Abstract
Selenium nanoparticles (SeNPs) have been receiving special attention in recent years due to their antioxidant capacity and antitumor properties. However, the mechanisms associated with these properties remain to be elucidated. For this reason, a global transcriptome analysis has been designed in this work and it was carried out using human hepatocarcinoma cells and chitosan-stabilized SeNPs (Ch-SeNPs) to identify new targets and pathways related to the antitumor mechanisms associated with Ch-SeNPs. The results obtained confirm the alteration of the cell cycle and the effect of Ch-SeNPs on different tumor suppressors and other molecules involved in key mechanisms related to cancer progression. Furthermore, we demonstrated the antioxidant properties of these nanoparticles and their capacity to induce senescence, which was further confirmed through the measurement of β-galactosidase activity.
1. Introduction
A correct balance between ROS generation by pro-oxidants and the action of antioxidants maintains the normal cellular redox status. Loss of this balance might play a critical role in cellular signaling pathways, sometimes evolving into an uncontrolled proliferation leading to neocarcinogenesis [1,2]. ROS are generated as a defense barrier against extracellular pathogens, including bacterial and viral infections, but an excessive production is also closely related to cancer pathologies. Cancer therapy is, in the great majority of the cases, based on chemotherapy, usually in combination with radiation therapy and immunotherapy. One of the main side effects of the cytotoxic agents used in chemotherapy is the generation of ROS, which induce DNA damage or affect the DNA machinery [3]. To avoid this problem, many chemotherapeutic strategies are focused on using antioxidants to deplete tumor cells from ROS-induced survival signaling pathways, such treatments also having potential preventive functions.
The emerging field of nanotechnology in recent years has opened up a new field of study for the development of novel drugs for a wide variety of diseases [4,5] and, in particular, for the development of promising nanoparticles and nanosystems with antitumor properties [6,7]. The antitumor effect of some of these nanoparticles is based on their antioxidant properties. Such is the case of gold nanoparticles, which have been successfully tested against human breast cancer (MCF-7) cells [8].
Selenium is a well-known essential micronutrient that participates in a large number of key physiological processes. It is also considered an element with antioxidant properties and, as a matter of fact, different selenospecies have been proposed as potential antitumor agents against different types of cancer [9,10]. In addition, and although further investigations are needed, it seems that SeNPs are biocompatible when exposing non-cancer cells such as human dermal fibroblasts [11]. In our previous work, we demonstrated the unique effect of chitosan-stabilized selenium nanoparticles (Ch-SeNPs) in comparison to other organic and inorganic selenospecies to induce cell cycle arrest while preventing uncontrolled generation of ROS that would have induced apoptosis [12]. Other authors have also demonstrated the use of the cytotoxicity exerted by SeNPs as a tool against different diseases [13,14,15,16,17]. Based on the above, there is strong evidence regarding the potential use of SeNPs as a very promising therapeutic alternative. However, before bringing the use of these nanoparticles to clinical models, it is necessary to elucidate the mechanisms of action at the molecular level. Although several efforts have been carried out in this way [11,12,18], further efforts are still needed to confirm the specific targets and mechanisms responsible for the observed antitumor effects of SeNPs.
Transcriptomics represent a high-throughput screening tool that provides in-depth understanding of cellular functions and the genomic landscape of transcription, thanks to its capability to mirror post-genomic variations. Thus, it represents an essential strategy for the study of such a dynamic pathology as cancer [19,20].
Based on all of the above, we had synthesized and characterized selenium nanoparticles stabilized with chitosan (Ch-SeNPs), which were further used to treat human hepatocellular carcinoma cells (HepG2 cells). The design and application of a whole transcriptome analysis to this system provided a wide number of altered targets and pathways, thus allowing us to delve deeper into the mechanisms associated with the antitumor effect of Ch-SeNPs.
2. Materials and Methods
2.1. Synthesis and Characterization of SeNPs
Chitosan, ascorbic acid, acetic acid and sodium selenite were purchased from Sigma-Aldrich (St. Louis, MO, USA) Chitosan-stabilized SeNPs (Ch-SeNPs) were synthesized following the procedure described by Bai et al. [21]. An aqueous chitosan polysaccharide solution (0.5% w/v) was prepared using 0.5 M acetic acid. Then, 10 mL of this chitosan solution were mixed with 7.5 mL of ascorbic acid, 0.23 M, and 5 mL of acetic acid, 2.4 M. To the resulting solution, 0.25 mL of sodium selenite, 0.51 M, were slowly added. The observed change of the solution from colorless to red was indicative of the reaction progression and the Ch-SeNPs formation. After the synthesis, the colloidal suspension was diluted to 50 mL with distilled water, resulting in final concentrations of 200 mg/L of Se and 0.1% of chitosan. Finally, the colloidal suspension was dialyzed for two hours at room temperature at a ratio of 10 mL against 2 L of distilled water and using a 12-kDa molecular weight cut-off (MWCO) membrane.
Ch-SeNPs were characterized by transmission electron microscopy (TEM) and energy dispersive X-ray spectroscopy (EDX) with a JEOL JEM 1400 PLUS operating at 120 kV and equipped with a charge-coupled device CCD camera (KeenView Camera) (JEOL Ltd., Tokyo, Japan). Sample preparation was performed by placing one or two drops of the Ch-SeNP colloidal suspension onto carbon-coated copper grids.
Electrophoretic mobility measurements for the Ch-SeNP colloidal suspension in water were used to calculate the zeta-potential (ζ-potential) values of the nanoparticles. Measurements were performed in a Zetasizer Nano ZS (Malvem Instruments Ltd., Malvern, UK) equipped with a 633 nm red laser. For this purpose, dilutions of the initial suspension were performed if needed. Measurements were recorded by placing ca. 1 mL of the suspension in disposable DTS1070 folded capillary cells (Malvern Instruments). The hydrodynamic size of the nanoparticles was measured by dynamic light scattering (DLS) with the same Malvern instrument. Values presented are means ± SD from quintuplicated measurements.
Stability of Ch-SeNPs was determined using an Agilent HP 7700x inductively coupled plasma mass spectrometer (ICP-MS) (Agilent, Santa Clara, CA, USA).
2.2. Cell Culture
A hepatocellular carcinoma cell line (Homo sapiens) known as HepG2 cells (HEPG2, ATCC HB-8065 TM, American Type Culture Collection, Manassas, VA, USA) was selected for this study. The cells were maintained in a Dulbecco’s modified Eagle´s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C and 5% CO2. Human breast cancer cells (MBA-MD-231) and human cervical cancer cells (HeLa) (American Type Culture Collection, Manassas, VA, USA) used to validate the senescence assay were cultured under the same conditions.
2.3. Cytotoxicity Assay
To evaluate the cytotoxicity induced by Ch-SeNPs, HepG2 cells were seeded into 96-well plates and incubated for 24 h. Then, the cells were exposed to ranging concentrations of Ch-SeNPs (0.1 to 5 mg/L) for 72 h. After this time, 20 μL of 3-(4,5-dimethyl-thiazol-2-yl)2,5-diphenyl tetrazolium bromide (MTT, 5 mg/mL) were added to each well and incubated for five hours at 37 °C. Then, the culture medium was removed, and 100 μL of dimethyl sulfoxide were added to dissolve the insoluble purple formazan products. Absorbance was measured at 595 nm using a microplate reader (TECAN). The final results were calculated on the basis of five replicates of the experiment.
2.4. Transcriptome Analysis
To evaluate potential alterations in the mRNA expression levels of HepG2 cells treated with Ch-SeNPs, a transcriptome microarray analysis was performed. The cells were seeded in culture plates for 24 h and then exposed to 1 mg/L of Ch-SeNPs for 72 h (37 °C and 5% CO2). Control (untreated) cells were seeded in parallel and incubated at 37 °C and 5% CO2 for 96 h (24 h + 72 h). After the exposure time, mRNA was extracted and purified using a commercial kit (PureLink®, Invitrogen, Carlsbad, CA, USA). Shortly thereafter, the cells were trypsinized and centrifuged and the supernatant was removed. The cells were lysed with a lysis buffer containing 2-mercaptoethanol. Lysates were then centrifuged and one volume of 70% ethanol was added to the samples. Seven hundred microliters of this volume were transferred to a conical tube and centrifuged. Afterwards, the samples were rinsed with a washing buffer and RNase-free water was added to the spin cartridge prior to centrifugation. The purified RNA was stored at −80 °C for further analysis. The samples were processed with a GeneChip® WT PLUS Reagent Kit (Applied Biosystems, Foster City, CA, USA), hybridized with Clariom™ D Array, human (Applied Biosystems) and scanned with a GeneChip® Scanner 3000 7 G (Applied Biosystems). Raw data were processed with the robust multi-array average (RMA) algorithm included in the Transcriptome Analysis Console (Applied Biosystems) for normalization and gene level analysis. For each experimental condition, three microarray experiments corresponding to three independent RNA replicates were processed and analyzed. Fold changes between experimental conditions were calculated as a quotient between the means of the gene expression signals. Statistical analysis was performed with ebayes limma included in the Transcriptome Analysis Console (Applied Biosystems). Those values with a false discovery rate (FDR) (adjusted p-value) ≤0.05 were considered significant.
