. Pattern Recognit. Lett. 2006, 27, 294–300. [Google Scholar] [CrossRef]
- Wolpert, D. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Naimi, A.I.; Balzer, L.B. Stacked generalization: An introduction to super learning. Eur. J. Epidemiol. 2018, 33, 459–464. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. Sigkdd Explor. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Valbuena, R.; Hernando, A.; Manzanera, J.A.; Görgens, E.B.; Almeida, D.R.A.; Mauro, F.; García-Abril, A.; Coomes, D.A. Enhancing of Accuracy Assessment for Forest Above-Ground Biomass Estimates Obtained from Remote Sensing via Hypothesis Testing and Overfitting Evaluation. Ecol. Model. 2017, 366, 15–26. [Google Scholar] [CrossRef]
- Zhiwei, X.; Xinghua, W. Research for information extraction based on wrapper model algorithm. In Proceedings of the Second International Conference on Computer Research and Development, Haiphong City, Vietnam, 7–10 May 2010; pp. 652–655. [Google Scholar]
- Hall, M.A.; Holmes, G. Benchmarking Attribute Selection Techniques for Discrete Class Data Mining. IEEE Trans. Knowl. Data Eng. 2003, 15, 3. [Google Scholar] [CrossRef]
- Packalén, P.; Maltamo, M. The k-MSN method for the prediction of species-specific stand attributes using airborne laser scanning and aerial photographs. Remote Sens. Environ. 2007, 109, 328–341. [Google Scholar] [CrossRef]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar] [CrossRef]
- Lee, J.; Im, J.; Kim, K.; Quackenbush, L.J. Machine Learning Approaches for Estimating Forest Stand Height Using Plot-Based Observations and Airborne LiDAR Data. Forests 2018, 9, 268. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Harding, D.J.; Keller, M.; Cohen, W.B.; Carabajal, C.C.; Del Bom Espirito-Santo, F.; Hunter, M.O.; De Oliveira, R. Estimates of forest canopy height and aboveground biomass using ICESat. Geophys. Res. Lett. 2005, 32, L22S02. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Harding, D.J.; Parker, G.G.; Acker, S.A.; Gower, S.T. LiDAR remote sensing of aboveground biomass in three biomes. Glob. Ecol. Biogeogr. 2002, 11, 393–400. [Google Scholar] [CrossRef]
- Li, M.; Im, J.; Quackenbush, L.; Liu, T. Forest biomass and carbon stock quantification using airborne lidar data: A case study over Huntington wildlife forest in the Adirondack Park. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3143–3156. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J.; Muller-Landau, H.C.; Vieilledent, G.; Vaudry, R.; Rasamoelina, M.; Hall, J.S.; Van Breugel, M. A universal airborne LiDAR approach for tropical forest carbon mapping. Oecologia 2012, 168, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne LiDAR data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Ni-Meister, W.; Lee, S.; Strahler, A.H.; Woodcock, C.E.; Schaaf, C.; Yao, T.; Ranson, K.J.; Sun, G.; Blair, J.B. Assessing general relationships between aboveground biomass and vegetation structure parameters for improved carbon estimate from lidar remote sensing. J. Geophys. Res. Biogeosci. 2010, 115. [Google Scholar] [CrossRef]
- Lekuona Zuazo, I.; Montealegre Gracia, A.L.; Lamelas Gracia, M.T. Cartografía de la biomasa aérea total en masas de Pinus radiata D. Don a partir de datos públicos LiDAR-PNOA e Inventario Forestal Nacional. GeoFocus 2017, 20, 87–107. [Google Scholar] [CrossRef]
- Tomé-Morán, J.L.; Esteban Cava, J.; Martín Alcón, S.; Escamochero, I.; Fernández-Landa, A. ForestMap, Online forest inventories using Murcia Regional Airborne LiDAR Data. In Proceedings of the XVII Congreso de la Asociación Española de Teledetección, Murcia, Spain, 3–7 Octubre 2017; pp. 147–150. [Google Scholar]
- González-Ferreiro, E.; Diéguez-Aranda, U.; Miranda, D. Estimation of stand variables in Pinus radiata D. Don plantations using different LiDAR pulse densities. Forestry 2012, 85, 281–292. [Google Scholar] [CrossRef]
- Gonzalez-Ferreiro, E.; Miranda, D.; Barreiro-Fernandez, L.; Bujan, S.; Garcia-Gutierrez, J.; Dieguez-Aranda, U. Modelling stand biomass fractions in Galician Eucalyptus globulus plantations by use of different LiDAR pulse densities. For. Syst. 2013, 22, 510–525. [Google Scholar] [CrossRef]
- Gonçalves-Seco, L.; González-Ferreiro, E.; Diéguez-Aranda, U.; Fraga-Bugallo, B.; Crecente, R.; Miranda, D. Assessing the attributes of high-density Eucalyptus globulus stands using airborne laser scanner data. Int. J. Remote Sens. 2011, 32. [Google Scholar] [CrossRef]
- García-Gutiérrez, J.; Gonzalez-Ferreiro, E.; Riquelme-Santos, J.C.; Miranda, D.; Dieguez-Aranda, U.; Navarro-Cerrillo, R.M. Evolutionary feature selection to estimate forest stand variables using LiDAR. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 119–131. [Google Scholar] [CrossRef]
- Navarro, J.A.; Fernández-Landa, A.; Tomé, J.L.; Guillén-Climent, M.L.; Ojeda, J.C. Testing the quality of forest variable estimation using dense image matching: A comparison with airborne laser scanning in a Mediterranean pine forest. Int. J. Remote Sens. 2018, 39. [Google Scholar] [CrossRef]
- Domingo, D.; Lamelas, M.T.; Montealegre, A.L.; García-Martín, A.; De la Riva, J. Estimation of Total Biomass in Aleppo Pine Forest Stands Applying Parametric and Nonparametric Methods to Low-Density Airborne Laser Scanning Data. Forests 2018, 9, 158. [Google Scholar] [CrossRef]
- Hernando, A.; Puerto, L.; Mola-Yudego, B.; Manzanera, J.A.; García-Abril, A.; Maltamo, M.; Valbuena, R. Estimation of forest biomass components using airborne LiDAR and multispectral sensors. iForest 2019, 12, 207–213. [Google Scholar] [CrossRef]
- Järnstedt, J.; Pekkarinen, A.; Tuominen, S.; Ginzler, C.; Holopainen, M.; Viitala, R. Forest variable estimation using a high-resolution digital surface model. ISPRS J. Photogramm. Remote Sens. 2012, 74, 78–84. [Google Scholar] [CrossRef]
- Kotivuori, E.; Korhonen, L.; Packalen, P. Nationwide airborne laser scanning based models for volume, biomass and dominant height in Finland. Silva Fenn. 2016, 50, 28. [Google Scholar] [CrossRef]
- Stephens, P.R.; Kimberley, M.O.; Beets, P.N.; Paul, T.S.H.; Searles, N.; Bell, A.; Brack, C.; Broadley, J. Airborne scanning LiDAR in a double sampling forest carbon inventory. Remote Sens. Environ. 2012, 117, 348–357. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).