Abstract
The gut microbiota is engaged in multiple interactions affecting host health, and gut dysbiosis can lead to many diseases. However, the effects of acetylcysteine (NAC) on the gut microbiome composition in pigs using metagenomic sequencing have not been reported. In this study, we used metagenome sequencing to study the effects of NAC on the pig gut microbiome. Sequencing results showed that microbial diversity was changed after NAC treatment. Antibiotic Resistance Genes Database (ARDB) analysis demonstrated that the main genes modified were macb, tsnr, norm, bl2be-per, vansb and pbp1b in the NAC group. Our data showed that NAC could affect microbial distribution at the phylum, gene and species levels. At the species level, NAC significantly increased the abundances of Megasphaera, Lactobacillus reuteri and Megasphaeraelsdenii and reduced the abundances of Phascolarctobacterium succinatutens, Prevotellacopri and Selenomonasbovis compared with the control group. In addition, Gene Ontology (GO) analysis revealed that in the NAC group, cellular process, metabolic process and single-organism process were the dominant terms. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis demonstrated that RNA transport, MAPK signaling pathway, cell cycle, glycosylphosphatidylinositol (GPI)-anchor biosynthesis and VEGF signaling pathway were the dominant signaling pathways in the NAC group. In conclusion, our results suggest that NAC may modify the piglet gut microbiome composition and these findings might provide a new strategy for maintaining animal and human health in the future.
1. Introduction
The “microbiota” is defined as all microbes, including their genomes and extrachromosomal components, both in and on the host animal [1]. The gut microbiota plays a critical role in maintaining the health of the host [2]. The gut microbiota is involved in several major physiological functions of the host, including digestion, metabolism and immunity [3]. Changes in the composition of gut microbiota may be associated with the pathogenesis of various neurodegenerative disorders such as Parkinson’s disease, Alzheimer’s disease, multiple sclerosis and amyotrophic lateral sclerosis [4]. Gut dysbiosis can cause inflammation and metabolic disorders, thus promoting the development of cardiovascular diseases [5]. The use of antibiotics has increased in recent years and recent research has shown that the antibiotics can affect the composition of the gut microbiota, reduce its biodiversity, change metabolic activity and select for antibiotic-resistant organisms [6]. Therefore, it is very important to study a new additive to reduce the use of antibiotics in order to maintain host health and prevent diseases.
Acetylcysteine (NAC) is a synthetic derivative of L-cysteine and a precursor of glutathione (GSH) that is a tripeptide containing a sulfhydryl group [7]. Previous research has shown that NAC has functions, including antioxidant, mucolytic and anti-inflammatory properties [8]. NAC has a strong scavenging activity for various free radicals, so it is often used in the treatment of acetaminophen overdose, mucosal lysis therapy and mental diseases [9]. In addition, NAC can suppress oxidative stress caused by Hcys through its antioxidant properties [10]. Previous studies have shown that NAC can change the gut microbiome composition in the treatment of hypertension [11]. NAC regulated the gut microbiota of mice by promoting the growth of beneficial bacteria and preventing the growth of diabetes-related genera; these results were obtained through 16S rRNA sequencing [12]. Furthermore, NAC supplementation played a positive role in regulating the gut microbiome during porcine epidemic eiarrhea virus (PEDV) infection [13]. Despite various clinical trials using NAC supplementation, there are no reports on using the drinking water method for metagenomic sequencing analysis to explore the effects of NAC on the pig gut microbiome composition.
The main purpose of this study was to investigate the effect of NAC on the compositional changes of the gut microflora in pigs. Our results showed that NAC could modulate the pig gut microbial composition. The findings of this study can be used as a new strategy to regulate the composition of gut microorganisms to prevent diseases and maintain host health.
2. Materials and Methods
2.1. Animals and Experimental Design
This study was conducted in strict accordance with the recommendations of the China Regulations for the Administration of Affairs Concerning Experimental Animals 1988 and the Hubei Regulations for the Administration of Affairs Concerning Experimental Animals 2005. All animal experiments and animal care in this study were approved by the Animal Care and Use Committee of Wuhan Polytechnic University, Hubei Province, China (EM951, 5 November 2020). Six 30-day-old healthy weaned piglets (Duroc × Landrace × Large White) weighing 8 to 10 kg were purchased from Wuhan Wannianqing Animal Husbandry Co., Ltd. (Wuhan, China) for in vivo experiments.