2.5. Senescence Assay
A cytochemical staining kit for the observation of β-galactosidase expression (Sigma-Aldrich) was selected to evaluate whether or not Ch-SeNPs induce senescence on HepG2, MBA-MD-231 and HeLa cells. The cells were seeded and exposed to either 1 mg/L of Ch-SeNPs, 10 μM of etoposide (positive senescence control) or 50 μM of etoposide (positive apoptosis control) during 72 h at 37 °C and 5% CO2. After the exposure time, cell culture solutions were removed and the cells were washed with PBS. The cells were then fixed with 20% formaldehyde, 2% glutaraldehyde, 70.4 mM Na2HPO4, 14.7 mM KH2PO4, 1.37 M NaCl and 26.8 mM KCl fixing buffer for seven minutes at room temperature. After removing the fixing buffer, the cells were washed with PBS and incubated with the staining mixture. Finally, the cells were incubated at 37 °C without CO2 overnight and then examined using a phase contrast microscope (Motic AE31).
3. Results and Discussion
3.1. Synthesis and Characterization of Ch-SeNPs
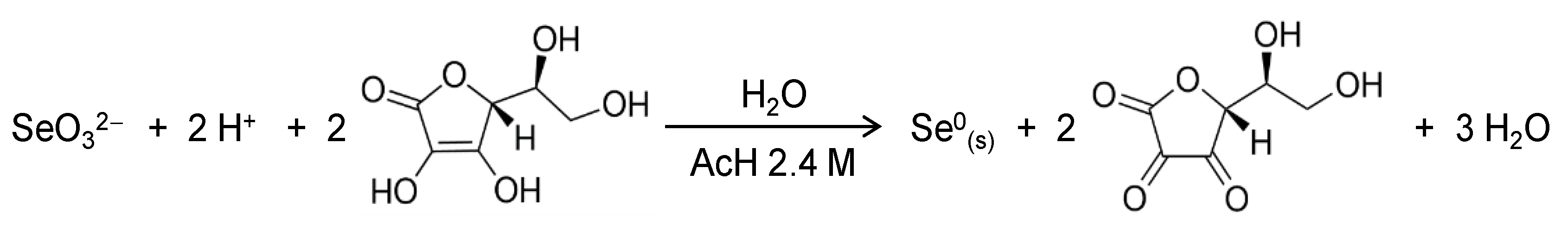
Selenium nanoparticles (SeNPs) were prepared using chemical reduction of selenite with ascorbic acid in the presence of chitosan as a soft template to control nucleation and growth of the inorganic selenium nanoparticles. Ascorbic acid acts as a reducing agent following the redox reaction (Scheme 1) in water where an acid medium is provided by acetic acid in our system [22]. The role of the polysaccharide chitosan in this redox system is to act as a stabilizer and a capping agent, affording red elemental selenium in colloidal state [23]. Furthermore, chitosan was selected because it has been shown to increase the bioavailability of SeNPs while improving the intrinsic antioxidant properties of Se [24,25].
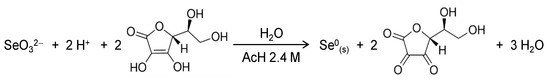
Scheme 1.
Redox reaction involved in the formation of SeNPs.
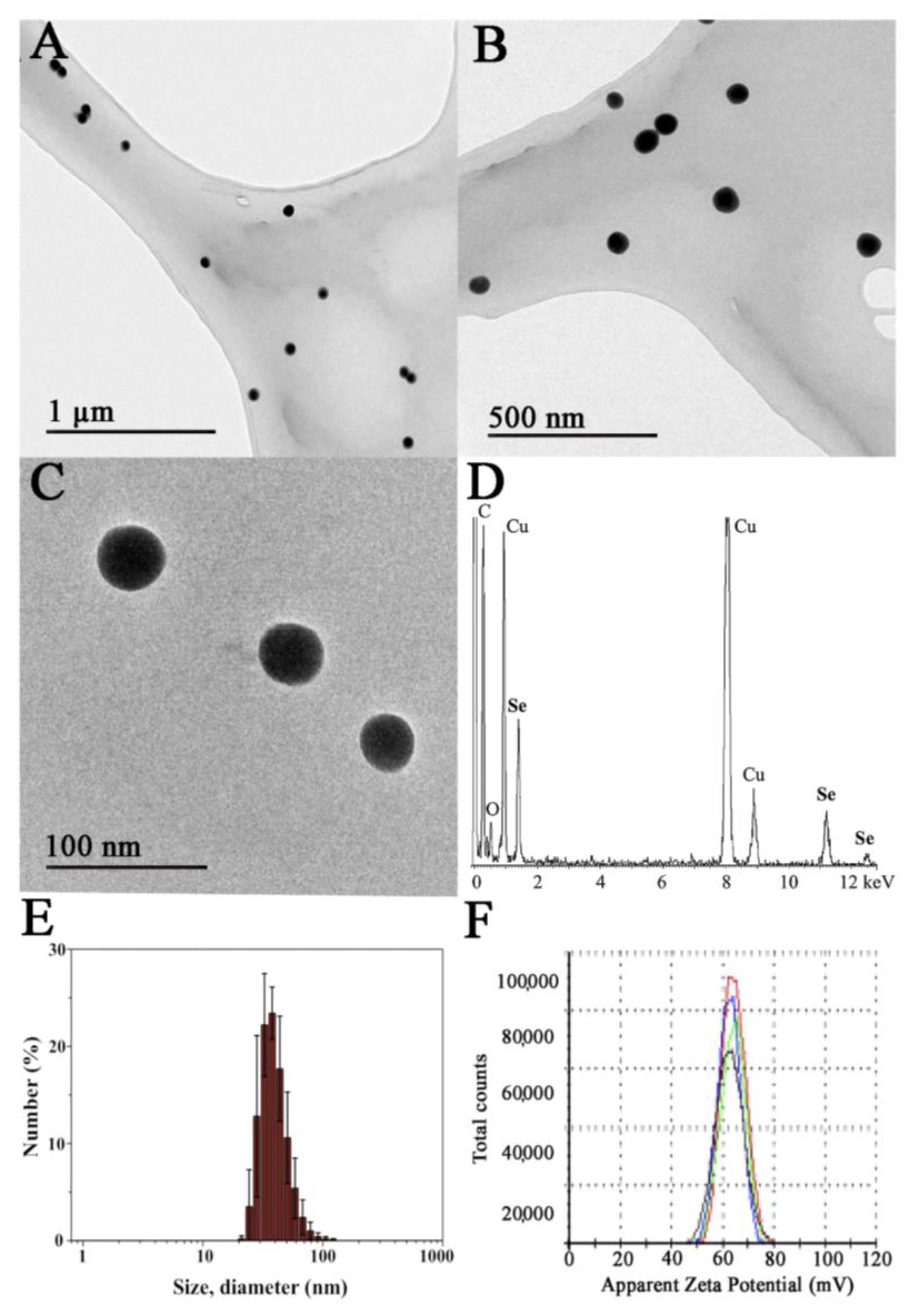
A final concentration of chitosan of 0.1% was employed to prepare the SeNPs since, under these conditions, the nanoparticles are colloidally stable and possess an adequate diameter range, as we described in previous works [11,26]. Well-dispersed nanoparticles were obtained as shown by transmission electron microscopy images, where Ch-SeNPs exhibit spherical morphology and homogenous sizes of around 40–60 nm (Figure 1A–C). Their composition in selenium was confirmed by energy dispersive spectroscopy analysis (Figure 1D). Signals for C and O from chitosan and the carbon-coated copper grid were also observed in the spectrum. The X-ray diffraction XRD pattern registered for Ch-SeNPs shows broad peaks and low signal-to-noise ratios that can be ascribed to small crystalline domain size in the range of few nanometres (see Figure S1 in the Supplementary Material).
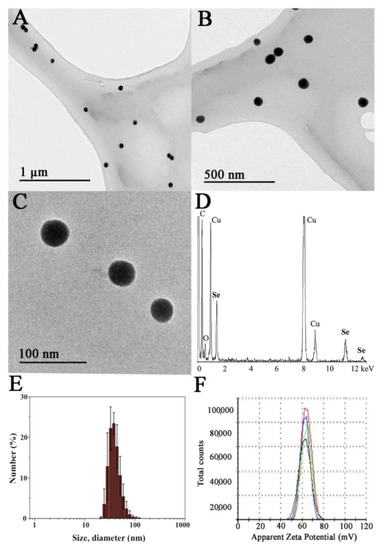
Figure 1.
