Abstract
An increasing number of patients in Africa are experiencing virological failure on a second-line antiretroviral protease inhibitor (PI)-containing regimen, even without resistance-associated mutations in the protease region, suggesting a potential role of other genes in PI resistance. Here, we investigated the prevalence of mutations associated with Lopinavir/Ritonavir (LPV/r) failure in the Envelope gene and the possible coevolution with mutations within the Gag-protease (gag-PR) region. Env and Gag-PR sequences generated from 24 HIV-1 subtype C infected patients failing an LPV/r inclusive treatment regimen and 344 subtype C drug-naïve isolates downloaded from the Los Alamos Database were analyzed. Fisher’s exact test was used to determine the differences in mutation frequency. Bayesian network probability was applied to determine the relationship between mutations occurring within the env and gag-PR regions and LPV/r treatment. Thirty-five mutations in the env region had significantly higher frequencies in LPV/r-treated patients. A combination of Env and Gag-PR mutations was associated with a potential pathway to LPV/r resistance. While Env mutations were not directly associated with LPV/r resistance, they may exert pressure through the Gag and minor PR mutation pathways. Further investigations using site-directed mutagenesis are needed to determine the impact of Env mutations alone and in combination with Gag-PR mutations on viral fitness and LPV/r efficacy.
1. Introduction
While the dispensing of HIV-1 protease inhibitors (PIs) as part of a routine second-line regimen has been a significant turning point in the management of HIV-1 [1], the development of resistance to PIs is also increasing [2].
PIs are known to inhibit the activity of the HIV-1 protease (PR) enzyme responsible for the proteolytic processing of HIV structural Gag and enzymatic Pol polyprotein components [3]. The viral protease (PR) enzyme cleaves the Gag precursor protein (Pr55Gag) and the Gag-Pol precursor protein (Pr160Gag-Pol), resulting in virion maturation [3,4,5]. This proteolytic process prevents the formation and maturation of infectious HIV particles. The efficacy of the PI is limited by the emergence of resistance mutations that are potentially caused by poor compliance, subtherapeutic systemic levels of the drug, or prolonged treatment with one PI-based regimen during virologic failure [6].
Interestingly, >20% of patients failing a PI-based regimen do not harbor any resistance-associated mutations in the protease (PR) domain [7,8,9]. While non-adherence has been identified as a key player, some studies have suggested that Gag mutations can indirectly affect the Gag cleavage site and drive PI resistance without any mutations in the protease region [10,11]. These mutations were commonly found close to the cleavage sites, although non-cleavage site mutations have also been linked with PI resistance [12,13]. Verheyen et al. (2006) have linked Gag precursor p7–p1 (A431V, K436R, and I437V) and p1–p6 (L449P, P452S, and P453L) cleavage site mutations with resistance to PIs [14,15,16]. These mutations are thought to compensate for any enzymatic impairment of protease caused by the loss of van der Waals interactions between the inhibitor and binding sites [12,14,15,17,18,19,20].
Others have suggested that the mechanism for PI failure may involve mutations in the Envelope (env) gene [5,21]. Coetzer et al. (2017) reported Gp41 mutations from both the Heptad repeat (HR) (607T and 641L) and cytoplasmic tail (CT) (721I) that potentially contribute to PI failure [21] Two studies that looked at virological failure during PI exposure both reported that mutations in the CT impacted PI susceptibility [22,23]. Furthermore, Env mutations have been suggested to promote cell to cell transmission, leading to high-level drug resistance mutations in ARV target genes; by doing so, they increase the level of resistance to a broad panel of ARVs in vitro [24,25]. The HIV-1 Envelope is said to concurrently evolve to escape from both neutralizing antibodies (NABs) and ARVs [25].
Although studies have reported the emergence of Gag and Env mutations in patients failing PIs [21,22,23], to date, no one has reported on the coevolution of Gag and Env in these patients. Here, we investigated the prevalence of Env mutations associated with PI treatment failure and the possibility of coevolution with mutations within Gag, and protease, in full-length Env and Gag-PR sequences from HIV-1 subtype C infected individuals failing a PI-inclusive regimen.
2. Materials and Methods
2.1. Study Cohort
This retrospective study used 24 stored plasma samples obtained from virologically failing PI-treated patients enrolled in the Protease Cleavage Site (PCS) study (2009–2013) at McCord and King Edward VIII hospitals Durban, South Africa [26]. All enrolled patients received Lopinavir/Ritonavir therapy for at least six months and had plasma HIV-1 RNA levels > 1000 copies/mL. In addition, sequences from 344 subtype C drug-naïve isolates were downloaded from the Los Alamos HIV-1 Database (http://hiv-web.lanl.gov) accessed on 10 September 2020. Written informed consent was obtained from all study participants. The Biomedical Research Ethics Committee of the University of KwaZulu-Natal (BREC NO: 678/17) approved the study.
