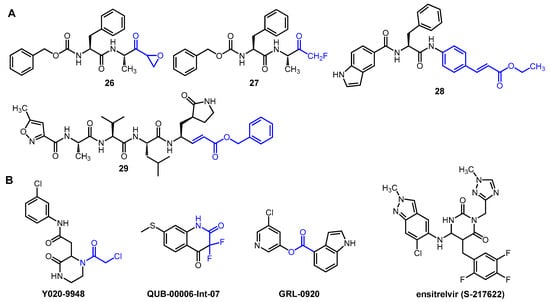
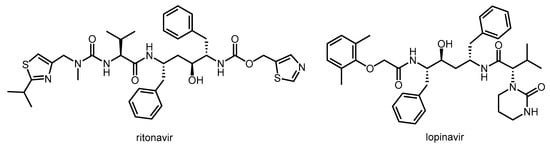
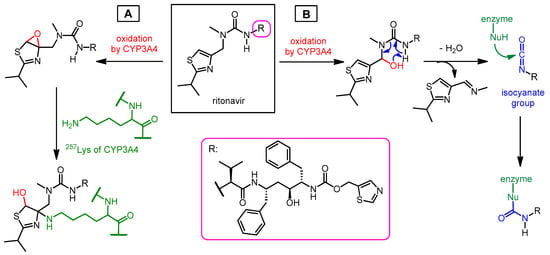
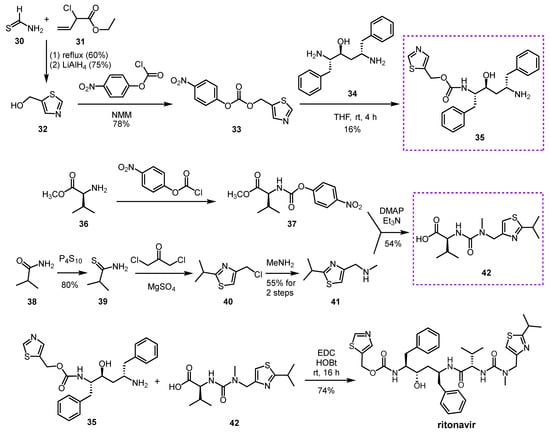
Abstract
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has presented an enormous challenge to health care systems and medicine. As a result of global research efforts aimed at preventing and effectively treating SARS-CoV-2 infection, vaccines with fundamentally new mechanisms of action and some small-molecule antiviral drugs targeting key proteins in the viral cycle have been developed. The most effective small-molecule drug approved to date for the treatment of COVID-19 is PaxlovidTM, which is a combination of two protease inhibitors, nirmatrelvir and ritonavir. Nirmatrelvir is a reversible covalent peptidomimetic inhibitor of the main protease (Mpro) of SARS-CoV-2, which enzyme plays a crucial role in viral reproduction. In this combination, ritonavir serves as a pharmacokinetic enhancer, it irreversibly inhibits the cytochrome CYP3A4 enzyme responsible for the rapid metabolism of nirmatrelvir, thereby increasing the half-life and bioavailability of nirmatrelvir. In this tutorial review, we summarize the development and pharmaceutical chemistry aspects of Paxlovid, covering the evolution of protease inhibitors, the warhead design, synthesis and the mechanism of action of nirmatrelvir, as well as the synthesis of ritonavir and its CYP3A4 inhibition mechanism. The efficacy of Paxlovid to novel virus mutants is also overviewed.
1. Introduction—Viral Proteases as Drug Targets
The genetics and reproduction of viruses differ significantly from what we are used to in eukaryotes in many respects. One important difference is that many viruses, including retroviruses, herpesviruses, flaviviruses and coronaviruses, do not encode functional proteins that are synthesized individually, but rather one or two large polyproteins that are then cleaved by viral proteases into functional proteins [1]. Proteases are a subgroup of hydrolases, enzymes that catalyze hydrolytic reactions. There are many mechanisms of proteolysis; a common method is the use of a nucleophilic group, generated from the side chain of serine (serine proteases) or cysteine (cysteine proteases), which can perform a nucleophilic attack on the partially positively polarized carbonyl carbon atom of the peptide bond. In aspartic proteases, a water molecule bound to aspartic acid in the active site of the enzyme acts as a nucleophile. In the case of serine and cysteine proteases, the nucleophile is generated by the amino acids in the active site of the enzyme. A common system for this is the so-called catalytic dyad or catalytic triad, which consists of two or three amino acids [1,2]. One of the three amino acids that make up the catalytic triad carries an acidic side chain, e.g., aspartic acid (Asp), the other carries an alkaline side chain, e.g., histidine (His), and the nucleophilic group is formed from the third amino acid, which is serine (Ser) or cysteine (Cys).
