Potential Associations of Mutations within the HIV-1 Env and Gag Genes Conferring Protease Inhibitor (PI) Drug Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Amplification and Sequencing Analyses of the Env Domain
2.3. Statistical Analysis
2.4. Bayesian Network
2.5. Coevolution Using CAPS
2.6. Positive Selection
3. Results
3.1. Participant Characteristics
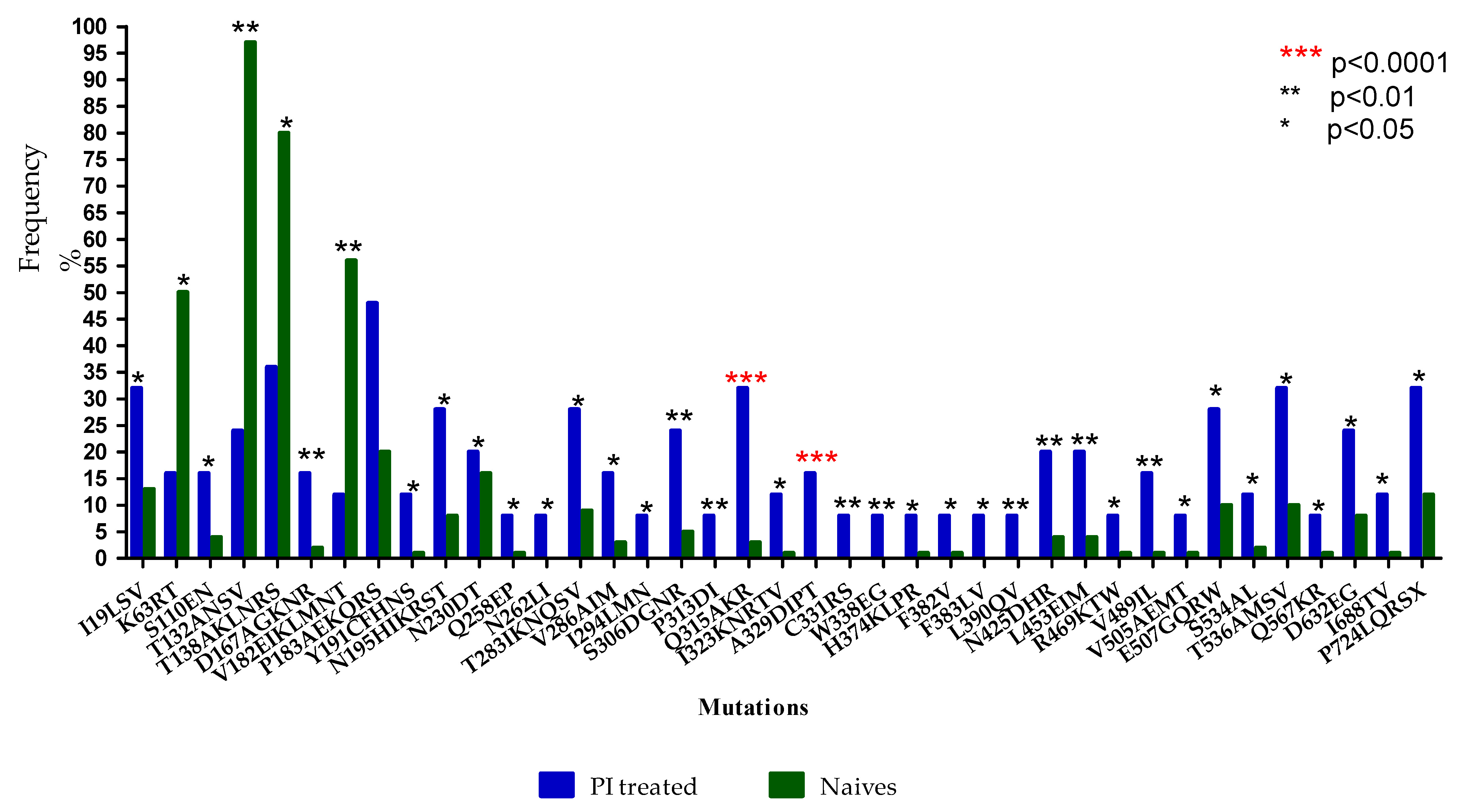
3.2. Prevalence of Envelope Mutations in LPV/r-Experienced versus ART-Naïve Subtype C Sequences
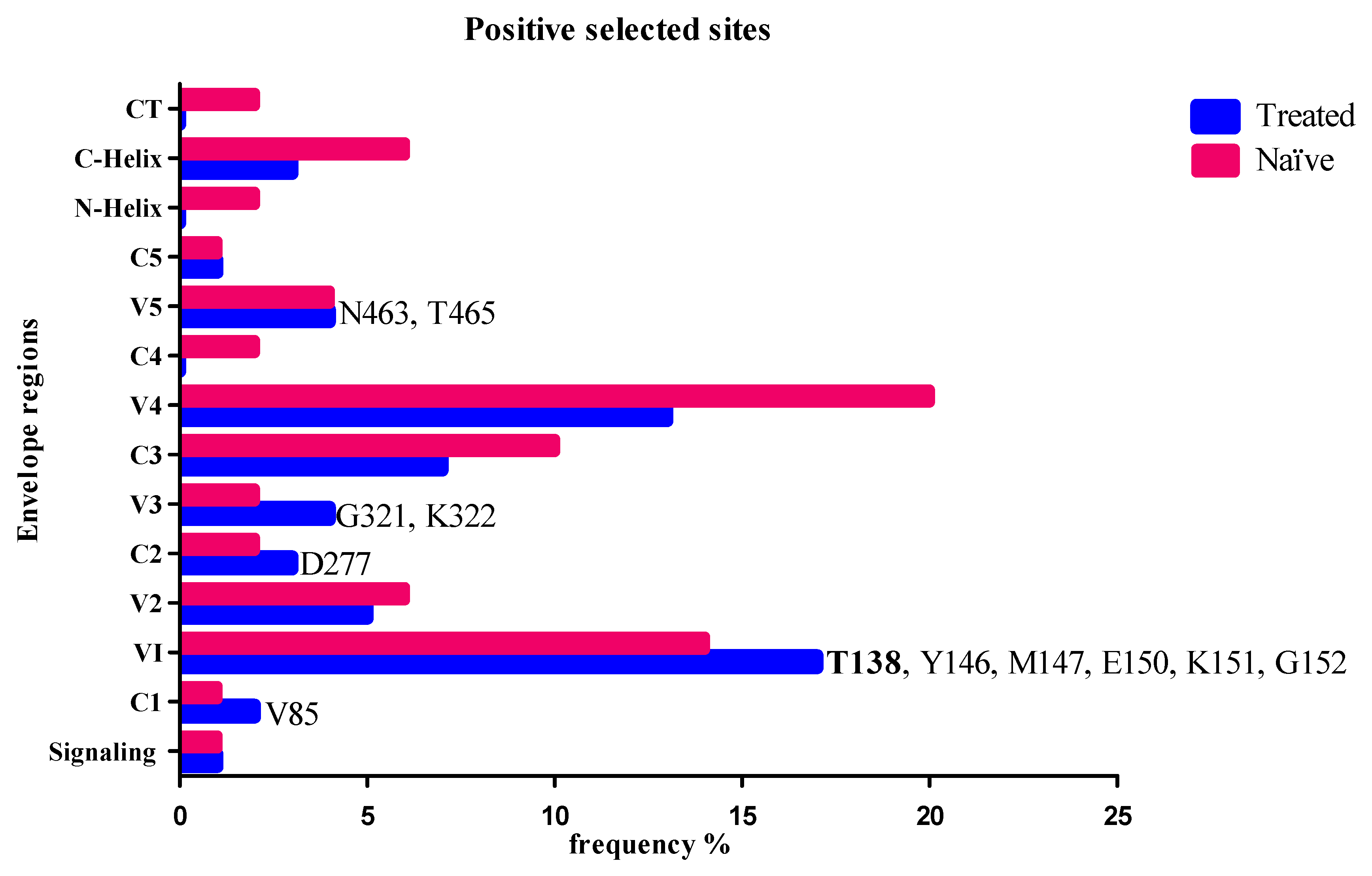
3.3. Positive Selected Sites in Envelope and Coevolution with Gag-PR Residues
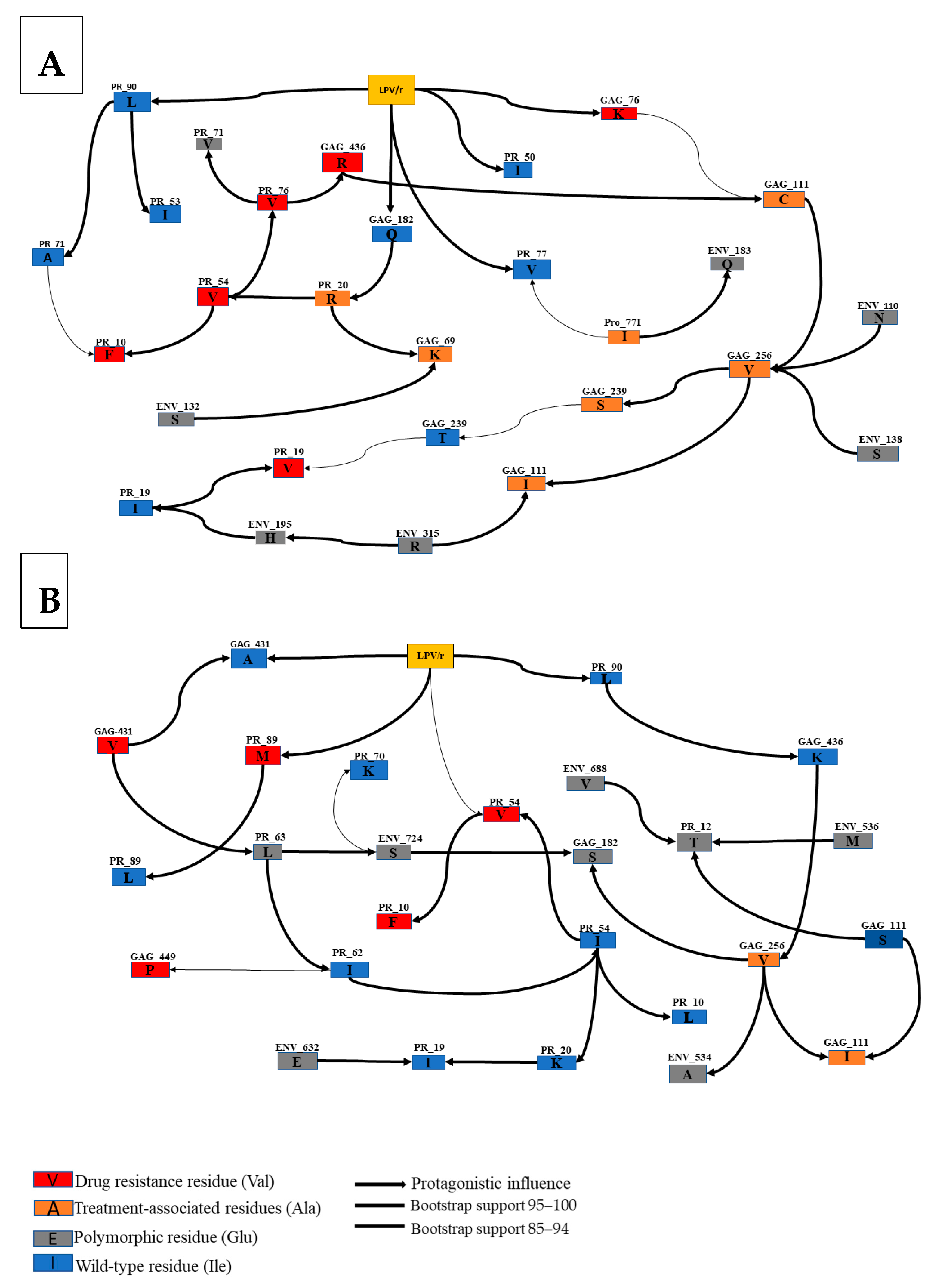
3.4. Interactions between Envelope and Gag-PR Mutations with LPV/r Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghosh, A.K.; Chapsal, B.D.; Weber, I.T.; Mitsuya, H. Design of HIV protease inhibitors targeting protein backbone: An effective strategy for combating drug resistance. Acc. Chem. Res. 2008, 41, 78–86. [Google Scholar] [CrossRef]
- Kim, R.; Baxter, J.D. Protease inhibitor resistance update: Where are we now? AIDS Patient Care STDs 2008, 22, 267–277. [Google Scholar] [CrossRef]
- Pettit, S.C.; Moody, M.D.; Wehbie, R.S.; Kaplan, A.H.; Nantermet, P.V.; Klein, C.A.; Swanstrom, R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 1994, 68, 8017–8027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lori, F.; Scovassi, A.I.; Zella, D.; Achilli, G.; Cattaneo, E.; Casoli, C.; Bertazzoni, U. Enzymatically active forms of reverse transcriptase of the human immunodeficiency virus. AIDS Res. Hum. Retrovir. 1988, 4, 393–398. [Google Scholar] [CrossRef]
- Rabi, S.A.; Laird, G.M.; Durand, C.M.; Laskey, S.; Shan, L.; Bailey, J.R.; Chioma, S.; Moore, R.D.; Siliciano, R.F. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J. Clin. Investig. 2013, 123, 3848–3860. [Google Scholar] [CrossRef] [PubMed]
- Zdanowicz, M.M. The pharmacology of HIV drug resistance. Am. J. Pharm. Educ. 2006, 70, 100. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O.; de Mendoza, C.; Pérez-Elías, M.J.; Guardiola, J.M.; Pedreira, J.; Dalmau, D.; Gónzalez, J.; Moreno, A.; Arribas, J.R.; Rubio, A. Drug resistance in patients experiencing early virological failure under a triple combination including indinavir. Aids 2001, 15, 1701–1706. [Google Scholar] [CrossRef]