2.2. Amplification and Sequencing Analyses of the Env Domain
Viral RNAs were extracted from 140 µL of plasma using the QIAamp RNA kit (QIAGEN Services, Inc., Germantown, MD, USA) and reverse-transcribed using a Thermoscript RT-PCR kit (Invitrogen, Carlsbad, CA, USA). A 1.7 kb Gag-Protease product was amplified by nested PCR using the Takara Ex Taq HS enzyme kit. Specific primers, Gag +1 5′-GAGATCTCTCGACGCAGGAC-3′ (HXB2 nucleotide: 675 to 697, forward primer) and 3″rvp 5′-GGAGTGTTATat GGATTTTCAGGCCCAATT-3′ (HXB2 positions: 2696 to 2725, reverse primer), were used for the first-round PCR, at 55 °C for 30 min (cDNA synthesis) and 94 °C for 2 min (initial denaturation), followed by 35 cycles of 94 °C for 15 s (denaturation), 55 °C for 30 s (annealing) and 68 °C for 2 min (extension), and ended with a 5 min incubation at 68 °C (final extension). Long fwd 5′-GAC TCG GCT TGC TGA AGC GCG CAC GGC AAG AGG CGA GGG GCG ACT GGT GAGTAC GCC AAA AAT TTT GAC TAG CGG AGG CTA GAA GGA GAGAGA TGG G-3′ (HXB2 nucleotide: 695 to 794, forward primer) and Long rev 5′-GGC CCA ATT TTT GAA ATT TTT CCT TCC TTT TCC ATT TCT GTA CAA ATT TCT ACT AAT GCT TTT ATT TTT TCT GTC AAT GGC CAT TGT TTA ACT TTT G-3′ (HXB2 nucleotide: 2706 to 2805, reverse primer) were used for nested PCR at the following conditions: 94 °C for 2 min (initial denaturation), 40 cycles of 94 °C for 30 s (denaturation), 60 °C for 30 s (annealing) and 72 °C for 2 min (extension) followed by a 7 min hold at 72 °C (final extension). Primers were complementary to subtype B, gag-protease-deleted NL4-3 plasmid (pNL4-3Δgag-protease) on either side. Amplicons were electrophoresed in a 1% agarose gel to confirm the presence of the 1.7 kb amplicon corresponding to the gag-PR gene. The size of the product was determined using the GeneRuler™ 1 kb DNA Ladder (Fermentas International Inc., Waltham, MA, USA), as shown in Supplementary Figure S1.
Next, we amplified an Env fragment spanning HXB2 positions 5580 to 8586 by nested PCR using first-round primers Rec2F 5′-GATAAAGCCACCTTTGCCTAGT-3′ (HXB2 positions: 5514) and Env2 5′-TTCTAGGTCTCGAGATACTGCT-3′ (HXB2 positions: 8889), second-round primers Env1 5′-AAGGGCCACAGAGGGAGCCATA-3′ (HXB2 positions: 5580) and E270R, 5′-GCGTCCCAGAAGTTCCACAA-3′ (HXB2 positions: 8566) and PCR conditions as previously described [27]. Sequences were generated using the Gag-Protease sequencing primers 5′-CTT GTC TAG GGC TTC CTT GGT-3′ (HXB2 position: 1078–1098), 5′-CTT CAG ACA GGA ACA GAGGA-3′ (991–1010), 5′-GGT TCT CTC ATC TGG CCT GG-3′ (1462–1481), 5′-CCT TGC CAC AGT TGA AAC ATT T-3′ (1960–1981), 5′-TAG AAG AAA TGA TGA CAG-3′ (1817–1834), 5′-CTA ATA CTGTatCAT CTG CTC CTG T-3′ (2328–2353), 5′-CCT GGC TTT AAT TTT TAC TGG-3′ (2196–2268). Cycling conditions were as follows: 94 °C for 2 min (initial denaturation), 35 cycles of 94 °C for 15 s (denaturation), 55 °C for 30 s (annealing) and 68 °C for 4 min (extension) followed by a 10 min hold at 68 °C (final extension). Amplification of the env region was confirmed by gel electrophoresis (shown in Supplementary Figure S2)
The gp160 Env was sequenced using population-based forward and reverse primers (shown in Supplementary Table S1) and the BigDye v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA), and run on an Applied Biosystems (ABI) 3130xl automated sequencer. Sequences were assembled and edited using the Applied Biosystems SeqScape® Software and aligned against subtype C reference.
2.3. Statistical Analysis
Multiple sequence alignments were generated using the ClustalX program (http://www.clustal.org/) accessed on 16 September 2020, and manually edited using BIOEDIT (Ibis Biosciences, An Abbott Company, Sylmar, CA, USA). The Virulign tool (https://github.com/rega-cev/virulign) accessed on 11 December 2020, was used to determine the amino acid substitution at each position in Gag-PR and Env on PI exposure and naïve isolate sequences. Fisher’s exact test was performed for all positions to identify mutations significantly more prevalent (p-value < 0.05) in PI-experienced patients.