The six piglets (three male piglets and three female piglets) were randomly divided into two groups: the control group (two male piglets and one female piglet) and the NAC group (one male piglet and two female piglets). The two groups were given equal amounts of the same feed every day. In the control group, the piglets were given basic drinking water. Piglets in the NAC group were given basic drinking water supplemented with 250 mg NAC/liter [14]. After 14 days, fresh stool samples were collected from the piglets of both groups. Each group of stool samples was taken for metagenomic sequencing.
2.2. DNA Extraction from the Stool Samples and Library Construction
The collected stool samples were stored in liquid nitrogen and placed at −80 °C for rapid freezing. The genomic DNA from the stool samples was extracted following the protocol from the Mini Stool DNA Isolation Kit (EMERTHER, Jiaxing, Shanghai, China, Cat#20200401). The extracted DNA was analyzed using a NanoDrop instrument (Thermo Scientific, Waltham, MA, USA). All DNA samples were used for library construction. DNA library construction was conducted using a DNA Library Fast Construction Kit (Illumina, San Diego, CA, USA, Cat#5067-4626) according to manufacturer’s instructions [15]. The quality of the library was verified by using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA, USA). Paired-end metagenomic sequencing was conducted on the Illumina platform with a 2 × 150 base pair (bp) read length.
2.3. Metagenomic Sequencing
Raw reads obtained by sequencing might contain unqualified reads with low overall quality and low terminal quality. These unqualified reads are likely to have a certain impact on the analysis quality; therefore, they must be filtered to obtain clean reads that can be used for data analysis. Raw reads with a Phred Score > 20 and sequencing fragments with lengths of less than 45 bp were removed. The raw data was submitted to the Sequence Read Archive (SRA) database under accession number “PRJNA818124” (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB11191947/overview (accessed on 30 May 2023)). The quality control reads were aligned to the host reference genome by Bowtie 2 (version 2.3.1) to obtain clean reads [16]. The clean reads were spliced by using metaSPAdes (SPAdes-3.10.1) software [17], and the contig sequences with lengths of less than 500 bp were filtered out. MetaGeneMark (GeneMark. HMM version 3.38) was used to predict the splicing results [18]. The databases commonly utilized to provide functional annotation include the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups (eggNOG) (v4.5) [19]. Antibiotic resistance genes were obtained by using the Antibiotic Resistance Genes (ARDB) database [20], and MetaPhlAn2 software https://huttenhower.sph.harvard.edu/metaphlan3/ (accessed on 30 May 2023) was employed to explore the species abundance of the microbial community [21]. Principal component analysis (PCA) was conducted using the ade4 tool in R to determine the species compositions of different samples. The differences between individuals or groups were observed through principal co-ordinates analysis (PCoA) using QIIM E2. The heatmaps at different levels were performed using the heatmap tool in R.
2.4. Statistical Analysis
The statistical analysis of the data was performed using R software https://www.r-project.org/ accessed on 30 May 2023. All comparisons in each group were pairwise. The Wilcoxon rank sum test was used to analyze the significant differences of the gene, eggNOG and KEGG pathways between the two groups of samples. A value of p ≤ 0.05 was considered statistically significant.
3. Results
3.1. Effects of NAC on the Pig Gut Microbiome
The effects of NAC supplementation in drinking water on the gut microbiome composition of pigs were studied by metagenomic sequencing. We identified 510,936,651 ± 20,934,839 clean reads from six pig stool samples, and 85,156,109 ± 20,934,839 sequences per stool sample were obtained (Table 1).

Table 1.
Statistical summary analysis of pig gut microbiome.
3.2. α-Diversity Analysis of Pig Gut Microbiome Composition
The Shannon index (H) was used to explore the diversity of community populations. Our data show that NAC supplementation (Group 2) could change the α-diversity of the gut microbial community relative to the control group (Group 1) (Figure 1). Overall, the α-diversity in the NAC group (Group 2) was lower than the control group (Group 1) (Figure 1).
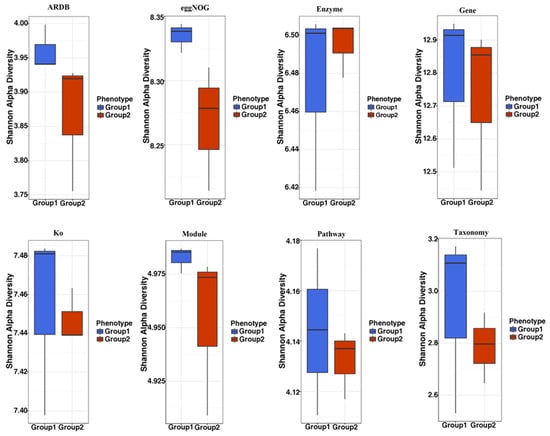
Figure 1.