- Wallis, C.L.; Mellors, J.W.; Venter, W.D.; Sanne, I.; Stevens, W. Protease inhibitor resistance is uncommon in HIV-1 subtype C infected patients on failing second-line lopinavir/r-containing antiretroviral therapy in South Africa. AIDS Res. Treat. 2011, 2011, 769627. [Google Scholar] [CrossRef] [Green Version]
- Obasa, A.E.; Mikasi, S.G.; Brado, D.; Cloete, R.; Singh, K.; Neogi, U.; Jacobs, G.B. Drug resistance mutations against protease, reverse transcriptase and integrase inhibitors in people living with HIV-1 receiving boosted protease inhibitors in South Africa. Front. Microbiol. 2020, 11, 438. [Google Scholar] [CrossRef] [Green Version]
- Parry, C.M.; Kolli, M.; Myers, R.E.; Cane, P.A.; Schiffer, C.; Pillay, D. Three residues in HIV-1 matrix contribute to protease inhibitor susceptibility and replication capacity. Antimicrob. Agents Chemother. 2011, 55, 1106–1113. [Google Scholar] [CrossRef] [Green Version]
- Alfadhli, A.; Mack, A.; Ritchie, C.; Cylinder, I.; Harper, L.; Tedbury, P.R.; Freed, E.O.; Barklis, E. Trimer Enhancement Mutation Effects on HIV-1 Matrix Protein Binding Activities. J. Virol. 2016, 90, 5657–5664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fun, A.; Wensing, A.M.; Verheyen, J.; Nijhuis, M. Human Immunodeficiency Virus Gag and protease: Partners in resistance. Retrovirology 2012, 9, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codoñer, F.M.; Peña, R.; Blanch-Lombarte, O.; Jimenez-Moyano, E.; Pino, M.; Vollbrecht, T.; Clotet, B.; Martinez-Picado, J.; Draenert, R.; Prado, J.G. Gag-protease coevolution analyses define novel structural surfaces in the HIV-1 matrix and capsid involved in resistance to Protease Inhibitors. Sci. Rep. 2017, 7, 3717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malet, I.; Roquebert, B.; Dalban, C.; Wirden, M.; Amellal, B.; Agher, R.; Simon, A.; Katlama, C.; Costagliola, D.; Calvez, V. Association of Gag cleavage sites to protease mutations and to virological response in HIV-1 treated patients. J. Infect. 2007, 54, 367–374. [Google Scholar] [CrossRef]