2.4. Bayesian Network
Bayesian network learning was performed using B-Course (http:/www.b-course) accessed on May 2021, which modeled direct and indirect associations between Gag, PR, and Env mutations and exposure to PI treatment. Only significant mutations identified using Fisher’s exact test were included in the model together with known PI-resistance-associated mutations. As previously described, associations between mutations learned and graphical representations were evaluated [28,29]. Non-parametric bootstrap analysis was calculated by running 5000 replicates to derive network robustness. Only interactions with bootstrap support over 65% were included in the network.
2.5. Coevolution Using CAPS
Gag-PR-Env coevolution analysis was performed using the Coevolution Analysis for Protein Sequences (CAPS, http://bioinf.gen.tcd.ie/caps/) accessed on 26 October 2020 program. CAPS identifies groups of coevolving pairs with a correlation coefficient > 0.5 by using amino acid (AA) sites to compare the correlated variance of their evolutionary rate using the probability scores between two pairs of aligned sequences [30]. The evolutionary rate was estimated using the blocks substitution matrix (BLOSUM). The distribution of 5000 randomly sampled values was used to identify coevolving codons.
2.6. Positive Selection
Positive selection pressure in Gag-PR-Env sites was assessed using the rate ratio of non-synonymous to synonymous (ω) substitutions in the CODEML program of the PAML software package (version 4.9) [31]. Analysis was performed using three classes ω ≤ 1, ω = 1, ω ≥ 1 assuming purifying, neutral, and positive selection, respectively. The ω value and likelihood estimates were calculated for three different codon-based ML pairs of site models: M0 (one ω) vs. M3 (discrete), M1a (nearly neutral) vs. M2a (positive selection), and M7 (beta) vs. M8 (beta and ω ≥ 1). Comparison of M0 vs. M3 is a test of site rate variation, while M1 vs. M2 is for positive selection. The likelihood ratio test was used to evaluate the best-fitting model for the data [32]. The Bayes Empirical Bayes method was used to identify specific sites under positive selection.
3. Results
3.1. Participant Characteristics
Twenty-four Gag-Prot-Env sequences from 24 patients failing a PI-inclusive treatment regimen were available for analysis, of whom 14 were females and 10 were males, with the median age of 35 years (interquartile range (IQR) 17–38 years). The median viral load was 4.84 log10 copies/mL (IQR 4.12–5.51). All patients were infected with HIV subtype C as established by the Virulign tool.
3.2. Prevalence of Envelope Mutations in LPV/r-Experienced versus ART-Naïve Subtype C Sequences
Twenty-four Env sequences were available for analysis. The prevalence of amino acids at each codon position was calculated by comparing sequences from LPV/r-treated and ART-naïve isolates. All sequences were confirmed as subtype C using the Rega subtyping tool. Figure 1 shows a total of thirty-five mutations identified in the env gene, with significantly higher frequencies in LPV/r-experienced patients compared to those that were ART-naïve. These were: I19LSV (32% vs. 13%), S110EN (16% vs. 4%), D167AGKNR (16% vs. 2%), P183AEKQRS (48% vs. 20%), Y191CFHNS (12% vs. 1%), N195HIKRST (28% vs. 8%), N230DT (20% vs. 16%), Q258EP (8% vs. 1%), N262LI (8% vs. 0%), T283IKNQSV (28% vs. 9%), V286AIM (16% vs. 3%), I294LMN (8% vs. 0%), S306DGNR (24% vs. 5%), P313DI (8% vs. 0%), Q315AKR (32% vs. 3%), I323KNRTV (12% vs. 1%), A329DIPT (16% vs. 0%), C331RS (8% vs. 0%), W338EG (8% vs. 0%), H374KLPR (8% vs. 1%), F382V (8% vs. 1%), F383LV (8% vs. 0%), L390QV (8% vs. 0%), N425DHR (20% vs. 4%), L453EIM (20% vs. 4%), R469KTW (8% vs. 1%), V489IL (16% vs. 1%), V505AEMT (8% vs. 1%), E507GQRW (28% vs. 10%), S534AL (12% vs. 2%), T536AMSV (32% vs. 10%), Q567KR (8% vs. 1%), D632EG (24% vs. 8%), I688TV (12% vs. 1%), P724LQRSX (32% vs. 12%). In contrast, the K63RT (16% vs. 50%), T132NQ (24% vs. 97%), T138NQ (36% vs. 80%), and V182EIKLMNT (12% vs. 56%) mutations were found at a lower frequency in the PI-experienced versus ART-naïve sequences.
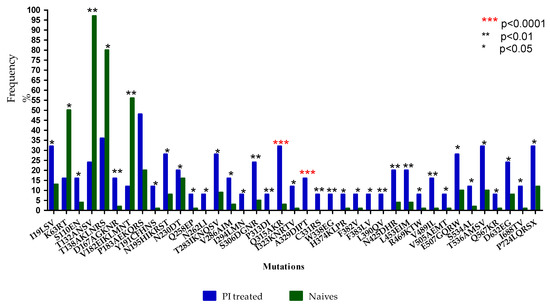
Figure 1.