Alpha diversity analysis of the effect of NAC on the pig gut microbiome composition. The stool samples were collected from the NAC group and the control group. The Shannon H index was adopted to explore the diversity by utilizing ARDB, eggNOG, Enzyme, Gene, Ko, Module, Pathway and Taxonomy. Group 1: the control group (CON0-a, CON0-b, CON0-c); Group 2: the NAC group (NAC-a, NAC-b, NAC-c).
3.3. β-Diversity Analysis of Pig Gut Microbiome Composition
PCA was used for analyzing the microbiome compositions of different samples. PCA analysis demonstrated that the NAC group (Group 2) and the control group (Group 1) formed different clusters, and most of the samples were distributed in their respective clusters (Figure 2). PCoA analysis showed that the different clusters were isolated in most of the samples from the NAC group (Group 2) and the control group (Group 1), which indicated that NAC could modify the gut microbiome composition (Figure 3).
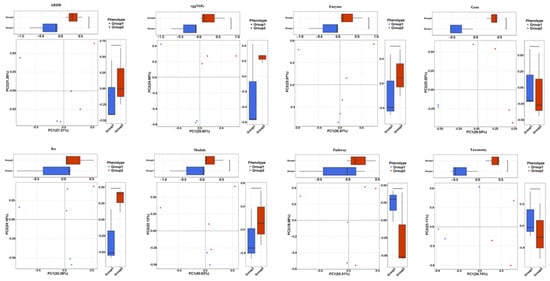
Figure 2.
Principal components analysis (PCA) of the influence of NAC on the piglet gut microbiome composition. PCA of piglet gut microbiome modifications of ARDB, eggNOG, Enzyme, Gene, KEGG orthology (Ko), Module, Pathway and Taxonomy in different groups. Group 1: the control group; Group 2: the NAC group.
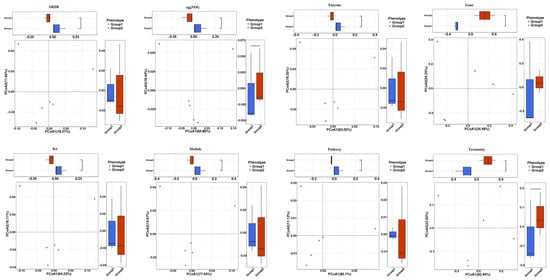
Figure 3.
Principal coordinates analysis (PCoA) of the influence of NAC on the piglet gut microbiome composition. PCoA of gut microbiome modifications of ARDB, eggNOG, Enzyme, Gene, KEGG orthology (Ko), Module, Pathway and Taxonomy in different groups. Group 1: the control group; Group 2: the NAC group.
The ARDB results showed that in the control group (Group 1), the main antibiotic resistance genes were vanrb [22], tetq [23], bl2b-tem [24], aac3iii, vanug and cml-e7 (Supplemental Figure S1). However, following NAC treatment (Group 2), the abundances of antibiotic resistance genes were significantly changed, and the main genes were macb [25], tsnr [26], norm, bl2be-per, vansb and pbp1b (Supplemental Figure S1).
3.4. Effects of NAC on Microbiome Composition at the Phylum Level
We explored the effects of NAC on piglet microbiome composition at the phylum level. The result demonstrated that the abundance of Firmicutes and Euryarchaeota was increased when the piglets were treated with NAC (Group 2) (Figure 4). NAC reduced the abundance of Bacteroidetes compared with the levels found in the control group (Group 1) (Figure 4).
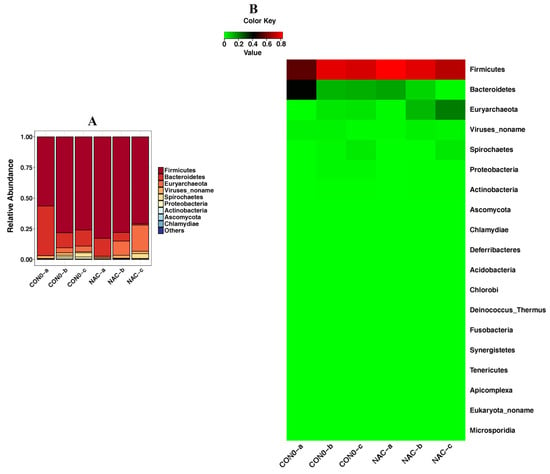
Figure 4.
Detection of phylum level according to piglet taxonomy analysis presented as a bar chart (A) and a heatmap (B). The abundance of a phylum is displayed on the y-axis. Group 1: the control group (CON0-a, CON0-b, CON0-c); Group 2: the NAC group (NAC-a, NAC-b, NAC-c).