- Nijhuis, M.; Van Maarseveen, N.M.; Lastere, S.; Schipper, P.; Coakley, E.; Glass, B.; Rovenska, M.; De Jong, D.; Chappey, C.; Goedegebuure, I.W. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med. 2007, 4, e36. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, J.; Litau, E.; Sing, T.; Daumer, M.; Balduin, M.; Oette, M.; Fatkenheuer, G.; Rockstroh, J.K.; Schuldenzucker, U.; Hoffmann, D. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir. Ther. 2006, 11, 879. [Google Scholar]
- Kletenkov, K.; Hoffmann, D.; Böni, J.; Yerly, S.; Aubert, V.; Schöni-Affolter, F.; Struck, D.; Verheyen, J.; Klimkait, T.; Study, S.H.C. Role of Gag mutations in PI resistance in the Swiss HIV cohort study: Bystanders or contributors? J. Antimicrob. Chemother. 2017, 72, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Larrouy, L.; Chazallon, C.; Landman, R.; Capitant, C.; Peytavin, G.; Collin, G.; Charpentier, C.; Storto, A.; Pialoux, G.; Katlama, C. Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naive HIV-infected patients. Antimicrob. Agents Chemother. 2010, 54, 2910–2919. [Google Scholar] [CrossRef] [Green Version]
- Gatanaga, H.; Suzuki, Y.; Tsang, H.; Yoshimura, K.; Kavlick, M.F.; Nagashima, K.; Gorelick, R.J.; Mardy, S.; Tang, C.; Summers, M.F. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 2002, 277, 5952–5961. [Google Scholar] [CrossRef] [Green Version]
- Kožíšek, M.; Henke, S.; Šašková, K.G.; Jacobs, G.B.; Schuch, A.; Buchholz, B.; Müller, V.; Kräusslich, H.-G.; Řezáčová, P.; Konvalinka, J. Mutations in HIV-1 gag and pol compensate for the loss of viral fitness caused by a highly mutated protease. Antimicrob. Agents Chemother. 2012, 56, 4320–4330. [Google Scholar] [CrossRef] [Green Version]
- Coetzer, M.; Ledingham, L.; Diero, L.; Kemboi, E.; Orido, M.; Kantor, R. Gp41 and Gag amino acids linked to HIV-1 protease inhibitor-based second-line failure in HIV-1 subtype A from Western Kenya. J. Int. AIDS Soc. 2017, 20, e25024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castain, L.; Perrier, M.; Charpentier, C.; Palich, R.; Desire, N.; Wirden, M.; Descamps, D.; Sayon, S.; Landman, R.; Valantin, M.