In some cysteine hydrolases, histidine and cysteine form a catalytic dyad in the active site; in such enzymes, the role of the third amino acid, Asp, is played by an activated water molecule (Scheme 1). In the first step of the enzyme’s catalytic mechanism, the imidazole ring of histidine as a base deprotonates the thiol group of cysteine, forming a thiolate–imidazolium ion pair. Next, the thiolate group performs a nucleophilic attack on the carbonyl C atom of the peptide bond of its substrate. As a result, the peptide bond is cleaved, the amine terminus of the peptide fragment (R-NH2) is released, while the acyl part forms a thioester with the cysteine, and the histidine is reestablished to its deprotonated form. Finally, the thioester bond of the acylated enzyme is hydrolyzed by an activated water molecule to generate a carboxylic acid group on the remaining substrate fragment (R’-COOH), regenerating the free enzyme.
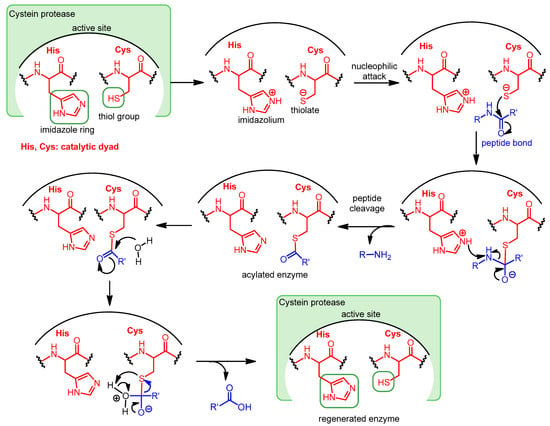
Scheme 1.
Catalytic mechanism of Cys proteases operating with a catalytic dyad.
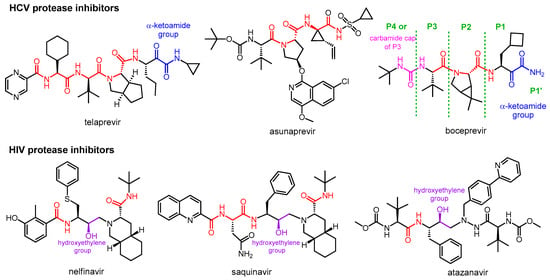
Viral proteases play a key role in viral replication for all positive single-stranded RNA viruses and some DNA viruses, such as herpesviruses. To treat infections caused by these viruses, proteases are considered excellent drug targets [3]. Protease inhibitors are now routinely used in antiviral therapy for human immunodeficiency virus (HIV) and hepatitis C virus (HCV) infections.
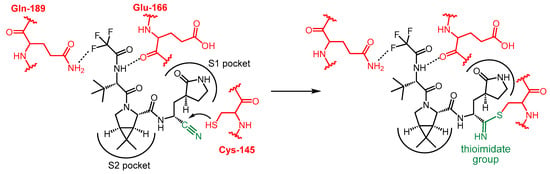
The pathogen responsible for the outbreak of the COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is an enveloped β-coronavirus. It belongs to the family of positive single-stranded RNA viruses and, as such, also encodes proteases. SARS-CoV-2 main protease (Mpro), also known as 3-chymotrypsin-like protease (3CLpro) or nonstructural protein 5 (NSP5), is a cysteine hydrolase that plays an essential role in the SARS-CoV-2 life cycle. Mpro is a homodimer with each monomer consisting of three distinct domains (I, II and III). Both monomers contain a catalytic site at the interface of domains I and II, but only one active site is functional in the dimeric form. Domain III is required for homodimerization, which is critical to the enzyme’s catalytic activity. The enzyme has six substrate binding sub-pockets in the active site, sub-pockets S1, S2 and S4 in the protein cavity, while sub-pockets S1’, S3 and S5 are located on the surface of the protein. The active site of the enzyme contains a catalytic dyad consisting of Cys145 and His41, and a catalytic water molecule H-bonded to His41 [4,5,6].