3.5. Effects of NAC on Microbiome Composition at the Genus Level
At the genus level, the relative abundances of Prevotella and Phascolarctobacterium were higher in the control group (Group 1) than in the NAC group (Group 2) (Figure 5). NAC could increase the abundances of Megasphaera, Lactobacillus and Methanobrevibacter compared with the levels found in the control group (Group 1) (Figure 5).
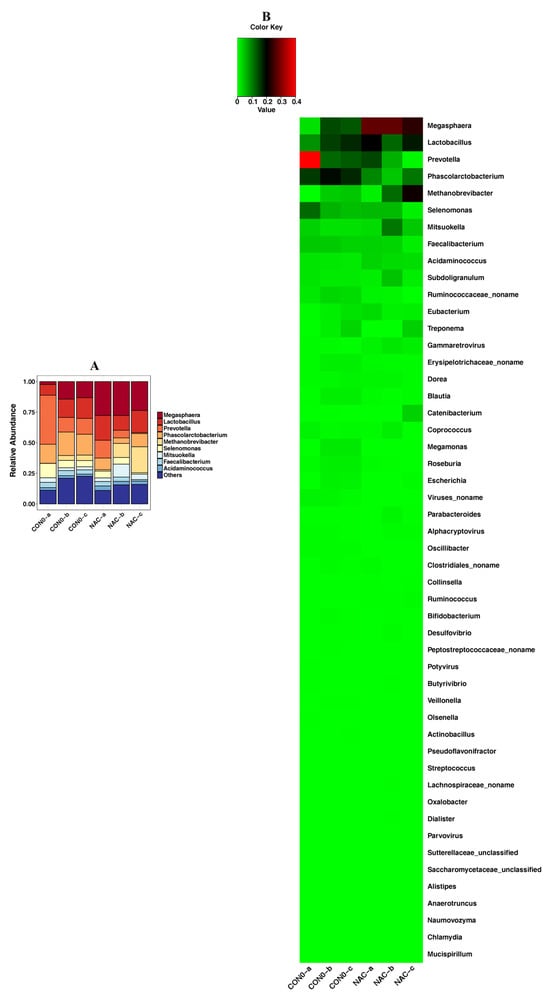
Figure 5.
Detection of genus level according to piglet taxonomy analysis presented as a bar chart (A) and a heatmap (B). The abundance of a genus is displayed on the y-axis. Group 1: the control group (CON0-a, CON0-b, CON0-c); Group 2: the NAC group (NAC-a, NAC-b, NAC-c).
3.6. Effects of NAC on Microbiome Composition at the Species Level
At the species level, the results showed that compared with the levels found in the control group (Group 1), NAC (Group 2) could significantly increase the abundances of Megasphaera, Lactobacillus reuteri, Methanobrevibacter and Megasphaeraelsdenii (Figure 6). However, the abundances of Phascolarctobacterium succinatutens, Prevotellacopri, Selenomonasbovis and Prevotellastercorea were significantly attenuated in the NAC group (Group 2) compared with the control group (Group 1) (Figure 6).

Figure 6.
Detection of species level according to piglet taxonomy analysis presented as a bar chart (A) and a heatmap (B). The abundance of a species is displayed on the y-axis. Group 1: the control group (CON0-a, CON0-b, CON0-c); Group 2: the NAC group (NAC-a, NAC-b, NAC-c).
3.7. Effects of NAC on Piglets Microbiome Composition Functional Changes
GO analysis showed that in the NAC group, cellular process, metabolic process and single-organism process were the dominant terms in the biological process category, whereas binding and catalytic activity were the most enriched in the molecular function category (Figure 7A). KEGG pathway analysis demonstrated that RNA transport, the MAPK signaling pathway, the cell cycle, RNA degradation, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, the tight junction and VEGF signaling pathways were the main signaling pathways in the NAC group (Figure 7B).
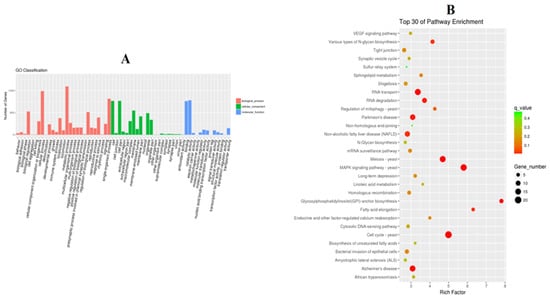
Figure 7.
Performance of GO enrichment (A) and the top 30 signaling pathways verified by KEGG when utilizing DAVID analysis (B).