-A. New mechanisms of resistance in virological failure to protease inhibitors: Selection of non-described protease, Gag and Gp41 mutations. J. Antimicrob. Chemother. 2019, 74, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Perrier, M.; Castain, L.; Regad, L.; Todesco, E.; Landman, R.; Visseaux, B.; Yazdanpanah, Y.; Rodriguez, C.; Joly, V.; Calvez, V. HIV-1 protease, Gag and gp41 baseline substitutions associated with virological response to a PI-based regimen. J. Antimicrob. Chemother. 2019, 74, 1679–1692. [Google Scholar] [CrossRef]
- Van Duyne, R.; Kuo, L.S.; Pham, P.; Fujii, K.; Freed, E.O. Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc. Natl. Acad. Sci. USA 2019, 116, 9040–9049. [Google Scholar] [CrossRef] [Green Version]
- Hikichi, Y.; Van Duyne, R.; Pham, P.; Groebner, J.L.; Wiegand, A.; Mellors, J.W.; Kearney, M.F.; Freed, E.O. Mechanistic Analysis of the Broad Antiretroviral Resistance Conferred by HIV-1 Envelope Glycoprotein Mutations. Mbio 2021, 12, e03134-20. [Google Scholar] [CrossRef]
- Marie, V.; Gordon, M. Gag-protease coevolution shapes the outcome of lopinavir-inclusive treatment regimens in chronically infected HIV-1 subtype C patients. Bioinformatics 2019, 35, 3219–3223. [Google Scholar] [CrossRef] [PubMed]
- Rangel, H.R.; Weber, J.; Chakraborty, B.; Gutierrez, A.; Marotta, M.L.; Mirza, M.; Kiser, P.; Martinez, M.A.; Este, J.A.; Quinones-Mateu, M.E. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 2003, 77, 9069–9073. [Google Scholar] [CrossRef] [Green Version]
- Deforche, K.; Camacho, R.; Grossman, Z.; Silander, T.; Soares, M.; Moreau, Y.; Shafer, R.W.; Van Laethem, K.; Carvalho, A.; Wynhoven, B. Bayesian network analysis of resistance pathways against HIV-1 protease inhibitors. Infect. Genet. Evol. 2007, 7, 382–390. [Google Scholar] [CrossRef]
- Theys, K.; Deforche, K.; Libin, P.; Camacho, R.J.; Van Laethem, K.; Vandamme, A.-M. Resistance pathways of human immunodeficiency virus type 1 against the combination of zidovudine and lamivudine. J. Gen. Virol. 2010, 91, 1898–1908. [Google Scholar] [CrossRef]