Since SARS-CoV-2 Mpro plays a key role in viral replication by cleaving viral polyproteins, inhibition of its catalytic activity represents an attractive therapeutic approach for the treatment of COVID-19. In addition, Mpro has two properties that make it an ideal target for antiviral drugs. First, its recognized sequence is Leu-Gln-Ser-Ala-Gly, and it cleaves the peptide chain after a glutamine (Gln) unit; since there are no known human Cys proteases that cleave the protein after Gln, the proteolytic action of SARS-CoV-2 Mpro can be specifically inhibited without inhibiting human proteases. Second, unlike spike protein, Mpro is a highly conserved protein, mutations in this protein could be fatal to the virus, which reduces the risk of developing drug resistance [5,6].
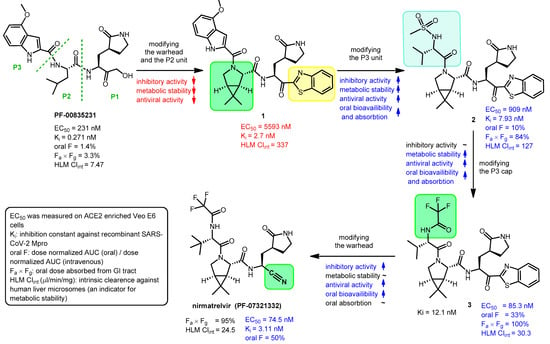
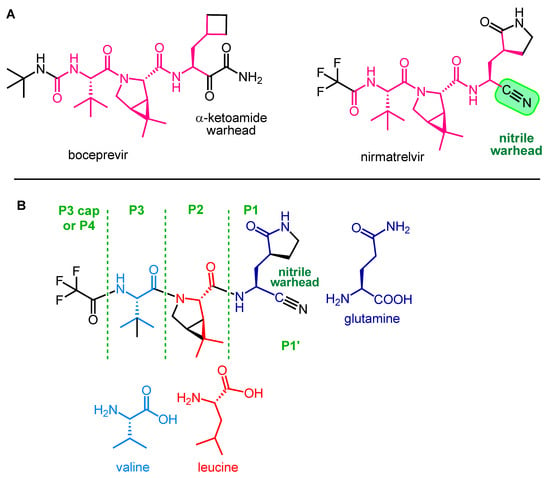
Nirmatrelvir is a newly developed potent inhibitor of the main protease of SARS-CoV-2. However, due to its rapid metabolism by the cytochrome CYP3A4 enzyme, it is not effective on its own in vivo. Nirmatrelvir became suitable for therapeutic use in com bination with ritonavir, originally designed as a HIV protease inhibitor, which can sufficiently increase the bioavailability of nirmatrelvir by inhibiting the CYP3A4 enzyme. The nirmatrelvir–ritonavir combination was developed by Pfizer and marketed under the brand name PaxlovidTM as an oral drug for the treatment of COVID-19.
Since the approval of Paxlovid for the emergency treatment of COVID-19 in 2021, many reviews have been published focusing primarily on the drug’s effectiveness, safety, side effects and drug–drug interactions [7,8,9,10,11,12]. In this tutorial review, we give a medicinal chemistry overview of the two pharmacologically active components of Paxlovid, focusing on the structural similarities of currently used protease inhibitor antivirals, the role of the warhead in the mechanism of action of nirmatrelvir, as well as the details of the booster effect of the ritonavir. We also briefly discuss drug–drug interactions and Paxlovid’s effectiveness against new virus variants.
4. Paxlovid—Application and Activity against Mutant Variants
PaxlovidTM is a product that contains co-packed nirmatrelvir (150 mg/tablet) and ritonavir (100 mg/tablet). The normal dose is 300 mg nirmatrelvir (two tablets) and 100 mg ritonavir (one tablet) two times per day. Nirmatrelvir dosage should be reduced in patients with moderate kidney dysfunction, and its use is not recommended in patients with severe kidney dysfunction. The treatment lasts 5 days [48].
In a phase II/III clinical trial (EPIC-HR = Evaluation of Protease Inhibition for COVID-19 in High-Risk Patients), Paxlovid reduced the combined risk of death and hospitalization related to COVID-19 by 89% compared to the placebo group [49]. A 2022 meta-analysis found that nirmatrelvir/ritonavir was successful in reducing hospitalizations and mortality in patients with COVID-19, but there was no difference between emergency department visits and intensive care unit admissions based on an analysis of 314,353 patient trials. [9] Unfortunately, the EPIC-SR trial, which was designed to test Paxlovid in a standard-risk population, was terminated due to low hospitalization/death rates in the standard-risk population.