4. Discussion
The purpose of this study was to explore the effect of NAC on the composition of the piglet gut microbiome. NAC is an effective antioxidant and a precursor of L-cysteine [27]. Previous studies have shown that the effectiveness of NAC in the treatment of clinical diseases mainly comes from its antioxidant or free radical scavenging properties [28]. Dietary supplementation with NAC could improve the intestinal dysfunction and animal health problems induced by lipopolysaccharide and heat stress in pigs and chickens [29,30]. NAC has been reported to promote intestinal cell growth and protein synthesis [31]. In addition, NAC has a beneficial effect on regulating the piglet gut microbiome in PEDV infection [13]. NAC supplementation in late gestation altered the fecal microbial communities in sows [32]. NAC could prevent glucose metabolic disturbances through reshaping the high-fat-diet-fed mouse gut microbiota structure [12]. In our study, our results demonstrated that NAC could modify the piglet gut microbiome composition; however, whether the change in the gut microbiomes of piglets affected the gut function needs further study.
In this study, due to the limitation of sequencing price, three samples from each group were used to estimate the effect of NAC on gut microbiome changes using the metagenome sequencing method. In previous studies, the modifications in the gut microbiome were usually measured using 16S rRNA sequencing [33]. 16S rRNA sequencing has certain limitations, such as the inability to identify most microorganisms at the strain and species levels and possible amplification bias [34,35]. In contrast, metagenomic sequencing can provide higher taxonomic map resolutions and the functional classification of the microbiome [36]. In addition, metagenomic sequencing can recognize not only bacteria but also viruses, fungi and protozoa [37]. Therefore, metagenome sequencing was employed in this study to comprehensively reflect the influence of NAC on the composition of the gut microbial community.
Probiotics are living microbial food supplements that are beneficial to health [38]. Some Lactobacillus species are widely used as probiotics for livestock and poultry due to their multiple benefits, such as improving the composition of the gastrointestinal tract microbiome [39] and improving the immunity of piglets [40]. Lactobacillus reuteriis was generally considered to be safe by the U.S. Food and Drug Administration [41]. Previous studies have shown that L. reuteri could enhance the intestinal mucosal barrier of newborn piglets through increasing the number of goblet cells and the mRNA expression levels of Muc2 and Lyz-1 [42]. Supplementation with L. reuteri increased plasma IL-2, IFN-α and IFN-β secretion from immune cells and improved the immunity of pigs [43]. L. reuteri also played anti-inflammatory roles by reducing the level of intestinal proinflammatory cytokines [44]. In addition, L. reuteri could reduce the production of inflammatory factors and oxidative stress in the intestinal mucosa of ETEC-infected piglets and improved diarrhea in weaned piglets [45]. In our study, we found that NAC could enhance the relative abundance of L. reuteri at the species level. Therefore, based on previous research, we speculate that L. reuteri might play roles involved in maintaining intestinal function and host health. However, the specific mechanism is still unclear and needs to be further assessed in our future study.
We also found that the abundance of Megasphaera was significantly upregulated in the NAC group. It has been documented that Megasphaera has many beneficial functions, such as synthesizing short chain fatty acids [46], regulating osmotic diarrhea [47], affecting the host immune response [48] and regulating intestinal homeostasis compounds [49]. In view of the fact that Megasphaeraelsdenii could convert lactic acid into short chain fatty acids, it is used as a probiotic for the treatment of diet-induced metabolic acidosis [50]. Therefore, Megasphaera might have effects in the enhanced host intestinal health elicited by NAC treatment.
Excitingly, our current study demonstrated that when piglets were administrated NAC, the abundance of Prevotellacopri was reduced. A previous study has shown that P. copri was closely related to carboplatin-induced intestinal mucositis and that reducing the abundance of P. copri could attenuate the inflammatory response [51]. P. copri could promote an inflammatory response via changing the mucus layer permeability and enhancing the intestinal inflammatory state [52]. In a mouse model, P. copri could promote weight loss and aggravate epithelial inflammation [53]. Thus, we inferred that the anti-inflammatory effect of NAC might be realized through changing the abundances of various microorganisms in the host gut.
Taken together, our data demonstrate that NAC modulated the composition of the piglet gut microbiome. Our study found that NAC increased the abundance of probiotics and reduced the abundance of harmful microorganisms, which might provide a novel way for maintaining host gut health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microbiolres14040132/s1, Figure S1: The influence of NAC on the ARDB changes.
Author Contributions
Y.Q. conceived and designed the experiments; S.F., X.T., J.L., Y.Y., J.H. and C.P. performed the experiments; S.F., L.G., C.Y., Y.L., B.Z. and Y.Q. analyzed the data; S.F. and X.T. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key Research and Development Plan of Hubei Province, China (2022BBA0055).