Characterization of the synthesized Ch-SeNPs. TEM images of a 200 mg/L suspension of Ch-SeNPs at different magnifications (A–C). Energy-dispersive X-ray (EDS) spectrum of Ch-SeNPs (D). Hydrodynamic size distribution of the Ch-SeNPs in an aqueous colloidal suspension measured by dynamic light scattering (E) and ζ-potential measurements of the same sample (F).
The hydrodynamic diameter of the Ch-SeNPs was measured in the aqueous colloidal suspension giving a monomodal distribution in the range of ca. 25 to 100 nm, with the maximum centered at 37.8 ± 2.7 nm (Figure 1E). These values are in concordance with the size of the inorganic selenium nanoparticles observed in the TEM images.
The surface stabilization of the SeNPs with chitosan was also confirmed by means of electrophoretic mobility measurements of the Ch-SeNP colloidal suspension in water (Figure 1F). The highly positive ζ-potential value of 62.5 ± 5.2 mV falls in the zone of colloidal stability and is ascribed to the equilibrium for the protonation of the amino groups of chitosan in water. As expected, chitosan allows SeNPs to form stable colloidal suspensions due to both electrostatic as well as steric stabilization.
The synthesized Ch-SeNPs were proven to be colloidally stable, with no flocculated material observed even two months after the synthesis. No significant differences in the SeNP size or shape were observed by TEM and the hydrodynamic size distribution did not undergo a displacement of its maximum in the DLS measurements. Moreover, the possible oxidation of Ch-SeNPs to ionic selenium was also discarded. The Ch-SeNP suspension was centrifuged weekly using a 10-KDa MWCO filter and the liquid fraction was analyzed by ICP-MS. The amount of Se found in all cases was less than 0.1%, which also demonstrates good stability of the synthesized Ch-SeNPs.
3.2. Cytotoxicity of Ch-SeNPs
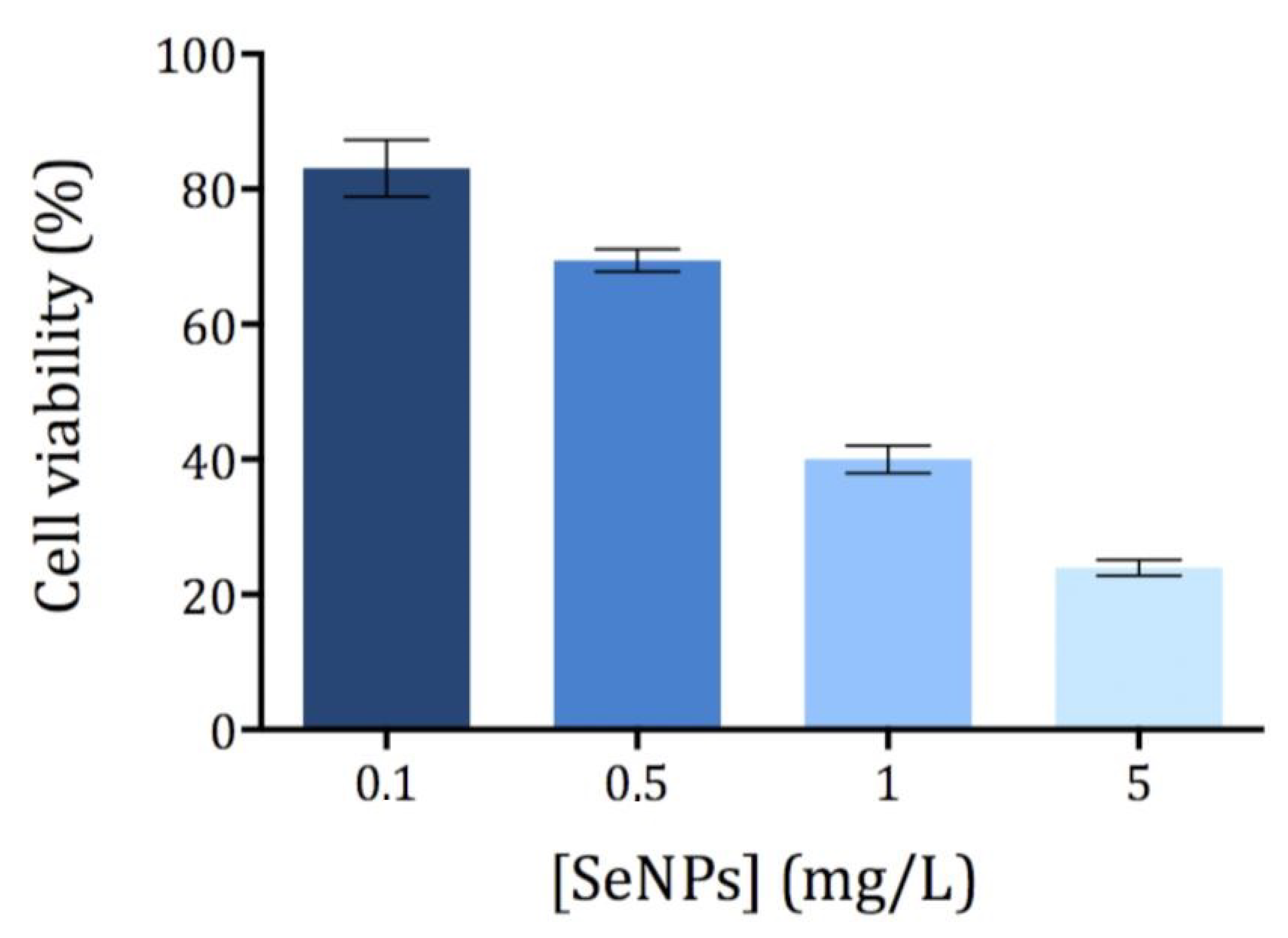
The cytotoxicity of Ch-SeNPs was evaluated by means of the MTT assay, which correlates the reduction potential of the cells with their viability. While healthy cells are able to reduce MTT to formazan (a colored compound), non-viable cells are not able to do so. Different concentrations of Ch-SeNPs ranging from 0.1 to 5 mg/L were tested using an exposure time of 72 h. All measurements showed a relative standard deviation below 10% (n = 5). The exposure time (72 h) was selected as optimal based on the previously published studies [11,17]. As expected, cell viability decreased with increasing concentrations of Ch-SeNPs (Figure 2). The cell viability of HepG2 was significantly compromised after exposure to Ch-SeNPs > 1.0 mg/L, falling below 30% at the highest concentration tested (5 mg/L). In order to investigate the molecular mechanisms involved in the potential antitumor effect exerted by Ch-SeNPs without drastically compromising the cell viability, 1 mg/L was selected for further experiments.
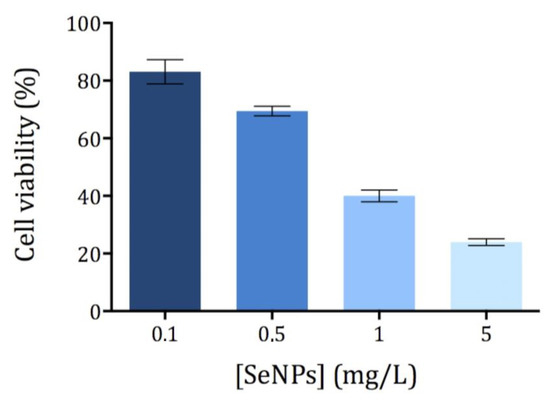
Figure 2.
Viability of the HepG2 cells exposed to Ch-SeNPs (n = 5).