- Fares, M.A.; Travers, S.A. A novel method for detecting intramolecular coevolution: Adding a further dimension to selective constraints analyses. Genetics 2006, 173, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, M.; Bielawski, J.P.; Yang, Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 2001, 18, 1585–1592. [Google Scholar] [CrossRef] [Green Version]
- Dang, L.V.P.; Pham, H.V.; Dinh, T.T.; Nguyen, T.H.; Vu, Q.T.H.; Vu, N.T.P.; Le, P.T.B.; Nguyen, L.V.; Le, H.T.; Vu, P.T. Characterization of envelope sequence of HIV virus in children infected with HIV in Vietnam. SAGE Open Med. 2020, 8, 2050312120937198. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.E.; Samal, A.B.; Vlach, J.; Saad, J.S. Solution structure and membrane interaction of the cytoplasmic tail of HIV-1 gp41 protein. Structure 2017, 25, 1708–1718.e1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, U. Acquired and Transmitted Drug Resistance in HIV-1 Subtype C: Implications of Novel Mutations on Replication Capacity, Cleavage and Drug Susceptibility. Ph.D. Thesis, University of Kwazulu-Natal, Durban, South Africa, 2015. [Google Scholar]
- Ntale, R.S. The Role of Early Cytotoxic Lymphocyte (CTL) Escape in the Pathogenesis of HIV-1 Subtype C Infection. Ph.D. Thesis, University of Cape Town, Cape Town, South Africa, 2012. [Google Scholar]
- Li, G.; Verheyen, J.; Rhee, S.-Y.; Voet, A.; Vandamme, A.-M.; Theys, K. Functional conservation of HIV-1 Gag: Implications for rational drug design. Retrovirology 2013, 10, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyser, J.M.; Zeng, C.Q.-Y.; Beharry, Z.; Palzkill, T.; Estes, M.K. Epitope mapping and use of epitope-specific antisera to characterize the VP5⁎ binding site in rotavirus SA11 NSP4. Virology 2008, 373, 211–228. [Google Scholar] [CrossRef] [Green Version]
- Ratcliff, A.N.; Shi, W.; Arts, E.J. HIV-1 resistance to maraviroc conferred by a CD4 binding site mutation in the envelope glycoprotein gp120. J. Virol. 2013, 87, 923–934. [Google Scholar] [CrossRef] [Green Version]