Institutional Review Board Statement
This study was conducted in strict accordance with the recommendations of the China Regulations for the Administration of Affairs Concerning Experimental Animals 1988 and the Hubei Regulations for the Administration of Affairs Concerning Experimental Animals 2005. All animal experiments and animal care in this study were approved by the Animal Care and Use Committee of Wuhan Polytechnic University, Hubei Province, China (EM951, 5 November 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Liu, L.; Yan, S.; Sun, W.; Jia, M.; Tian, S.; Huang, S.; Zhou, Z.; Zhu, W. Gut Microbiota: A Key Factor in the Host Health Effects Induced by Pesticide Exposure? J. Agric. Food. Chem. 2020, 68, 10517–10531. [Google Scholar] [CrossRef]
- Bin, P.; Tang, Z.; Liu, S.; Chen, S.; Xia, Y.; Liu, J.; Wu, H.; Zhu, G. Intestinal microbiota mediates Enterotoxigenic Escherichia coli-induced diarrhea in piglets. BMC Vet. Res. 2018, 14, 385. [Google Scholar] [CrossRef]
- Sarkar, R.S.; Banerjee, S. Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 2019, 328, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Qian, Z.; Yin, J.; Xu, W.; Zhou, X. The role of intestinal microbiota in cardiovascular disease. J. Cell. Mol. Med. 2019, 23, 2343–2350. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Fernandez, B.L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Raghu, G.; Berk, M.; Campochiaro, P.A.; Jaeschke, H.; Marenzi, G.; Richeldi, L.; Wen, F.-Q.; Nicoletti, F.; Calverley, P.M.A. The Multifaceted Therapeutic Role of N-Acetylcysteine (NAC) in Disorders Characterized by Oxidative Stress. Curr. Neuropharmacol. 2021, 19, 1202–1224. [Google Scholar] [CrossRef] [PubMed]
- Craver, B.M.; Ramanathan, G.; Hoang, S.; Chang, X.; Mendez Luque, L.F.; Brooks, S.; Lai, H.Y.; Fleischman, A.G. N-acetylcysteine inhibits thrombosis in a murine model of myeloproliferative neoplasm. Blood Adv. 2020, 4, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, V.; Afsharian, P.; Shahhoseini, M.; Kalantar, S.M.; Moini, A. A Review on Various Uses of N-Acetyl Cysteine. Cell J. 2017, 19, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Leon, M.; Sawmiller, D.; Shytle, R.D.; Tan, J. Therapeutic Cocktail Approach for Treatment of Hyperhomocysteinemia in Alzheimer’s Disease. Cell Med. 2018, 10, 2155179017722280. [Google Scholar] [CrossRef]
- Hsu, C.N.; Hou, C.Y.; Chang-Chien, G.P.; Lin, S.; Tain, Y.L. Maternal N-Acetylcysteine Therapy Prevents Hypertension in Spontaneously Hypertensive Rat Offspring: Implications of Hydrogen Sulfide-Generating Pathway and Gut Microbiota. Antioxidants 2020, 9, 856. [Google Scholar] [CrossRef]
- Zheng, J.; Yuan, X.; Zhang, C.; Jia, P.; Jiao, S.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. N-Acetylcysteine alleviates gut dysbiosis and glucose metabolic disorder in high-fat diet-fed mice. J. Diabetes 2019, 11, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Lyu, Y.; Li, X.; Wu, M.; Yu, K.; Li, S.; Ji, C.; Zhang, Q.; Zhang, Y.; Zhao, D.; et al. Impact of N-Acetylcysteine on the Gut Microbiota in the Piglets Infected with Porcine Epidemic Diarrhea Virus. Front. Vet. Sci. 2020, 7, 582338. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, J.; Hou, Y.; Yi, D.; Ding, B.; Xie, J.; Zhang, Y.; Chen, H.; Wu, T.; Zhao, D.; et al. N-Acetylcysteine supplementation alleviates intestinal injury in piglets infected by porcine epidemic diarrhea virus. Amino Acids 2017, 49, 1931–1943. [Google Scholar] [CrossRef]
- Kapp, J.D.; Green, R.E.; Shapiro, B. A Fast and Efficient Single-stranded Genomic Library Preparation Method Optimized for Ancient DNA. J. Hered. 2021, 112, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Meera, S.P.; Sreeshan, A.; Augustine, A. Leaf tissue specific transcriptome sequence and de novo assembly datasets of Asiatic mangrove Rhizophora mucronata Lam. Data Brief. 2020, 31, 105747. [Google Scholar] [CrossRef] [PubMed]