Paxlovid was approved for emergency use authorization by the USA in 2021, for the treatment of patients with mild/moderate COVID-19 with high risk of progression to severe disease, with no requirement of oxygen supply. It gained conditional authorization in 2021 in the UK and in 2022 in the EU [48].
Nirmatrelvir has proven to be highly active against the Omicron variant of SARS-CoV-2 and its sub-variants. However, there are already circulating strains with mutations that may give the virus some level of resistance to nirmatrelvir [50,51]. Fortunately, such known mutations are not very common. In a study based on the GISAID database, among more than 13 million sequences, the occurrence of resistance-causing mutations was 0.5%, and no increasing trend was observed; however, there are strains in which certain mutations are dominant [52]. Most of the omicron subvariants are still sensitive to nirmatrelvir, but it is important to monitor the emergence of new potentially resistant strains [53].
5. Conclusions
Although there are several effective and safe vaccines against COVID-19, this virus is unlikely to disappear anytime soon, so there is a need for effective therapeutic agents, especially oral medicines that people can take at home. There are open questions about Paxlovid, e.g., the therapeutic advantage for vaccinated and standard-risk patients, or the phenomenon of rebound, which means that symptoms reappear in some patients after the end of therapy [54,55,56]. Nevertheless, Paxlovid is a valuable antiviral agent against SARS-CoV-2 with high efficacy and safety. At the same time, despite the success of the nirmatrelvir–ritonavir combination, research into alternative MPro inhibitors should not be stopped; fortunately, research efforts aimed at developing novel MPro inhibitors are ongoing [34], as was briefly presented in Section 2.5.
The development of pan-coronavirus antivirals, based on the screening of natural compounds or the design of new molecules, is emerging as a new strategy to combat potential future pandemics. Fusion inhibitors are typically considered pan-coronavirus agents that target the heptad repeat 1 (HR-1) domain of the spike protein S2 subunit. Fusion inhibitors can effectively inhibit the infection of SARS-CoV-2 variants and other human coronaviruses, their broad-spectrum effect is based on the fact that the spike protein (S protein) responsible for viral entry plays a key role in viral infections, and the HR-1 domain is highly conserved region between coronaviruses [57,58,59].
Author Contributions
Conceptualization, A.B. and M.B. writing—original draft preparation, M.B.; writing—review and editing, A.B.; funding acquisition, A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Laboratory of Virology, Hungary, project no. RRF-2.3.1-21-2022-00010.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sharma, A.; Gupta, S.P. Fundamentals of Viruses and Their Proteases. In Viral Proteases and Their Inhibitors; Gupta, S.P., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 1–24. ISBN 978-0-12-809712-0. [Google Scholar]