3.3. Revealing the Mechanisms Associated with the Antitumor Effect of Ch-SeNPs by Transcriptomic Analysis
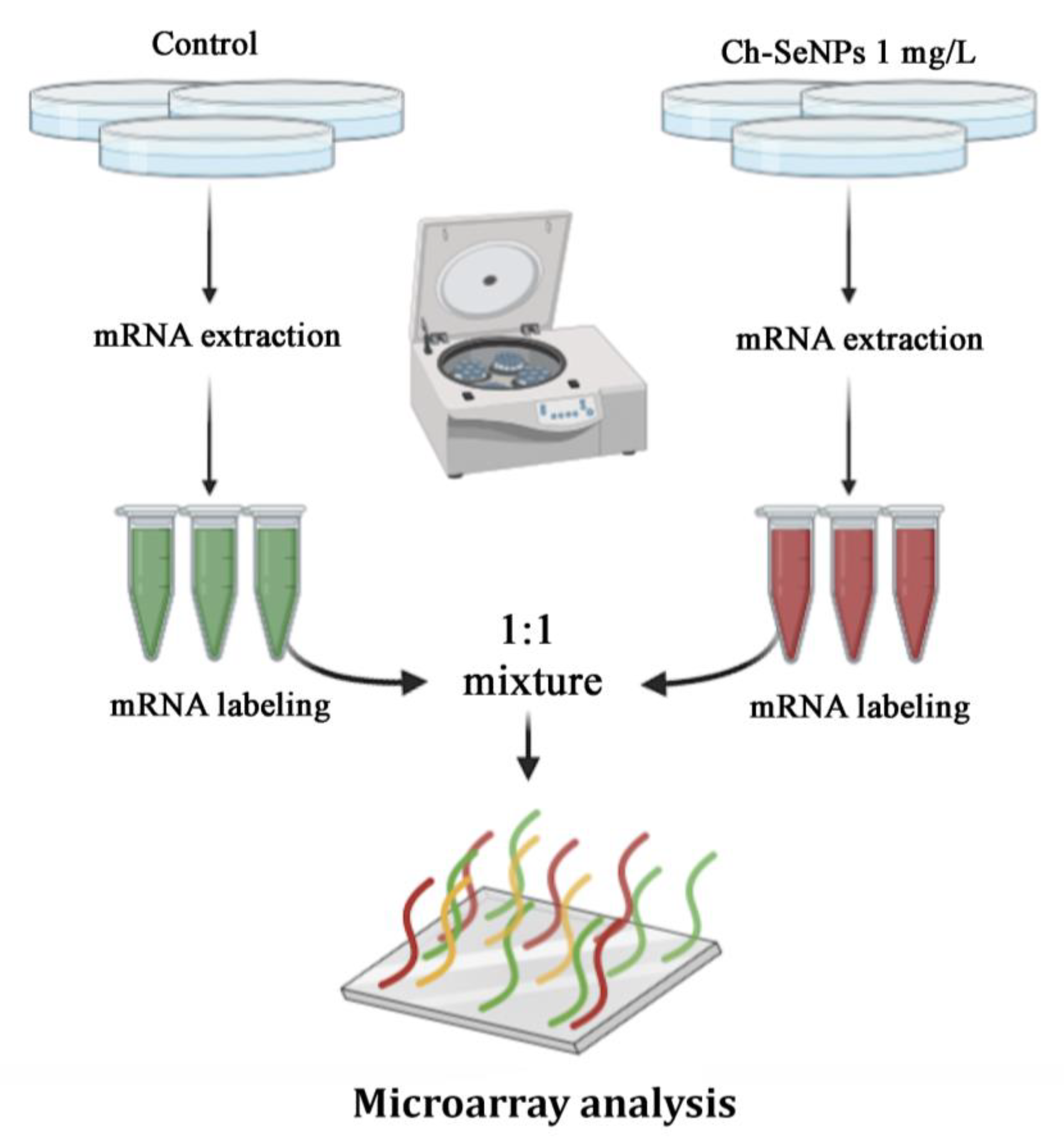
In a previous study, we confirmed the ability of Ch-SeNPs to inhibit proliferation and migration of HepG2 cells and demonstrated that Ch-SeNPs do not induce apoptosis but cell cycle arrest by partially inhibiting the expression of CDK1, which regulates the promotion of the cell cycle from the S/G2 phase to mitosis [11]. This effect was unique for Ch-SeNPs as compared to other selenospecies including Se (IV), Se (VI), SeCys2, SeMet and Se-MeSeCys [11]. Furthermore, we demonstrated that the observed effect was due exclusively to SeNPs, as the cells treated with chitosan (Ch) alone did not experience any effect compared to the control cells [17]. Since partial inhibition of CDK1 represents a way of precluding tumor cells progression, we carried out a pairwise differential gene expression analysis to compare the control cells with the cells exposed to Ch-SeNPs (Figure 3) to get a deeper insight into the molecular mechanisms underlying the potential antitumor effect of Ch-SeNPs.
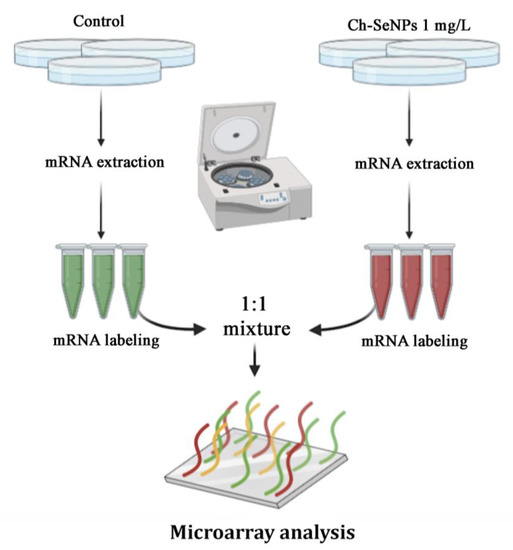
Figure 3.
General scheme of the transcriptome analysis.
Among the more than 20,000 well-annotated human genes analyzed (see Table S1 in the Supplementary Material), 279 genes were found differentially expressed with a log2 fold change of 2.0 and 0.5 (with a p-value < 0.05) (Table A1). From those differentially expressed genes, 221 were found upregulated (ratios above 2.0), while 58 were found downregulated (ratios below 0.5).
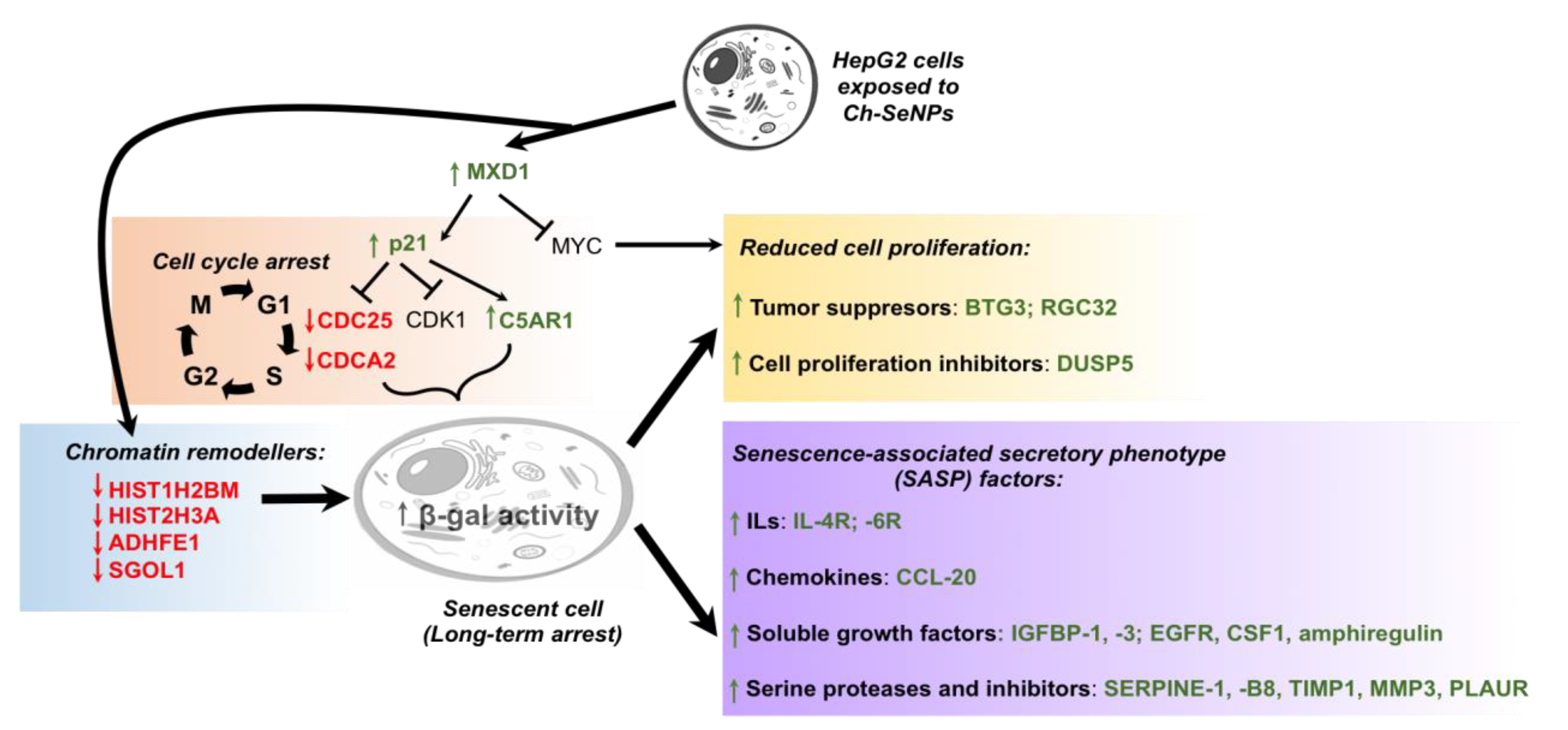
Supporting the inhibition of CDK1 found in the previous work [11], overexpression of CDKN1A (fold change (FC) = 2.67), also known as p21, was observed after Ch-SeNP exposure. This upregulation correlated with reduced expression levels of other cell cycle participant genes such as cyclin CDK1 (FC = 0.80), CDC25 (FC = 0.47) and CDCA2 (FC = 0.50) [27,28]. While CDC25 is essential for the G1–S transition associated with the activation of the cell cycle kinase cyclin E-CDK2 [27,29], knockdown of CDCA2 significantly inhibits cellular proliferation by arresting cell cycle progression at the G1 phase, effect that has also been observed when using a well-known antitumor compound such as cisplatin [30,31]. The activation of p21 upon Ch-SeNP exposure is supported by the fact that this gene, together with p15 and p27, is directly activated by MXD1, which was overexpressed in our experiment (FC = 2.42). The tumor suppressor MXD family encompasses a group of transcriptional repressors that antagonize the activation of genes mediated by oncogenic MYC [32]. Diminishing expression of angiopoietin-like 3 protein (ANGPTL3) has been correlated with reduced cellular growth and cell cycle arrest at the G1 phase [33]. Interestingly, ANGPTL3, a target downstream of p21, showed strong downregulation (FC = 0.49) upon Ch-SeNP exposure. Moreover, FAM111B, a gene, the silencing whereof induces cell cycle arrest at the G2/M phase through the p53-signaling pathway [34], was also found downregulated (FC = 0.50) in our analysis. Taken together and as expected, alteration of the genes involved in cell cycle regulation was confirmed upon Ch-SeNP exposure, thus supporting our previous studies and suggesting the cell cycle as an interesting target on which Ch-SeNPs exert their antitumor action (Figure 4).