- Vosloo, S.; Huo, L.; Anderson, C.L.; Dai, Z.; Sevillano, M.; Pinto, A. Evaluating de Novo Assembly and Binning Strategies for Time Series Drinking Water Metagenomes. Microbiol. Spectr. 2021, 9, e0143421. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Meng, X.; Liu, S.; Zhou, P.; Zeng, C.; Fu, C.; Dou, Q.; Wu, A.; Li, C. Gut Microbiota Composition Associated with Clostridium difficile-Positive Diarrhea and C. difficile Type in ICU Patients. Front. Cell Infect. Microbiol. 2020, 10, 190. [Google Scholar] [CrossRef]
- Lu, J.; Huang, R.; Peng, Y.; Wang, H.; Feng, Z.; Fan, Y.; Zeng, Z.; Wang, Y.; Wei, J.; Wang, Z. Effects of DISC1 on Alzheimer’s disease cell models assessed by iTRAQ proteomics analysis. Biosci. Rep. 2022, 42, BSR20211150. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Tang, N.; Lei, H.; Fang, Q.; Liu, L.; Zhou, Q.; Song, C. Metagenomic Analysis of Antibiotic Resistance Genes in Untreated Wastewater from Three Different Hospitals. Front. Microbiol. 2021, 12, 709051. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Kakuta, M.; Hasegawa, T.; Yamaguchi, R.; Uchino, E.; Murashita, K.; Nakaji, S.; Imoto, S.; Yanagita, M.; Okuno, Y. Metagenomic profiling of gut microbiome in early chronic kidney disease. Nephrol. Dial. Transplant. 2021, 36, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.R.; Androga, G.O.; Ballard, S.A.; Howden, B.P.; Riley, T.V. A Phenotypically Silent vanB2 Operon Carried on a Tn1549-Like Element in Clostridium difficile. mSphere 2016, 1, e00177-16. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, Q.; Lou, S.; Tu, J.; Yin, W.; Li, X.; Jin, Y.; Radnaeva, L.D.; Nikitina, E.; Makhinov, A.N.; et al. Spatiotemporal distributions of sulfonamide and tetracycline resistance genes and microbial communities in the coastal areas of the Yangtze River Estuary. Ecotoxicol. Environ. Saf. 2023, 259, 115025. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Yu, P.; Ge, P.; Jiang, Y.; Xu, R.; Chen, R.; Liu, X. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int. J. Mol. Med. 2017, 39, 364–372. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, Z.; Wang, S.; Song, L. Occurrence and prevalence of antibiotic resistance genes and pathogens in an industrial park wastewater treatment plant. Sci. Total Environ. 2023, 880, 163278. [Google Scholar] [CrossRef]
- Gan, W.C.; Ng, H.F.; Ngeow, Y.F. Mechanisms of Linezolid Resistance in Mycobacteria. Pharmaceuticals 2023, 16, 784. [Google Scholar] [CrossRef]
- Zhitkovich, A. N-Acetylcysteine: Antioxidant, Aldehyde Scavenger, and More. Chem. Res. Toxicol. 2019, 32, 1318–1319. [Google Scholar] [CrossRef]
- Yi, D.; Hou, Y.; Tan, L.; Liao, M.; Xie, J.; Wang, L.; Ding, B.; Yang, Y.; Gong, J. N-acetylcysteine improves the growth performance and intestinal function in the heat-stressed broilers. Anim. Feed. Sci. Technol. 2016, 220, 83–92. [Google Scholar] [CrossRef]
- Yi, D.; Hou, Y.; Xiao, H.; Wang, L.; Zhang, Y.; Chen, H.; Wu, T.; Ding, B.; Hu, C.-A.A.; Wu, G. N-Acetylcysteine improves intestinal function in lipopolysaccharides-challenged piglets through multiple signaling pathways. Amino Acids 2017, 49, 1915–1929. [Google Scholar] [CrossRef]
- Yi, D.; Hou, Y.; Wang, L.; Long, M.; Hu, S.; Mei, H.; Yan, L.; Hu, C.-A.A.; Wu, G. N-acetylcysteine stimulates protein synthesis in enterocytes independently of glutathione synthesis. Amino Acids 2016, 48, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lin, H.; Lin, W.; Xu, X. Exercise Ameliorates Insulin Resistance of Type 2 Diabetes through Motivating Short-Chain Fatty Acid-Mediated Skeletal Muscle Cell Autophagy. Biology 2020, 9, 203. [Google Scholar] [CrossRef]