Frequency (%) of Env mutations in ART-naïve versus LPV/r-treated subtype C isolates. Asterisks (*): *** represents p < 0.0001, ** represents p < 0.01, and * represents p < 0.05.
3.3. Positive Selected Sites in Envelope and Coevolution with Gag-PR Residues
Here, we determined which amino acid changes were under positive selection pressure following treatment failure using PAML [30]. Figure 2 shows PAML analysis of amino acid changes from participants failing an LPV/r-inclusive treatment regimen and ART-naïve participants that were positively selected. A total of twelve sites were positively selected: V85, T138, Y146, M147, E150, K151, G152, D277, G321, K322, N463, and T465. None of these sites were previously associated with entry and fusion inhibitor or LPV/r failure.
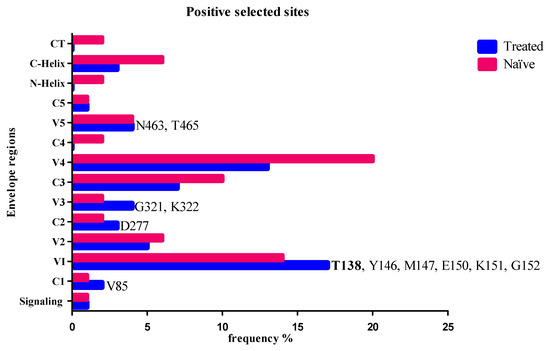
Figure 2.
Positively selected amino acid codons in Env in LPV/r-treated and naïve isolates with the Bayesian probability of p > 99%. Blue shows the positively selected codons in the treated isolates, while maroon indicates the positively selected codons in the naïve isolates. Codon positions identified only in treated and not in naïve isolates are shown at the relevant env region. T138 is indicated in bold font and has a higher frequency in the treated isolates.
Coevolution Analyses for Protein Sequences (CAPS) was used to determine whether amino acids in Gag, PR, and Env sites coevolved in patients failing an LPV/r-inclusive treatment regimen. All positions with amino acid variation greater than 1% were included in the analysis. The coevolution analysis identified seven amino acid coevolving pairs, but these did not correlate significantly. The coevolving pairs were mostly seen between Env and Gag sites (Env: 11, 23, 29,142, 147, 336, 389, 400, 496, 534, 617, and 668; Gag: 108, 225, 256, 335, 372, 388, 478, and 480), while only one was observed between Env and PR (400-Env and 76-PR) sites.
3.4. Interactions between Envelope and Gag-PR Mutations with LPV/r Treatment
Using Bayesian network learning, we explored the interactions between Gag, PR, and Env mutations in isolates exposed to LPV/r treatment. Only statistically significant positions according to Fisher’s exact test were included in the analysis. Figure 3A shows mutations in the env Gp120 region (S110N-Env, T132S-Env, T138S-Env, P183Q-Env, and Q315R-Env) that were indirectly associated with exposure to LPV/r treatment via known Gag-PR mutations (Q69K-Gag, S111I-Gag, I256V-Gag, and V77I-PR). More specifically, P183Q-Env was indirectly associated with LPV/r treatment via interaction with V77I-PR. T132S-Env also showed a robust connection with Q69K-Gag mutation, which is further related to K20R-PR mutation. Both T138S-Env and S110N-Env showed a strong association with I256V-Gag. Interestingly, a robust direct connection with LPV/r exposure was observed between a known Gag-PR mutation (R76K-Gag) and wildtype variants at codon positions in Gag (Q182-Gag-PR-WT) and PR that have been linked to drug resistance (I50-PR-WT, V77-PR-WT, and L90-PR-WT).
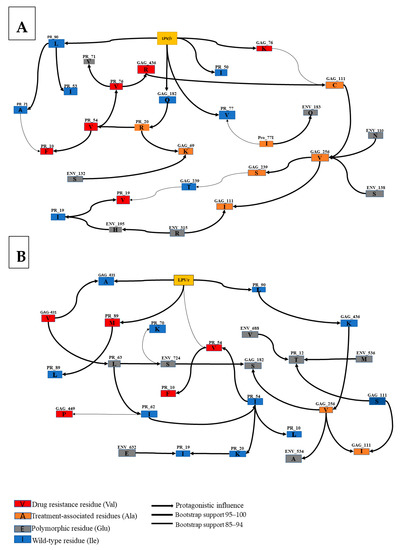
Figure 3.
Annotated Bayesian network based on 5000 bootstraps, showing the association between nodes indicating Env and Gag mutations in subtype C LPV/r-treated sequences. Here, we determined the association between (A) Env Gp120, Gag, PR mutations, and treatment experience and (B) Gp41, Gag, PR, and treatment experience. The color code of nodes is defined based on their link with LPV/r: drug resistance variants (red), treatment associated variants (orange) and natural occurring: Polymorphic variants (gray), wild-type variants (blue). The arcs represent a direct dependency between corresponding nodes, and the thickness of an arc is in proportion to bootstrap support. The arc direction does not represent the accumulation of mutations or causal meaning but may indicate a multivariable effect in the network.