- Testa, B.; Mayer, J.M. Hydrolysis in Drug and Prodrug Metabolism Chemistry Biochemistry and Enzymology; Verlag Helvetica Chimica Acta and WILEY-VCH GmbH et Co.: Weinheim, Germany, 2003; ISBN 3-906390-25-X. [Google Scholar]
- Majerová, T.; Konvalinka, J. Viral proteases as therapeutic targets. Mol. Asp. Med. 2022, 88, 101159. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffmann, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef] [PubMed]
- Citarella, A.; Dimasi, A.; Moi, D.; Passarella, D.; Scala, A.; Piperno, A.; Micale, N. Recent Advances in SARS-CoV-2 Main Protease Inhibitors: From Nirmatrelvir to Future Perspectives. Biomolecules 2023, 13, 1339. [Google Scholar] [CrossRef]
- Wen, W.; Chen, C.; Tang, J.; Wang, C.; Zhou, M.; Cheng, Y.; Zhou, X.; Wu, Q.; Zhang, X.; Feng, Z.; et al. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: A meta-analysis. Ann. Med. 2022, 54, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Pesko, B.; Deng, A.; Chan, J.D.; Neme, S.; Dhanireddy, S.; Jain, R. Safety, and tolerability of paxlovid (nirmatrelvir/ritonavir) in high risk patients. Clin. Infect. Dis. 2022, 75, 2049–2050. [Google Scholar] [CrossRef]
- Amani, B.; Amani, B. Efficacy and safety of nirmatrelvir/ritonavir (Paxlovid) for COVID-19: A rapid review and meta-analysis. J. Med. Virol. 2023, 95, e28441. [Google Scholar] [CrossRef] [PubMed]
- Paltra, S.; Conrad, T. Effectiveness of Paxlovid—A review. medRxiv 2023. [Google Scholar] [CrossRef]
- Zhu, C.-T.; Yin, J.-Y.; Chen, X.-H.; Liu, M.; Yang, S.-G. Appraisal of evidence reliability and applicability of Paxlovid as treatment for SARS-COV-2 infection: A systematic review. Rev. Med. Virol. 2023, 33, e2476. [Google Scholar] [CrossRef]
- Gerhart, J.; Cox, D.S.; Singh, R.S.P.; Chan, P.L.S.; Rao, R.; Allen, R.; Shi, H.; Masters, J.C.; Damle, B. A Comprehensive Review of the Clinical Pharmacokinetics, Pharmacodynamics, and Drug Interactions of Nirmatrelvir/Ritonavir. Clin. Pharmacokinet. 2024, 63, 27–42. [Google Scholar] [CrossRef]
- de Leuw, P.; Stephan, C. Protease inhibitor therapy for hepatitis C virus-infection. Expert Opin. Pharmacother. 2018, 19, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Halfon, P.; Locarnini, S. Hepatitis C virus resistance to protease inhibitors. J. Hepatol. 2011, 55, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV/AIDS (Auckl.) 2015, 7, 95–104. [Google Scholar] [PubMed]
- Ghosh, A.K.; Osswald, H.L.; Prato, G. Recent Progress in the Development of HIV-1 Protease Inhibitors for the Treatment of HIV/AIDS. J. Med. Chem. 2016, 59, 5172–5208. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.L.; Kania, R.S.; Brothers, M.A.; Davies, J.F.; Ferre, R.A.; Gajiwala, K.S.; He, M.; Hogan, R.J.; Kozminski, K.; Li, L.Y.; et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020, 63, 12725–12747. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hui, D.; Wu, A.; Chan, P.; Cameron, P.; Joynt, G.M.; Ahuja, A.; Yung, M.Y.; Leung, C.B.; To, K.F.; et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1986–1994. [Google Scholar] [CrossRef]