- Giandhari, J.; Basson, A.E.; Sutherland, K.; Parry, C.M.; Cane, P.A.; Coovadia, A.; Kuhn, L.; Hunt, G.; Morris, L. Contribution of Gag and protease to HIV-1 phenotypic drug resistance in pediatric patients failing protease inhibitor-based therapy. Antimicrob. Agents Chemother. 2016, 60, 2248–2256. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.M.; Perno, C.F. HIV-1 genetic variability and clinical implications. Int. Sch. Res. Not. 2013, 2013, 481314. [Google Scholar] [CrossRef] [Green Version]
- Mata-Munguía, C.; Escoto-Delgadillo, M.; Torres-Mendoza, B.; Flores-Soto, M.; Vázquez-Torres, M.; Gálvez-Gastelum, F.; Viniegra-Osorio, A.; Castillero-Manzano, M.; Vázquez-Valls, E. Natural polymorphisms and unusual mutations in HIV-1 protease with potential antiretroviral resistance: A bioinformatic analysis. BMC Bioinform. 2014, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Doualla-Bell, F.; Avalos, A.; Gaolathe, T.; Mine, M.; Gaseitsiwe, S.; Ndwapi, N.; Novitsky, V.A.; Brenner, B.; Oliveira, M.; Moisi, D. Impact of human immunodeficiency virus type 1 subtype C on drug resistance mutations in patients from Botswana failing a nelfinavir-containing regimen. Antimicrob. Agents Chemother. 2006, 50, 2210–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maphumulo, N.F.; Gordon, M.L. Potential Associations of Mutations within the HIV-1 Env and Gag Genes Conferring Protease Inhibitor (PI) Drug Resistance. Microbiol. Res. 2021, 12, 967-977. https://doi.org/10.3390/microbiolres12040071
Maphumulo NF, Gordon ML. Potential Associations of Mutations within the HIV-1 Env and Gag Genes Conferring Protease Inhibitor (PI) Drug Resistance. Microbiology Research. 2021; 12(4):967-977. https://doi.org/10.3390/microbiolres12040071
Chicago/Turabian StyleMaphumulo, Ntombikhona F., and Michelle L. Gordon. 2021. "Potential Associations of Mutations within the HIV-1 Env and Gag Genes Conferring Protease Inhibitor (PI) Drug Resistance" Microbiology Research 12, no. 4: 967-977. https://doi.org/10.3390/microbiolres12040071
APA StyleMaphumulo, N. F., & Gordon, M. L. (2021). Potential Associations of Mutations within the HIV-1 Env and Gag Genes Conferring Protease Inhibitor (PI) Drug Resistance. Microbiology Research, 12(4), 967-977. https://doi.org/10.3390/microbiolres12040071