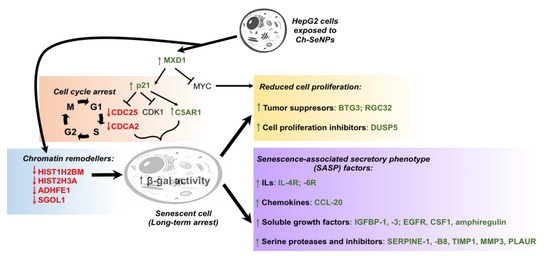
Figure 4.
Proposed mechanism leading to cell cycle arrest and senescence in the HepG2 cells exposed to Ch-SeNPs.Since senescence plays a key role in cell cycle arrest, we investigated the pattern expression of key senescence targets after exposure to Ch-SeNPs. Senescence consists of a stable cell cycle arrest in which cells become resistant to any growth stimuli, generally in response to DNA damage. This state implies several morphological and metabolic changes, chromatin reorganization, deregulations in gene expression and even production of an inflammatory phenotype known as the senescence-associated secretory phenotype (SASP) [35]. Expression patterns for some of those genes were indeed found inhibited in the cells exposed to Ch-SeNPs; such were the cases of HIST1H2BM (FC = 0.43), HIST2H3A (FC = 0.44), ADHFE1 (FC = 0.49) and SGO1 (FC = 0.42) [36,37,38]. In addition, previous studies also demonstrated that inhibition of C5AR1 can attenuate acquisition of the senescent phenotype [39]. Remarkably, C5AR1 (FC = 5.64) was found highly overexpressed in our study. This result, together with the fact that overexpression of C5AR1 has been related to the overexpression of p21 and other genes associated with the appearance of a senescent phenotype [40] such as IL-6 (FC = 2.83) and PAI-1 (FC = 7.03), also found overexpressed after Ch-SeNPs treatment, supports the potential of Ch-SeNPs to induce senescence in HepG2 cells. Particularly interesting is the overexpression of PAI-1, also known as SERPINE 1, a serine protease inhibitor that functions as the main inhibitor of the tissue plasminogen activator (tPA) (Figure 4).
Besides the effect of Ch-SeNPs on cell cycle arrest and their potential to induce senescence in HepG2 cells, well-known tumor suppressors were also found altered. That is the case of RASD1 (FC = 3.42), a key inhibitory player in glioma tumor progression [41]. Similarly, EGR1, which controls a network of suppressor gene products and is even considered a tumor suppressor itself [42], was also found significantly overexpressed (FC = 4.22). In addition, it has been described that EGR1 induces the expression of PAI-1 that, as stated above, was found clearly upregulated in the cells exposed to Ch-SeNPs [43]. Another tumor suppressor that was found overexpressed in the transcriptome analysis was CSTA (FC = 4.4), which is considered a potential biomarker for lung cancer and tumor differentiation. Although the role of CSTA in cancer has not yet been fully elucidated, Ma et al. [44] confirmed that CSTA exerts a tumor suppressive function by inhibiting the MAPK and the AKT pathways. CSTA has been found downregulated in several lung cancers and its restoration after chemotherapy is considered a sign of good prognosis [44]. DUSP5 (FC = 14.23), which is also considered a tumor suppressor, was highly overexpressed in cells treated with Ch-SeNPs. DUSP5 has also been related to the inhibition of the ERK2/MAPK pathways. Loss of DUSP5 expression has been detected in advanced gastric and prostate cancer; furthermore, the reexpression of DUSP5 in gastric cancer cell lines has been demonstrated to reduce both cell proliferation and colony-forming ability in vitro [45]. Additional tumor suppressors that were found upregulated upon Ch-SeNPs treatment were GPRC5A (FC = 5.90) and AKAP12 (FC = 8.11). Deletion of these transcripts has been demonstrated to promote tumor initiation and progression [46,47]. In addition to these overexpressed tumor suppressors, other interesting transcripts that are currently being considered as therapeutic targets of special interest against cancer and that were found downregulated after exposing HepG2 cells to Ch-SeNPs were ROR1 (FC = 0.50), PRLR (FC = 0.44), HMMR (FC = 0.40) and CENPI (FC = 0.29). While ROR1 knockdown resulted in decreased proliferation and migration but enhanced resistance to apoptosis and anoikis [48], PRLR overexpression has been linked with increased proliferation, viability, clonogenicity, chemoresistance and matrix metalloproteinase activity [49]. As for HMMR, its overexpression promotes germline stem cell self-renewal and intracranial tumor propagation [50], while upregulation of CENPI has been associated with tumorigenesis and drug resistance through chromosome instability [51]. Considering other potential therapeutic targets that have been found to be altered after exposure of HepG2 cells to Ch-SeNPs, the inhibition observed in the ALDOB transcript (FC = 0.20) is noteworthy. Metabolic reprogramming has been proposed as an alternative mechanism used by metastatic cells to obtain the high levels of energy they need. In particular, it has been described that metastatic cells from the liver upregulate the enzyme ALDOB, which provides fuel for several pathways of the central carbon metabolism, thus promoting the fructose metabolism. Based on this, targeting the expression of ALDOB could represent a powerful chemotherapeutic strategy against metastatic cells [52]. Interestingly, treatment with Ch-SeNPs was able to drastically inhibit the expression of ALDOB in HepG2 cells.
Oxidative stress is defined by the difference between the production of reactive oxygen species (ROS) and their elimination through metabolism. Hypoxia and oxidative stress are involved in cell cycle progression and proliferation, cell survival and apoptosis, energy metabolism, cell morphology, cell–cell adhesion, cell motility and angiogenesis and thereby can promote tumorigenesis [53,54]. Many chemotherapeutic strategies focus on the use of antioxidants to deplete tumor cells from ROS-induced survival signaling pathways. Such treatment may also have preventive functions. Since selenium has been considered as an antioxidant element, we sought to investigate the potential role of Ch-SeNPs as a promising candidate to be used in this type of chemotherapeutic strategies; thus, expression of the transcripts potentially involved in these mechanisms was also investigated. Our analysis showed downregulation of ANGPTL8 (FC = 0.30) and CD24 (FC = 0.48) in the cells exposed to Ch-SeNPs. While ANGPTL8 is considered an inflammatory marker [55], CD24 is also a much-highlighted marker for some types of cancers (e.g., breast cancer). Interestingly, it has been demonstrated that their suppression correlates with lower cell proliferation rates and reduced ROS production [56]. In addition, several transcripts from the p450 cytochrome (CYP) family, one of the main enzymatic sources of ROS production [57,58], were found downregulated in our assay at different levels: CYP7A1 (FC = 0.5), CYP4F2 (FC = 0.5), CYP3A7 (FC = 0.46), CYP3A5 (FC = 0.44) and CYPA43 (FC = 0.41). Furthermore, ADHFE1 (FC = 0.49) was also found inhibited. ADHFE1 is considered an oncoprotein associated with disease survival and known for promoting reduction of the glutamine metabolism while increasing formation of D-2-hydroxyglutarate and mitochondrial reactive oxygen species (ROS) [59]. The fact that Ch-SeNP exposure inhibited the expression of ADHFE1 also supports the antioxidant role of Ch-SeNPs. Similarly, the transcription factor SOX6 (FC = 0.48), which also promotes elevated levels of oxidative stress and is normally overexpressed in tumors such as the Ewing sarcoma, was found inhibited in HepG2 cells treated with Ch-SeNPs [60].