Figure 3B shows the association between Gag, PR, Env Gp41 mutations, and LPV/r treatment experience. There was a strong interaction between T536M-Env, S543A-Env, I688V-Env, and P724S-Env and Gag-PR mutations (S12T-PR, P63L-PR, Q182S-Gag, and I256V-Gag). Specifically, P724S-Env was directly associated with P63L-PR and Q182S-Gag and indirectly associated with A431V found at a Gag cleavage site. Another direct association was seen between S534A-Env and I256V-Gag. Both T536M-Env and I688V-Env were directly associated with S12T-PR. There was no direct interaction between treatment experience and Env Gp41 mutations.
4. Discussion
Mutations in the env gene have previously been associated with resistance to PIs [5]; however, no studies have been conducted using subtype C PI failures, nor have they investigated their role in combination with the Gag. In this study, LPV/r-resistance-associated mutations in Gag, PR, and Env were characterized in HIV-1 subtype C patients from KZN, South Africa, who were failing an LPV/r-inclusive treatment regimen. We identified several amino acids in the env gene that were associated with LPV/r failure. We also found potential pathways leading to LPV/r resistance that involved a combination of Gag, PR, and Env mutations.
Significantly higher frequencies of AA changes in the GP120 region of the LPV/r-experienced group were commonly identified. These were mostly in the variable loops (V2: D167AR, V182I, P183QS, Y191F and N195H, and V3: S306R, P313DI, Q315R, I323KNR, A329DT, and C331RS); however, increased frequencies were also observed in the constant regions (C2: N230DT, Q258EP, N262LI, T283I, V286I and I294N, and C3: W338EG, H374KR, F382V, and F383LV). Amino acid changes in these regions can, therefore, potentially increase the infectivity and replication capacity of the virus [33].
In GP41, sequences from LPV/r failures harbored significantly higher frequencies of Heptad repeat (HR) mutations at codons 534, 536, 567, and 632. Consistent with our findings, an HIV-1 subtype A study also observed higher levels of mutations in HR in LPV/r failures [21]. Furthermore, higher mutation frequencies were also seen in the cytoplasmic tail (CT); this is in line with other studies that reported CT mutations associated with virological failure in PI-experienced participants [21,22,23]. As the CT plays a functional role in Env incorporation during virion assembly, this suggests that mutations in this region may influence the efficiency of Env incorporation [34].
Using Bayesian network learning, we found potential pathways leading to LPV/r resistance that involved a combination of Env and Gag mutations. I256V appeared to be an essential mutation as it was shown to be directly associated with three Env mutations (S110N-Env, T138S-Env, and S543A-Env) and indirectly associated with Q315R-Env via S111I-Gag and P724S-Env via Q182S-Gag. Although less is known about the I256V-Gag mutation, it has been previously associated with LPV/r failure in subtype C studies [35,36]. It has also been associated with reducing drug susceptibility to benzodiazepine and benzimidazole in Gag subtype B studies [37]. Mutations T138S-Env and S110N-Env showed a strong interaction with I256V-Gag, forming a pathway (T138S-Env + S110N-Env + I256V + S111C + R76K) to LPV/r exposure. A study by Singh et al. (2015) identified I256V, S111C, and R76K in Gag following LPV/r treatment in subtype C patients in South Africa [35]. Interestingly, T138 was also positively selected in our PAML analysis. Although a wildtype amino acid, T138, is known to play an essential role in antibody binding, and although the change from Threonine (T) to Serine (S) weakened the binding, T138S did not inhibit antibody binding [38].
Another potential pathway for LPV/r resistance included Env mutations Q315R-Env and the same Gag mutations I256V, S111C, and R76K, with the addition of S111I. While Q315R has been identified as a naturally occurring polymorphism in subtype A [39], its role as a drug-resistance-associated mutation in subtype C requires more mechanistic studies. Mutation T132S-Env showed a robust connection with Q69K-Gag, which has previously been associated with reduced susceptibility to PIs [40].
Interestingly, known PI-resistance-associated mutations K20R-PR, I54V-PR, L10F-PR, and L76V-PR were not directly connected to LPV/r exposure in the network, suggesting that immune selection pressure may also be in play.
While not directly associated with LPV/r resistance, Env Gp41 (S543A-Env and P724S-Env) mutations were associated with Gag (Q182S-Gag and I256V-Gag) mutations. Env mutation P724S was strongly associated with Q182S-Gag, which was further associated with I256V. S534A-Env was also directly associated with I256V-Gag, further supporting the role of I256V-Gag in connecting Env in the pathway to drug resistance. Interestingly, most of the mutations in Gag connected to Env mutations were located in the capsid domain (such as I256V), which is similar to other studies [21].