- Menéndez, J.C. Approaches to the Potential Therapy of COVID-19: A General Overview from the Medicinal Chemistry Perspective. Molecules 2022, 27, 658–683. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Chia, C.S.B. Novel Nitrile Peptidomimetics for Treating COVID-19. Med. Chem. Lett. 2022, 13, 330–331. [Google Scholar] [CrossRef]
- Joyce, R.P.; Hu, V.W.; Wang, J. The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): An orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Med. Chem. Res. 2022, 31, 1637–1646. [Google Scholar] [CrossRef]
- Li, J.; Lin, C.; Zhou, X.; Zhong, F.; Zheng, P.; Yang, Y.; Zhang, Y.; Yu, B.; Fan, X.; McCormick, P.J.; et al. Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332. Virol. J. 2022, 96, e0201321. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.T.; Nguyen, T.H.; Tung, N.T.; Mai, B.K. Insights into the binding and covalent inhibition mechanism of PF-07321332 to SARS-CoV-2 Mpro. RSC Adv. 2022, 12, 3729–3737. [Google Scholar] [CrossRef] [PubMed]
- Marzi, M.; Vakil, M.K.; Bahmanyar, M.; Zarenezhad, E. Paxlovid: Mechanism of Action, Synthesis, and In Silico Study. Biomed. Res. Int. 2022, 2022, 7341493. [Google Scholar] [CrossRef] [PubMed]
- Kallam, S.R.; Eda, V.R.; Sen, S.; Datrika, R.; Rapolu, R.K.; Khobare, S.; Gajare, V.; Banda, M.; Khan, R.A.R.; Singh, M.; et al. A diastereoselective synthesis of boceprevir’s gem-dimethyl bicyclic [3.1.0] proline intermediate from an insecticide ingredient cis-cypermethric acid. Tetrahedron 2017, 73, 4285–4294. [Google Scholar] [CrossRef]
- Zhao, Y.; Fang, C.; Zhang, Q.; Zhang, R.; Zhao, X.; Duan, Y.; Wang, H.; Zhu, Y.; Feng, L.; Zhao, J.; et al. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell 2022, 13, 689–693. [Google Scholar] [CrossRef] [PubMed]
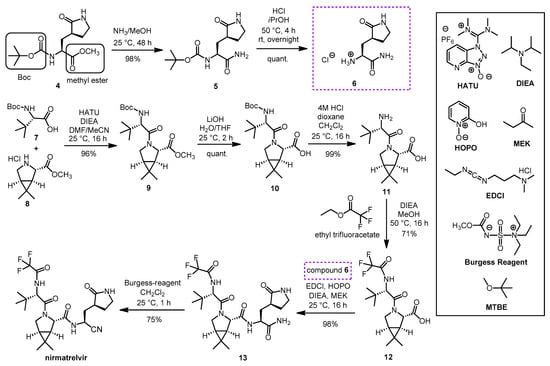
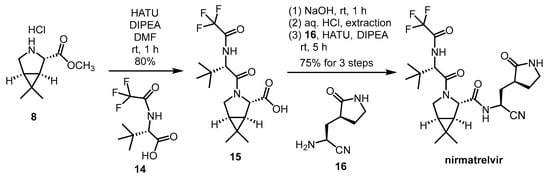
- Preschel, H.D.; Otte, R.T.; Zhuo, Y.; Ruscoe, R.E.; Burke, A.J.; Kellerhals, R.; Horst, B.; Hennig, S.; Janssen, E.; Green, A.P.; et al. Multicomponent Synthesis of the SARS-CoV-2 Main Protease Inhibitor Nirmatrelvir. J. Org. Chem. 2023, 88, 12565–12571. [Google Scholar] [CrossRef] [PubMed]
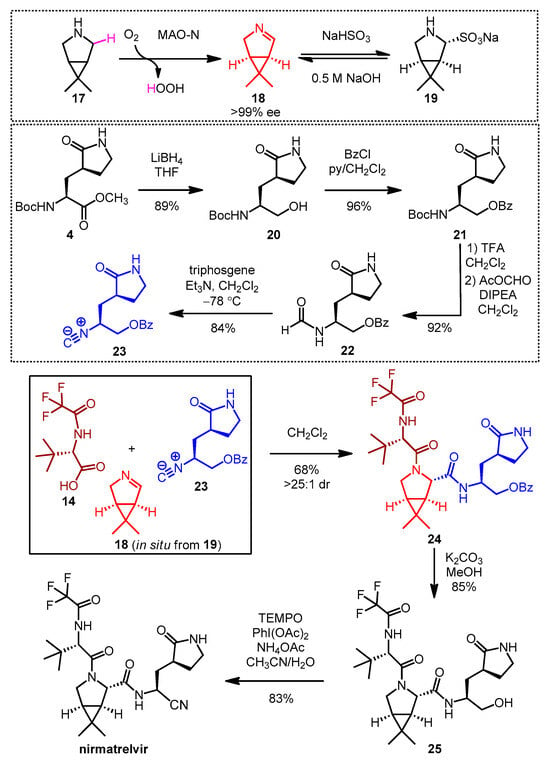
- Köhler, V.; Bailey, K.R.; Znabet, A.; Raftery, J.; Helliwell, M.; Turner, N.J. Enantioselective Biocatalytic Oxidative Desymmetrization of Substituted Pyrrolidines. Angew. Chem. Int. Ed. 2010, 49, 2182–2184. [Google Scholar] [CrossRef]
- Flores-Reyes, J.C.; Islas-Jácome, A.; González-Zamora, E. The Ugi three-component reaction and its variants. Org. Chem. Front. 2021, 8, 5460–5514. [Google Scholar] [CrossRef]
- Li, T.; Liang, J.; Ambrogelly, A.; Brennan, T.; Gloor, G.; Huisman, G.; Lalonde, J.; Lekhal, A.; Mijts, B.; Muley, S.; et al. Efficient, Chemo-enzymatic Process for Manufacture of the Boceprevir Bicyclic [3.1.0] Proline Intermediate Based on Amine Oxidase-Catalyzed Desymmetrization. J. Am. Chem. Soc. 2012, 134, 6467–6472. [Google Scholar] [CrossRef]
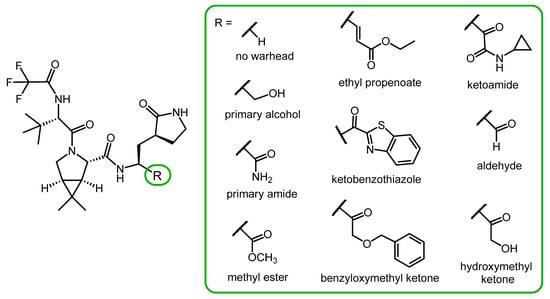