3.4. Ch-SeNPs Induce Senescence in a Preclinical Human Cell Model
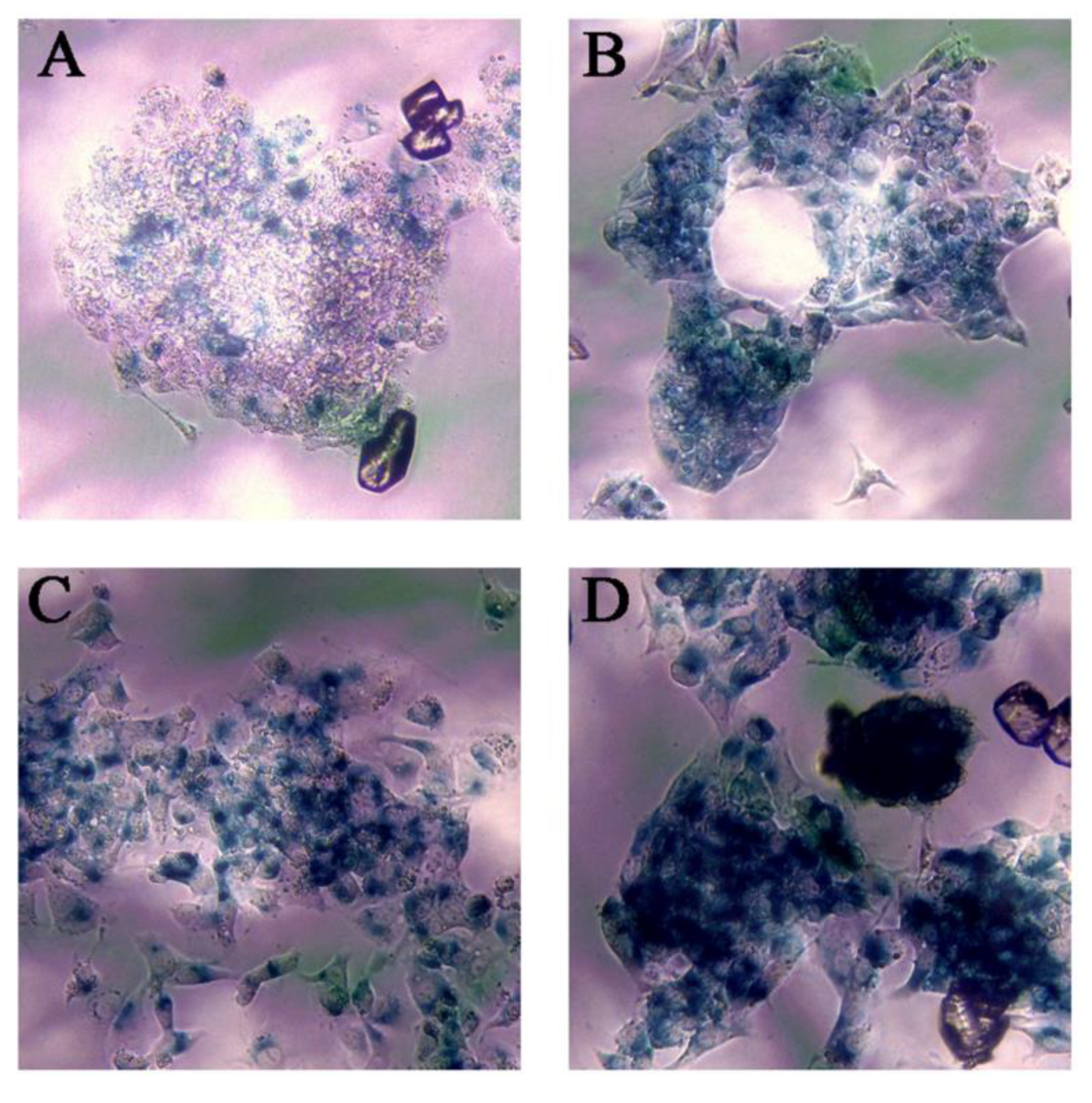
One of the aspects considered most relevant after studying the results of the transcriptomic analysis was the ability of Ch-SeNPs to induce senescence. In order to validate such interesting results, we performed an experiment to measure the activity of β-galactosidase, a well-known marker of senescence, in HepG2 cells after exposure to Ch-SeNPs (Figure 5). The results demonstrated that mock HepG2 cells showed background levels of senescence (Figure 5A), whilst the Ch-SeNP-exposed cells did show a significant expression of β-galactosidase staining (Figure 5B). These levels were similar to those observed in cells treated with 10 μM etoposide (Figure 5C), a bona fide inducer of senescence in HepG2 cells. On the contrary, cells treated with 50 μM etoposide (Figure 5D), which is a positive control for apoptosis, showed stronger staining compared to cells treated with Ch-SeNPs, thus suggesting that Ch-SeNPs induce senescence in human hepatocarcinoma cells rather than apoptosis. In order to investigate whether or not Ch-SeNPs were able to induce senescence in different cancer cell lines, additional experiments to measure the activity of β-galactosidase in MBA-MD-231 and HeLa cells (see Figures S2 and S3 in the Supplementary Material) were also carried out. In both cases, the results were comparable to those obtained for HepG2 cells. The expression levels of β-galactosidase were similar in the cells exposed to Ch-SeNPs and the positive control of senescence (10 mM etoposide).
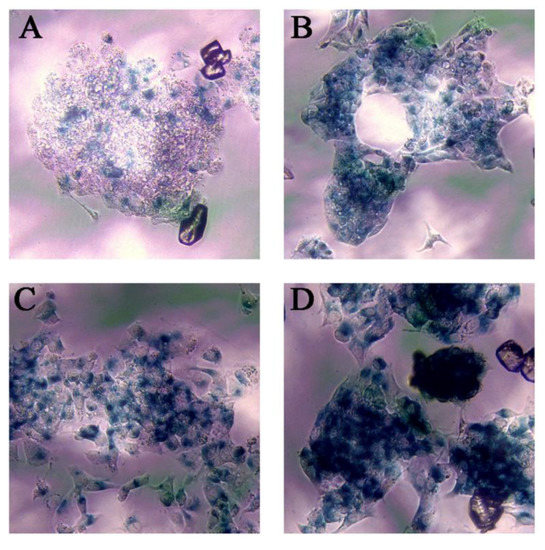
Figure 5.
Senescence assay. Blue staining shows β-galactosidase activity. Control cells (A), cells exposed to 1 mg/L of Ch-SeNPs (B), cells exposed to 10 μM of etoposide—positive senescence control (C), and cells exposed to 50 μM of etoposide—positive apoptosis control (D).
4. Conclusions
This study was carried out with the aim of identifying new mechanisms associated with the antitumor potential of the use of Ch-SeNPs. For this purpose, a model of human hepatocellular carcinoma cells (HepG2 cells) was selected and a whole transcriptome analysis was carried out. This analysis showed the alteration of different groups of transcripts involved in the arrest of the cell cycle and in the induction of senescence. Likewise, it has been proven that exposure to Ch-SeNPs induces the alteration of tumor suppressors, which supports the capacity of Ch-SeNPs to inhibit tumor proliferation. In addition, several transcripts related to ROS generation and the induction of oxidative stress were also found altered, demonstrating the antioxidant capacity of Ch-SeNPs. Overall, the set of altered transcripts allowed confirming the antitumor potential of Ch-SeNPs through their effect on several key mechanisms related to cancer progression. Especially relevant was the discovery of the capacity of Ch-SeNPs to induce senescence, which was confirmed by measuring β-galactosidase activity. Based on the promising results obtained and the proven capability of Ch-SeNPs as potential antitumor agents, the focus of future work will be on the evaluation of the effect of Ch-SeNPs in other cell lines and in pre-clinical animal models. Additionally, efforts will be made to design novel hybrid nanosystems for cancer cell targeting and selective delivery of SeNPs.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4923/13/3/356/s1, Figure S1: TEM image and XRD pattern of Ch-SeNPs, Figure S2: Senescence assay of MBA-MD-231 cells exposed to Ch-SeNPs, Figure S3: Senescence assay of HeLa cells exposed to Ch-SeNPs, Table S1: Whole transcriptome analysis of HepG2 cells exposed to Ch-SeNPs.
Author Contributions
Conceptualization, J.L.L.-G. and J.R.-T.; Data curation, H.E., E.G.-C. and B.G.; Formal analysis, E.G.-C., J.R.-T. and J.L.L.-G.; Funding acquisition, M.V.-R.; Investigation, H.E. and B.G.; Methodology, H.E.; Supervision, M.V.-R., B.G. and J.L.L.-G.; Writing—original draft, H.E. and E.G.-C.; Writing—review & editing, J.R.-T., B.G., M.V.-R. and J.L.L.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants of the Ministerio de Ciencia e Innovación (CTQ2017-85673-R) and the European Research Council (ERC-2015-AdG; VERDI; proposal No. 694160). CIBER is a public research consortium created by ISCIII whose actions are co-funded by the European Regional Development Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
H.E. thanks the Ministerio de Ciencia e Innovación for a pre-doctoral fellowship (PRE2018-084196). We would also like to thank Arancha Rivera Pedrosa for editing Figure 4.