Another potential network to LPV/r resistance involved mutations found in the env in combination with the gag and PR region. Env mutation P183Q-Env was directly associated with V77I-PR, previously associated with PI drug resistance [41,42,43]. In addition, Env mutations T536M and I688V showed a strong direct association with PR mutation S12T-PR, which is known to be selected in LPV/r- and RTV-treated patients in subtype C [40]. Interestingly, the P724S Env mutation was linked to LPV/r failure via the P63L-PR and A431V-Gag pathways. The A431V Gag mutation located in the CS of Gag is known to confer resistance to all PIs except Darunavir (DRV) [15], while P63L-PR was observed at baseline in patients initiating antiretroviral therapy (Lopinavir/r, Lamivudine, and Zidovudine) [15]. These findings suggest that there might be other pathways to LPV/r treatment failure that involve Gag (Matrix and Capsid), minor PR, and Env mutations.
5. Conclusions
We found a high prevalence of Env mutations in HIV-1 subtype C associated with LPV/r treatment failure. The majority of these associations were in combination with a Gag (Matrix and Capsid) and/or PR mutation. Further investigations using site-directed mutagenesis need to be conducted to determine whether Env mutations alone can affect LPV/r efficacy.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microbiolres12040071/s1, Figure S1: Gel picture, showing the amplification of Gag-PR. Gag-PR size 1.7 kb. Figure S2: Gel picture showing the amplification of envelope. Envelope size 3.5 kb. Table S1: Sequencing Primers.
Author Contributions
Conceptualization, N.F.M., M.L.G.; data curation, N.F.M., M.L.G.; formal analysis, N.F.M.; funding acquisition, M.L.G.; investigation, N.F.M.; methodology, N.F.M.; resources, M.L.G.; writing of the manuscript, N.F.M., M.L.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Funding, Poliomyelitis Research Foundation, and the UKZN College of Health Sciences.
Institutional Review Board Statement
This study was approved by the University of KwaZulu-Natal Biomedical Research Ethics Committee (BREC NO. 678/17).
Informed Consent Statement
Consent was not applicable since the study was conducted using the stored plasma samples.
Data Availability Statement
Data are contained within the article and the Supplementary Material; further information can be obtained from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ghosh, A.K.; Chapsal, B.D.; Weber, I.T.; Mitsuya, H. Design of HIV protease inhibitors targeting protein backbone: An effective strategy for combating drug resistance. Acc. Chem. Res. 2008, 41, 78–86. [Google Scholar] [CrossRef]
- Kim, R.; Baxter, J.D. Protease inhibitor resistance update: Where are we now? AIDS Patient Care STDs 2008, 22, 267–277. [Google Scholar] [CrossRef]
- Pettit, S.C.; Moody, M.D.; Wehbie, R.S.; Kaplan, A.H.; Nantermet, P.V.; Klein, C.A.; Swanstrom, R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994, 68, 8017–8027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lori, F.; Scovassi, A.I.; Zella, D.; Achilli, G.; Cattaneo, E.; Casoli, C.; Bertazzoni, U. Enzymatically active forms of reverse transcriptase of the human immunodeficiency virus. AIDS Res. Hum. Retrovir. 1988, 4, 393–398. [Google Scholar] [CrossRef]
- Rabi, S.A.; Laird, G.M.; Durand, C.M.; Laskey, S.; Shan, L.; Bailey, J.R.; Chioma, S.; Moore, R.D.; Siliciano, R.F. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J. Clin. Investig. 2013, 123, 3848–3860. [Google Scholar] [CrossRef] [PubMed]
- Zdanowicz, M.M. The pharmacology of HIV drug resistance. Am. J. Pharm. Educ. 2006, 70, 100. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O.; de Mendoza, C.; Pérez-Elías, M.J.; Guardiola, J.M.; Pedreira, J.; Dalmau, D.; Gónzalez, J.; Moreno, A.; Arribas, J.R.; Rubio, A. Drug resistance in patients experiencing early virological failure under a triple combination including indinavir. Aids 2001, 15, 1701–1706. [Google Scholar] [CrossRef]
- Wallis, C.L.; Mellors, J.W.; Venter, W.D.; Sanne, I.; Stevens, W. Protease inhibitor resistance is uncommon in HIV-1 subtype C infected patients on failing second-line lopinavir/r-containing antiretroviral therapy in South Africa. AIDS Res. Treat. 2011, 2011, 769627. [Google Scholar] [CrossRef] [Green Version]
- Obasa, A.E.; Mikasi, S.G.; Brado, D.; Cloete, R.; Singh, K.; Neogi, U.; Jacobs, G.B. Drug resistance mutations against protease, reverse transcriptase and integrase inhibitors in people living with HIV-1 receiving boosted protease inhibitors in South Africa. Front. Microbiol. 2020, 11, 438. [Google Scholar] [CrossRef] [Green Version]