- Vankadara, S.; Dawson, M.D.; Fong, J.Y.; Oh, Q.Y.; Ang, Q.A.; Liu, B.; Chang, H.Y.; Koh, J.; Koh, X.; Tan, Q.W.; et al. A Warhead Substitution Study on the Coronavirus Main Protease Inhibitor Nirmatrelvir. Med. Chem. Lett. 2022, 13, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Citarella, A.; Moi, D.; Pedrini, M.; Pérez-Peña, H.; Pieraccini, S.; Dimasi, A.; Stagno, C.; Micale, N.; Schirmeister, T.; Sibille, G.; et al. Synthesis of SARS-CoV-2 Mpro inhibitors bearing a cinnamic ester warhead with in vitro activity against human coronaviruses. Org. Biomol. Chem. 2023, 21, 3811–3824. [Google Scholar] [CrossRef] [PubMed]
- Ashraf-Uz-Zaman, M.; Chua, T.K.; Li, X.; Yao, Y.; Moku, B.K.; Mishra, C.B.; Avadhanula, V.; Piedra, P.A.; Song, Y. Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease. ACS Infect. Dis. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, S.; Convertino, I.; Cappello, E.; Valdiserra, G.; Bonasoa, M.; Tuccori, M. Lessons learnt from the preclinical discovery and development of ensitrelvir as a COVID-19 therapeutic option. Expert Opin. Drug Discov. 2024, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Cooper, C.L.; van Heeswijk, R.P.G.; Gallicano, K.; Cameron, D.W. A Review of Low-Dose Ritonavir in Protease Inhibitor Combination Therapy. Clin. Infect. Dis. 2003, 36, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, R.S.; Goa, K.L. Lopinavir/Ritonavir A Review of its Use in the Management of HIV Infection. Drugs 2003, 63, 769–802. [Google Scholar] [CrossRef]
- Klein, K.; Zanger, U.M. Pharmacogenomics of cytochrome P450 3A4: Recent progress toward the “missing heritability” problem. Front. Genet. 2013, 4, 12. [Google Scholar] [CrossRef]
- Patel, T.K.; Patel, P.B.; Barvaliya, M.; Saurabh, M.K.; Bhalla, H.L.; Khosla, P.P. Efficacy and safety of lopinavir-ritonavir in COVID-19: A systematic review of randomized controlled trials. J. Infect. Public Health 2021, 14, 740–748. [Google Scholar] [CrossRef]
- Marzolini, C.; Kuritzkes, D.R.; Marra, F.; Boyle, A.; Gibbons, S.; Flexner, C.; Pozniak, A.; Boffito, M.; Waters, L.; Burger, D.; et al. Recommendations for the Management of Drug-Drug Interactions Between the COVID-19 Antiviral Nirmatrelvir/Ritonavir (Paxlovid) and Comedications. Clin. Pharmacol. Ther. 2022, 112, 1191–1200. [Google Scholar] [CrossRef]
- Loos, N.H.C.; Beijnen, J.H.; Schinkel, A.H. The Mechanism-Based Inactivation of CYP3A4 by Ritonavir: What Mechanism? Int. J. Mol. Sci. 2022, 23, 9866–9890. [Google Scholar] [CrossRef]
- Rock, B.M.; Hengel, S.M.; Rock, D.A.; Wienkers, L.C.; Kunze, K.L. Characterization of ritonavir-mediated inactivation of cytochrome P450 3A4. Mol. Pharmacol. 2014, 86, 665–674. [Google Scholar] [CrossRef]
- Kempf, D.J.; Sham, H.L.; Marsh, K.C.; Flentge, C.A.; Betebenner, D.; Green, B.E.; McDonald, E.; Vasavanonda, S.; Saldivar, A.; Wideburg, N.E.; et al. Discovery of ritonavir, a potent inhibitor of HIV protease with high oral bioavailability and clinical efficacy. J. Med. Chem. 1998, 41, 602–617. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Bilcer, G.; Schiltz, G. Syntheses of FDA Approved HIV Protease Inhibitors. Synthesis 2001, 2001, 2203–2229. [Google Scholar] [CrossRef] [PubMed]