Conflicts of Interest
Authors declare there is no conflict of interest.
Appendix A

Table A1.
Differentially expressed transcripts in the HepG2 cells exposed to Ch-SeNPs.
Table A1.
Differentially expressed transcripts in the HepG2 cells exposed to Ch-SeNPs.
| Fold Change (FC) | Gene Code | Gene Name |
|---|---|---|
| 14.23 | DUSP5 | dual specificity phosphatase 5 |
| 13.90 | RGCC | regulator of cell cycle |
| 9.70 | PAQR5 | progestin and AdipoQ receptor family member V |
| 9.01 | TMC7 | transmembrane channel-like 7 |
| 8.82 | AQP3 | aquaporin 3 (Gill blood group) |
| 8.11 | AKAP12 | A-kinase (PRKA) anchor protein 12 |
| 7.11 | PALLD | palladin, cytoskeletal associated protein |
| 7.03 | PAI-1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| 5.96 | LURAP1L | leucine-rich adaptor protein 1-like |
| 5.90 | CSGALNACT2 | chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| 5.90 | GPRC5A; MIR614 | G protein-coupled receptor, class C, group 5, member A; microRNA 614 |
| 5.64 | C5AR1 | complement component 5a receptor 1 |
| 5.42 | EMP3 | epithelial membrane protein 3 |
| 5.39 | GPAT3 | glycerol-3-phosphate acyltransferase 3 |
| 5.19 | TM4SF19-TCTEX1D2 | TM4SF19-TCTEX1D2 readthrough (NMD candidate) |
| 5.02 | RBM24 | RNA-binding motif protein 24 |
| 4.88 | TM4SF19 | transmembrane 4 L six family member 19 |
| 4.88 | SERPINB8 | serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| 4.82 | IER3 | immediate early response 3 |
| 4.75 | PDGFA | platelet-derived growth factor alpha polypeptide |
| 4.65 | MMP3 | matrix metallopeptidase 3 |
| 4.40 | CSTA | cystatin A (stefin A) |
| 4.38 | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| 4.31 | CAPN2 | calpain 2, (m/II) large subunit |
| 4.25 | SLC51B | solute carrier family 51, beta subunit |
| 4.22 | EGR1 | early growth response 1 |
| 4.15 | SLC16A6 | solute carrier family 16, member 6 |
| 4.11 | JAG1 | jagged 1 |
| 4.09 | ESAM | endothelial cell adhesion molecule |
| 4.09 | AXL | AXL receptor tyrosine kinase |
| 4.04 | PAEP | progestogen-associated endometrial protein |
| 3.95 | HEY1 | hes-related family bHLH transcription factor with YRPW motif 1 |
| 3.93 | FHL2 | four and a half LIM domains 2 |
| 3.88 | TIMP1 | TIMP metallopeptidase inhibitor 1 |
| 3.85 | IGFBP3 | insulin-like growth factor binding protein 3 |
| 3.69 | SPRY4 | sprouty RTK signaling antagonist 4 |
| 3.65 | F2RL1 | coagulation factor II (thrombin) receptor-like 1 |
| 3.56 | ITPR3 | inositol 1,4,5-trisphosphate receptor, type 3 |
| 3.49 | PLEKHH2 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| 3.43 | S100A11 | S100 calcium-binding protein A11 |
| 3.42 | RASD1 | RAS, dexamethasone-induced 1 |
| 3.38 | TUSC3 | tumor suppressor candidate 3 |
| 3.37 | CD55 | CD55 molecule, decay-accelerating factor for complement (Cromer blood group) |
| 3.36 | HKDC1 | hexokinase domain containing 1 |
| 3.35 | SPIRE1 | spire-type actin nucleation factor 1 |
| 3.32 | ARG2 | arginase 2 |
| 3.31 | NKAP | NFKB-activating protein |
| 3.26 | GDF15 | growth differentiation factor 15 |
| 3.25 | ACSL5 | acyl-CoA synthetase long-chain family member 5 |
| 3.24 | AREG | amphiregulin |
| 3.23 | EGFR | epidermal growth factor receptor |
| 3.21 | SLC1A2 | solute carrier family 1 (glial high-affinity glutamate transporter), member 2 |
| 3.21 | ARHGEF2 | Rho/Rac guanine nucleotide exchange factor 2 |
| 3.18 | SFN | stratifin |
| 3.13 | IGFBP1 | insulin-like growth factor-binding protein 1 |
| 3.09 | LGALS3 | lectin, galactoside-binding, soluble, 3 |
| 3.07 | ID1 | inhibitor of DNA binding 1, dominant negative helix–loop–helix protein |
| 3.06 | SRGAP1 | SLIT-ROBO Rho GTPase-activating protein 1 |
| 3.05 | NTSR1 | neurotensin receptor 1 (high affinity) |
| 3.05 | ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) |
| 3.00 | CACNA2D4 | calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| 2.99 | HPCAL1 | hippocalcin-like 1 |
| 2.99 | GTPBP2 | GTP-binding protein 2 |
| 2.95 | MCTP1 | multiple C2 domains, transmembrane 1 |
| 2.93 | TCP11L2 | t-complex 11, testis-specific-like 2 |
| 2.92 | SH3RF1 | SH3 domain containing ring finger 1 |
| 2.91 | FOSL1 | FOS-like antigen 1 |
| 2.90 | ATP6V0D2 | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| 2.87 | DYNC2H1 | dynein, cytoplasmic 2, heavy-chain 1 |
| 2.85 | CLIP4 | CAP-GLY domain-containing linker protein family, member 4 |
| 2.85 | RAP1GAP2 | RAP1 GTPase-activating protein 2 |
| 2.83 | SERPINE2 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| 2.83 | IL6R | interleukin 6 receptor |
| 2.81 | KLF5 | Kruppel-like factor 5 (intestinal) |
| 2.80 | NDE1; MIR484 | nudE neurodevelopment protein 1; microRNA 484 |
| 2.80 | ATF6 | activating transcription factor 6 |
| 2.78 | CCPG1; MIR628 | cell cycle progression 1; microRNA 628 |
| 2.78 | ARAP2 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| 2.77 | TRIB1 | tribbles pseudokinase 1 |
| 2.74 | ERRFI1 | ERBB receptor feedback inhibitor 1 |
| 2.74 | GCLC | glutamate-cysteine ligase, catalytic subunit |
| 2.73 | SGMS2 | sphingomyelin synthase 2 |
| 2.73 | CD22; MIR5196 | CD22 molecule; microRNA 5196 |
| 2.71 | MT2A | metallothionein 2A |
| 2.69 | GLIPR1 | GLI pathogenesis-related 1 |
| 2.67 | CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| 2.67 | CEMIP | cell migration inducing protein, hyaluronan-binding |
| 2.66 | TGFB1 | transforming growth factor beta 1 |
| 2.65 | CD58 | CD58 molecule |
| 2.63 | MEP1A | meprin A, alpha (PABA peptide hydrolase) |
| 2.62 | BHLHE40 | basic helix–loop–helix family, member e40 |
| 2.62 | TIMM9 | translocase of inner mitochondrial membrane 9 homolog (yeast) |
| 2.62 | BMP6 | bone morphogenetic protein 6 |
| 2.62 | IL4R | interleukin 4 receptor |
| 2.60 | TMEM2 | transcript identified by AceView, Entrez Gene ID(s) 23670 |
| 2.58 | DMRTA1 | DMRT-like family A1 |
| 2.58 | ANKRD1 | ankyrin repeat domain 1 (cardiac muscle) |
| 2.55 | MT1B; MT1CP | metallothionein 1B; metallothionein 1C, pseudogene |
| 2.51 | AKR1B10 | aldo-keto reductase family 1, member B10 (aldose reductase) |
| 2.49 | CD109 | CD109 molecule |
| 2.48 | SLC20A1 | solute carrier family 20 (phosphate transporter), member 1 |
| 2.46 | ATF3 | activating transcription factor 3 |
| 2.46 | TAGLN3 | transgelin 3 |
| 2.45 | MOSPD1 | motile sperm domain-containing 1 |
| 2.45 | IL11 | interleukin 11 |
| 2.43 | KDM7A | lysine (K)-specific demethylase 7A |
| 2.42 | CCR7 | chemokine (C-C motif) receptor 7 |
| 2.42 | MXD1 | MAX dimerization protein 1 |
| 2.40 | PITPNC1 | phosphatidylinositol transfer protein, cytoplasmic 1 |