- Parry, C.M.; Kolli, M.; Myers, R.E.; Cane, P.A.; Schiffer, C.; Pillay, D. Three residues in HIV-1 matrix contribute to protease inhibitor susceptibility and replication capacity. Antimicrob. Agents Chemother. 2011, 55, 1106–1113. [Google Scholar] [CrossRef] [Green Version]
- Alfadhli, A.; Mack, A.; Ritchie, C.; Cylinder, I.; Harper, L.; Tedbury, P.R.; Freed, E.O.; Barklis, E. Trimer Enhancement Mutation Effects on HIV-1 Matrix Protein Binding Activities. J. Virol. 2016, 90, 5657–5664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fun, A.; Wensing, A.M.; Verheyen, J.; Nijhuis, M. Human Immunodeficiency Virus Gag and protease: Partners in resistance. Retrovirology 2012, 9, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codoñer, F.M.; Peña, R.; Blanch-Lombarte, O.; Jimenez-Moyano, E.; Pino, M.; Vollbrecht, T.; Clotet, B.; Martinez-Picado, J.; Draenert, R.; Prado, J.G. Gag-protease coevolution analyses define novel structural surfaces in the HIV-1 matrix and capsid involved in resistance to Protease Inhibitors. Sci. Rep. 2017, 7, 3717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malet, I.; Roquebert, B.; Dalban, C.; Wirden, M.; Amellal, B.; Agher, R.; Simon, A.; Katlama, C.; Costagliola, D.; Calvez, V. Association of Gag cleavage sites to protease mutations and to virological response in HIV-1 treated patients. J. Infect. 2007, 54, 367–374. [Google Scholar] [CrossRef]
- Nijhuis, M.; Van Maarseveen, N.M.; Lastere, S.; Schipper, P.; Coakley, E.; Glass, B.; Rovenska, M.; De Jong, D.; Chappey, C.; Goedegebuure, I.W. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007, 4, e36. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, J.; Litau, E.; Sing, T.; Daumer, M.; Balduin, M.; Oette, M.; Fatkenheuer, G.; Rockstroh, J.K.; Schuldenzucker, U.; Hoffmann, D. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir. Ther. 2006, 11, 879. [Google Scholar]
- Kletenkov, K.; Hoffmann, D.; Böni, J.; Yerly, S.; Aubert, V.; Schöni-Affolter, F.; Struck, D.; Verheyen, J.; Klimkait, T.; Study, S.H.C. Role of Gag mutations in PI resistance in the Swiss HIV cohort study: Bystanders or contributors? J. Antimicrob. Chemother. 2017, 72, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Larrouy, L.; Chazallon, C.; Landman, R.; Capitant, C.; Peytavin, G.; Collin, G.; Charpentier, C.; Storto, A.; Pialoux, G.; Katlama, C. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 2010, 54, 2910–2919. [Google Scholar] [CrossRef] [Green Version]
- Gatanaga, H.; Suzuki, Y.; Tsang, H.; Yoshimura, K.; Kavlick, M.F.; Nagashima, K.; Gorelick, R.J.; Mardy, S.; Tang, C.; Summers, M.F. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 2002, 277, 5952–5961. [Google Scholar] [CrossRef] [Green Version]
- Kožíšek, M.; Henke, S.; Šašková, K.G.; Jacobs, G.B.; Schuch, A.; Buchholz, B.; Müller, V.; Kräusslich, H.-G.; Řezáčová, P.; Konvalinka, J. Mutations in HIV-1 gag and pol compensate for the loss of viral fitness caused by a highly mutated protease. Antimicrob. Agents Chemother. 2012, 56, 4320–4330. [Google Scholar] [CrossRef] [Green Version]
- Coetzer, M.; Ledingham, L.; Diero, L.; Kemboi, E.; Orido, M.; Kantor, R. Gp41 and Gag amino acids linked to HIV-1 protease inhibitor-based second-line failure in HIV-1 subtype A from Western Kenya. J. Int. AIDS Soc. 2017, 20, e25024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castain, L.; Perrier, M.; Charpentier, C.; Palich, R.; Desire, N.; Wirden, M.; Descamps, D.; Sayon, S.; Landman, R.; Valantin, M.-A. New mechanisms of resistance in virological failure to protease inhibitors: Selection of non-described protease, Gag and Gp41 mutations. J. Antimicrob. Chemother. 2019, 74, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Perrier, M.; Castain, L.; Regad, L.; Todesco, E.; Landman, R.; Visseaux, B.; Yazdanpanah, Y.; Rodriguez, C.; Joly, V.; Calvez, V. HIV-1 protease, Gag and gp41 baseline substitutions associated with virological response to a PI-based regimen. J. Antimicrob. Chemother. 2019, 74, 1679–1692. [Google Scholar] [CrossRef]
- Van Duyne, R.; Kuo, L.S.; Pham, P.; Fujii, K.; Freed, E.O. Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc. Natl. Acad. Sci. USA 2019, 116, 9040–9049. [Google Scholar] [CrossRef] [Green Version]