- Luo, Z.; Xu, X.; Sho, T.; Luo, W.; Zhang, J.; Xu, W.; Yao, J.; Xu, J. Effects of n-acetyl-cysteine supplementation in late gestational diet on maternal-placental redox status, placental NLRP3 inflammasome, and fecal microbiota in sows. J. Anim. Sci. 2019, 97, 1757–1771. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr. Opin. Microbiol. 2007, 10, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.J.; Raskin, L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 2012, 7, e43093. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kang, M.J.; Lee, S.Y.; Lee, E.; Kim, K.; Won, S.; Suh, D.I.; Kim, K.W.; Sheen, Y.H.; Ahn, K.; et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J. Allergy Clin. Immunol. 2018, 141, 1310–1319. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Ma, S.; Franzosa, E.A.; Vatanen, T.; Morgan, X.C.; Huttenhower, C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017, 18, 228. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Yuan, L.; Deng, J.; Yang, Q. Lactobacillus protects the integrity of intestinal epithelial barrier damaged by pathogenic bacteria. Front. Cell. Infect. Microbiol. 2015, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Yan, F.; Cao, H.; Cover, T.L.; Whitehead, R.; Washington, M.K.; Polk, D.B. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 2007, 132, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Dell’Anno, M.; Callegari, M.L.; Reggi, S.; Caprarulo, V.; Giromini, C.; Spalletta, A.; Coranelli, S.; Rossi, C.A.S.; Rossi, L. Lactobacillus plantarum and Lactobacillus reuteri as Functional Feed Additives to Prevent Diarrhoea in Weaned Piglets. Animals 2021, 11, 1766. [Google Scholar] [CrossRef]
- Yang, J.; Wang, C.; Liu, L.; Zhang, M. Lactobacillus reuteri KT260178 Supplementation Reduced Morbidity of Piglets Through Its Targeted Colonization, Improvement of Cecal Microbiota Profile, and Immune Functions. Probiotics Antimicrob. Proteins 2020, 12, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, Q.; Wang, J.; Qiu, X.; Qi, R.; Huang, J. Identification of differentially expressed miRNAs after Lactobacillus reuteri treatment in the ileum mucosa of piglets. Genes. Genom. 2020, 42, 1327–1338. [Google Scholar] [CrossRef]
- Xie, W.; Song, L.; Wang, X.; Xu, Y.; Liu, Z.; Zhao, D.; Wang, S.; Fan, X.; Wang, Z.; Gao, C.; et al. A bovine lactoferricin-lactoferrampin-encoding Lactobacillus reuteri CO21 regulates the intestinal mucosal immunity and enhances the protection of piglets against enterotoxigenic Escherichia coli K88 challenge. Gut Microbes 2021, 13, 1956281. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.A.; Marathe, N.P.; Lanjekar, V.; Ranade, D.; Shouche, Y.S. Comparative genome analysis of Megasphaera sp. reveals niche specialization and its potential role in the human gut. PLoS ONE 2013, 8, e79353. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.J. Role of colonic short-chain fatty acid transport in diarrhea. Annu. Rev. Physiol. 2010, 72, 297–313. [Google Scholar] [CrossRef] [PubMed]
- Bachem, A.; Makhlouf, C.; Binger, K.J.; de Souza, D.P.; Tull, D.; Hochheiser, K.; Whitney, P.G.; Fernandez-Ruiz, D.; Dähling, S.; Kastenmüller, W.; et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity 2019, 51, 285–297.e285. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Sedighi, R.; Alipour, D. Assessment of probiotic effects of isolated Megasphaeraelsdenii strains in Mehraban sheep and Holstein lactating cows. Anim. Feed. Sci. Technol. 2019, 248, 126–131. [Google Scholar] [CrossRef]
- Yu, C.; Zhou, B.; Xia, X.; Chen, S.; Deng, Y.; Wang, Y.; Wu, L.; Tian, Y.; Zhao, B.; Xu, H.; et al. Prevotellacopri is associated with carboplatin-induced gut toxicity. Cell Death Dis. 2019, 10, 714. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, N.; Chassaing, B.; Nahori, M.A.; de Bodt, J.; Moura, A.; Lecuit, M.; Dussurget, O.; Bérard, M.; Marzorati, M.; Fehlner-Peach, H.; et al. A Listeria monocytogenes Bacteriocin Can Target the CommensalPrevotellacopri and Modulate Intestinal Infection. Cell Host Microbe 2019, 26, 691–701.e695. [Google Scholar] [CrossRef] [PubMed]
- Hofer, U. Microbiome: Anelloviridae go viral. Nat. Rev. Microbiol. 2014, 12, 4–5. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).