- Kempf, D.J.; Marsh, K.C.; Fino, L.C.; Bryant, P.; Craig-Kennard, A.; Sham, H.L.; Zhao, C.; Vasavanonda, S.; Kohlbrenner, W.E.; Wideburg, N.E.; et al. Design of Orally Bioavailable, Symmetry-Based Inhibitors of HIV Protease. Bioorg. Med. Chem. 1994, 2, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; McKee, S.P.; Thompson, W.J.; Darke, P.L.; Zugay, J.C. Potent HIV-1 Protease Inhibitors: Stereoselective Synthesis of a Dipeptide Mimic. J. Org. Chem. 1993, 58, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Nirmatrelvir Plus Ritonavir: First Approval. Drugs 2022, 82, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.L.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N. Eng. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lewandowski, E.M.; Tan, H.; Zhang, X.; Morgan, R.T.; Zhang, X.; Jacobs, L.M.C.; Butler, S.G.; Gongora, M.V.; Choy, J.; et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. ACS Cent. Sci. 2023, 9, 1658–1669. [Google Scholar] [CrossRef]
- Mótyán, J.A.; Mohamed Mahdi, M.; Hoffka, G.Y.; Tőzsér, J. Potential Resistance of SARS-CoV-2 Main Protease (Mpro) against Protease Inhibitors: Lessons Learned from HIV-1 Protease. Int. J. Mol. Sci. 2022, 23, 3507. [Google Scholar] [CrossRef]
- Ip, J.D.; Chu, A.W.; Chan, W.; Leung, R.C.; Abdullah, S.M.U.; Sun, Y.; To, K.K. Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance. eBioMedicine 2023, 91, 104559. [Google Scholar] [CrossRef]
- Chatterjee, S.; Bhattacharya, M.; Dhama, K.; Lee, S.; Chakraborty, C. Resistance to nirmatrelvir due to mutations in the Mpro in the subvariants of SARS-CoV-2 Omicron: Another concern? Mol. Ther. Nucleic Acids 2023, 13, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Epling, B.P.; Rocco, J.M.; Boswell, K.L.; Laidlaw, E.; Galindo, F.; Kellogg, A.; Das, S.; Roder, A.; Ghedin, E.; Kreitman, A.; et al. COVID-19 redux: Clinical, virologic, and immunologic evaluation of clinical rebound after nirmatrelvir/ritonavir. medRxiv 2022. [Google Scholar] [CrossRef]
- Antonelli, G.; Focosi, D.; Turriziani, O.; Tuccori, M.; Brandi, R.; Fillo, S.; Ajassa, C.; Lista, F.; Mastroianni, C.M. Virological and clinical rebounds of COVID-19 soon after nirmatrelvir/ritonavir discontinuation. Clin. Microbiol. Infect. 2022, 28, 1657–1658. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; McConnell, S.; Shoham, S.; Casadevall, A.; Maggi, F.; Antonelli, G. Nirmatrelvir and COVID-19: Development, pharmacokinetics, clinical efficacy, resistance, relapse, and pharmacoeconomics. Int. J. Antimicrob. Agents 2023, 61, 106708. [Google Scholar] [CrossRef]
- Mori, M.; Quaglio, D.; Calcaterra, A.; Ghirga, F.; Sorrentino, L.; Cammarone, S.; Fracella, M.; D’Auria, A.; Frasca, F.; Criscuolo, E.; et al. Natural Flavonoid Derivatives Have Pan-Coronavirus Antiviral Activity. Microorganisms 2023, 11, 314–332. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zheng, A.; Tang, Y.; Chai, Y.; Chen, J.; Cheng, L.; Hu, Y.; Qu, J.; Lei, W.; Liu, W.J.; et al. A pan-coronavirus peptide inhibitor prevents SARS-CoV-2 infection in mice by intranasal delivery. Sci. China Life Sci. 2023, 66, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Wang, L.; Jiao, F.; Lu, L.; Xia, S.; Jiang, S. Pan-coronavirus fusion inhibitors to combat COVID-19 and other emerging coronavirus infectious diseases. J. Med. Virol. 2023, 95, e28143. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).