- Hikichi, Y.; Van Duyne, R.; Pham, P.; Groebner, J.L.; Wiegand, A.; Mellors, J.W.; Kearney, M.F.; Freed, E.O. Mechanistic Analysis of the Broad Antiretroviral Resistance Conferred by HIV-1 Envelope Glycoprotein Mutations. Mbio 2021, 12, e03134-20. [Google Scholar] [CrossRef]
- Marie, V.; Gordon, M. Gag-protease coevolution shapes the outcome of lopinavir-inclusive treatment regimens in chronically infected HIV-1 subtype C patients. Bioinformatics 2019, 35, 3219–3223. [Google Scholar] [CrossRef] [PubMed]
- Rangel, H.R.; Weber, J.; Chakraborty, B.; Gutierrez, A.; Marotta, M.L.; Mirza, M.; Kiser, P.; Martinez, M.A.; Este, J.A.; Quinones-Mateu, M.E. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 2003, 77, 9069–9073. [Google Scholar] [CrossRef] [Green Version]
- Deforche, K.; Camacho, R.; Grossman, Z.; Silander, T.; Soares, M.; Moreau, Y.; Shafer, R.W.; Van Laethem, K.; Carvalho, A.; Wynhoven, B. Bayesian network analysis of resistance pathways against HIV-1 protease inhibitors. Infect. Genet. Evol. 2007, 7, 382–390. [Google Scholar] [CrossRef]
- Theys, K.; Deforche, K.; Libin, P.; Camacho, R.J.; Van Laethem, K.; Vandamme, A.-M. Resistance pathways of human immunodeficiency virus type 1 against the combination of zidovudine and lamivudine. J. Gen. Virol. 2010, 91, 1898–1908. [Google Scholar] [CrossRef]
- Fares, M.A.; Travers, S.A. A novel method for detecting intramolecular coevolution: Adding a further dimension to selective constraints analyses. Genetics 2006, 173, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, M.; Bielawski, J.P.; Yang, Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 2001, 18, 1585–1592. [Google Scholar] [CrossRef] [Green Version]
- Dang, L.V.P.; Pham, H.V.; Dinh, T.T.; Nguyen, T.H.; Vu, Q.T.H.; Vu, N.T.P.; Le, P.T.B.; Nguyen, L.V.; Le, H.T.; Vu, P.T. Characterization of envelope sequence of HIV virus in children infected with HIV in Vietnam. SAGE Open Med. 2020, 8, 2050312120937198. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Saad, J.S. Solution structure and membrane interaction of the cytoplasmic tail of HIV-1 gp41 protein. Structure 2017, 25, 1708–1718.e1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, U. Acquired and Transmitted Drug Resistance in HIV-1 Subtype C: Implications of Novel Mutations on Replication Capacity, Cleavage and Drug Susceptibility. Ph.D. Thesis, University of Kwazulu-Natal, Durban, South Africa, 2015. [Google Scholar]
- Ntale, R.S. The Role of Early Cytotoxic Lymphocyte (CTL) Escape in the Pathogenesis of HIV-1 Subtype C Infection. Ph.D. Thesis, University of Cape Town, Cape Town, South Africa, 2012. [Google Scholar]
- Li, G.; Verheyen, J.; Rhee, S.-Y.; Voet, A.; Vandamme, A.-M.; Theys, K. Functional conservation of HIV-1 Gag: Implications for rational drug design. Retrovirology 2013, 10, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyser, J.M.; Zeng, C.Q.-Y.; Beharry, Z.; Palzkill, T.; Estes, M.K. Epitope mapping and use of epitope-specific antisera to characterize the VP5⁎ binding site in rotavirus SA11 NSP4. Virology 2008, 373, 211–228. [Google Scholar] [CrossRef] [Green Version]
- Ratcliff, A.N.; Shi, W.; Arts, E.J. HIV-1 resistance to maraviroc conferred by a CD4 binding site mutation in the envelope glycoprotein gp120. J. Virol. 2013, 87, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Giandhari, J.; Basson, A.E.; Sutherland, K.; Parry, C.M.; Cane, P.A.; Coovadia, A.; Kuhn, L.; Hunt, G.; Morris, L. Contribution of Gag and protease to HIV-1 phenotypic drug resistance in pediatric patients failing protease inhibitor-based therapy. Antimicrob. Agents Chemother. 2016, 60, 2248–2256. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.M.; Perno, C.F. HIV-1 genetic variability and clinical implications. Int. Sch. Res. Not. 2013, 2013, 481314. [Google Scholar] [CrossRef] [Green Version]
- Mata-Munguía, C.; Escoto-Delgadillo, M.; Torres-Mendoza, B.; Flores-Soto, M.; Vázquez-Torres, M.; Gálvez-Gastelum, F.; Viniegra-Osorio, A.; Castillero-Manzano, M.; Vázquez-Valls, E. Natural polymorphisms and unusual mutations in HIV-1 protease with potential antiretroviral resistance: A bioinformatic analysis. BMC Bioinform. 2014, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Doualla-Bell, F.; Avalos, A.; Gaolathe, T.; Mine, M.; Gaseitsiwe, S.; Ndwapi, N.; Novitsky, V.A.; Brenner, B.; Oliveira, M.; Moisi, D. Impact of human immunodeficiency virus type 1 subtype C on drug resistance mutations in patients from Botswana failing a nelfinavir-containing regimen. Antimicrob. Agents Chemother. 2006, 50, 2210–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).