| 2.40 | PLAUR | plasminogen activator, urokinase receptor |
| 2.40 | RABGGTB; SNORD45B; SNORD45A; SNORD45C | Rab geranylgeranyltransferase, beta subunit; small nucleolar RNA, C/D box 45B; small nucleolar RNA, C/D box 45A; small nucleolar RNA, C/D box 45C |
| 2.39 | LETM2 | leucine zipper-EF-hand-containing transmembrane protein 2 |
| 2.36 | C6orf48; SNORD52; SNORD48 | chromosome 6 open reading frame 48; small nucleolar RNA, C/D box 52; small nucleolar RNA, C/D box 48 |
| 2.36 | AP1S3 | adaptor-related protein complex 1 sigma 3 subunit |
| 2.36 | PPP1R15A | protein phosphatase 1, regulatory subunit 15A |
| 2.36 | ARL8A | ADP-ribosylation factor like GTPase 8A |
| 2.36 | PAQR3 | progestin and AdipoQ receptor family member III |
| 2.35 | LCP1 | lymphocyte cytosolic protein 1 (L-plastin) |
| 2.34 | HMGA1 | high-mobility group AT-hook 1 |
| 2.33 | RCL1 | RNA terminal phosphate cyclase-like 1 |
| 2.33 | ANTXR2 | anthrax toxin receptor 2 |
| 2.31 | KPNA5 | karyopherin alpha 5 (importin alpha 6) |
| 2.30 | RRAS2 | related RAS viral (r-ras) oncogene homolog 2 |
| 2.30 | CCL20 | chemokine (C–C motif) ligand 20 |
| 2.30 | TUBE1 | tubulin, epsilon 1 |
| 2.30 | LRRC8B | leucine-rich repeat-containing 8 family, member B |
| 2.29 | DFNA5 | deafness, autosomal dominant 5 |
| 2.28 | ERN1 | endoplasmic reticulum to nucleus signaling 1 |
| 2.28 | ZADH2 | zinc binding alcohol dehydrogenase domain-containing 2 |
| 2.27 | PPP1R18 | protein phosphatase 1, regulatory subunit 18 |
| 2.26 | ASB2 | ankyrin repeat and SOCS box-containing 2 |
| 2.26 | IDS | iduronate 2-sulfatase |
| 2.25 | MAGI2 | membrane-associated guanylate kinase, WW and PDZ domain-containing 2 |
| 2.25 | MT1A | metallothionein 1A |
| 2.25 | ANXA3 | annexin A3 |
| 2.25 | ZAK | sterile alpha motif and leucine zipper-containing kinase |
| 2.24 | CSF1 | colony-stimulating factor 1 (macrophage) |
| 2.24 | PIM1 | Pim-1 proto-oncogene, serine/threonine kinase |
| 2.23 | MSANTD3 | Myb/SANT-like DNA-binding domain containing 3 |
| 2.22 | NAGS | N-acetylglutamate synthase |
| 2.22 | REXO2 | RNA exonuclease 2 |
| 2.22 | VASP | vasodilator-stimulated phosphoprotein |
| 2.21 | CREB5 | cAMP responsive element-binding protein 5 |
| 2.21 | STEAP2 | STEAP family member 2, metalloreductase |
| 2.21 | MRGPRX4 | MAS-related GPR, member X4 |
| 2.20 | ITPRIP | inositol 1,4,5-trisphosphate receptor-interacting protein |
| 2.20 | DIEXF | digestive organ expansion factor homolog (zebrafish) |
| 2.20 | MT1X | metallothionein 1X |
| 2.19 | PTGR1 | prostaglandin reductase 1 |
| 2.19 | C9orf72 | chromosome 9 open reading frame 72 |
| 2.19 | ABR | active BCR-related |
| 2.18 | HIVEP2 | human immunodeficiency virus type I enhancer-binding protein 2 |
| 2.18 | KLF11 | Kruppel-like factor 11 |
| 2.18 | VNN1 | Vanin 1 |
| 2.17 | LAPTM5 | lysosomal protein transmembrane 5 |
| 2.17 | MT1IP | metallothionein 1I, pseudogene |
| 2.17 | CAMSAP2 | calmodulin regulated spectrin-associated protein family, member 2 |
| 2.16 | DPY19L4 | dpy-19-like 4 (C. elegans) |
| 2.16 | SLC22A15 | solute carrier family 22, member 15 |
| 2.16 | PTPN3 | protein tyrosine phosphatase, non-receptor type 3 |
| 2.16 | SPP1 | secreted phosphoprotein 1 |
| 2.16 | NLRC4 | NLR family, CARD domain-containing 4 |
| 2.16 | CMSS1 | cms1 ribosomal small subunit homolog (yeast) |
| 2.15 | HBEGF | heparin-binding EGF-like growth factor |
| 2.15 | TGM2 | transglutaminase 2 |
| 2.15 | NLN | neurolysin (metallopeptidase M3 family) |
| 2.14 | APOPT1 | apoptogenic 1, mitochondrial |
| 2.14 | UPP1 | uridine phosphorylase 1 |
| 2.13 | LAMP3 | lysosomal-associated membrane protein 3 |
| 2.13 | C5orf28 | chromosome 5 open reading frame 28 |
| 2.13 | P2RX5-TAX1BP3 | P2RX5-TAX1BP3 readthrough (NMD candidate) |
| 2.13 | PHF21A | PHD finger protein 21A |
| 2.12 | CTSB | cathepsin B |
| 2.12 | S100P | S100 calcium-binding protein P |
| 2.11 | ANKRA2 | ankyrin repeat, family A (RFXANK-like), 2 |
| 2.11 | FLOT1 | transcript identified by AceView, Entrez Gene ID(s) 10211 |
| 2.11 | CEP290 | centrosomal protein 290kDa |
| 2.11 | MT1H | metallothionein 1H |
| 2.10 | RIT1 | Ras-like without CAAX 1 |
| 2.09 | CEP295NL; TIMP2 | CEP295 N-terminal like; TIMP metallopeptidase inhibitor 2 |
| 2.09 | CES1 | carboxylesterase 1 |
| 2.09 | ASF1A | anti-silencing function 1A histone chaperone |
| 2.09 | OPTN | optineurin |
| 2.09 | GRB10 | growth factor receptor-bound protein 10 |
| 2.09 | AKR1B15 | aldo-keto reductase family 1, member B15 |
| 2.08 | MTMR6 | myotubularin-related protein 6 |
| 2.08 | FAN1 | FANCD2/FANCI-associated nuclease 1 |
| 2.08 | ACTR10 | actin-related protein 10 homolog (S. cerevisiae) |
| 2.08 | RNF19B | ring finger protein 19B |
| 2.08 | NFIL3 | nuclear factor, interleukin 3-regulated |
| 2.08 | NQO1 | NAD(P)H dehydrogenase, quinone 1 |
| 2.08 | DHRS7 | dehydrogenase/reductase (SDR family) member 7 |
| 2.08 | MT1L | metallothionein 1L (gene/pseudogene) |
| 2.07 | LIF | leukemia inhibitory factor |
| 2.07 | DUSP12 | dual specificity phosphatase 12 |
| 2.07 | KCNMB3 | potassium channel subfamily M regulatory beta subunit 3 |
| 2.06 | PXK | PX domain-containing serine/threonine kinase |
| 2.06 | CD9 | CD9 molecule |
| 2.06 | H1F0 | H1 histone family, member 0 |
| 2.06 | ADORA2B | adenosine A2b receptor |
| 2.06 | KRT23 | keratin 23, type I |
| 2.06 | BTG3 | BTG family, member 3 |
| 2.05 | AOX1 | aldehyde oxidase 1 |
| 2.04 | SNORA17A; SNORA17B; SNHG7 | small nucleolar RNA, H/ACA box 17A; small nucleolar RNA, H/ACA box 17B; small nucleolar RNA host gene 7 |
| 2.04 | CPNE8 | copine VIII |
| 2.04 | EIF4A2; SNORA63; SNORD2; SNORA4; SNORA81; MIR1248 | eukaryotic translation initiation factor 4A2; small nucleolar RNA, H/ACA box 63; small nucleolar RNA, C/D box 2; small nucleolar RNA, H/ACA box 4; small nucleolar RNA, H/ACA box 81; microRNA 1248 |
| 2.03 | SMAD6 | SMAD family member 6 |
| 2.03 | YIPF4 | Yip1 domain family member 4 |
| 2.03 | FASTKD1 | FAST kinase domains 1 |
| 2.03 | TMTC3 | transmembrane and tetratricopeptide repeat containing 3 |
| 2.02 | ADAT2 | adenosine deaminase, tRNA-specific 2 |
| 2.02 | ago-02 | argonaute RISC catalytic component 2 |
| 2.02 | TMEM167B | transmembrane protein 167B |
| 2.01 | DCAF10 | DDB1 and CUL4 associated factor 10 |
| 2.01 | RAB3GAP1 | RAB3 GTPase-activating protein subunit 1 (catalytic) |
| 2.00 | GBE1 | glucan (1,4-alpha-), branching enzyme 1 |
| 2.00 | CLIP2 | CAP-GLY domain-containing linker protein 2 |
| 2.00 | SOWAHC | sosondowah ankyrin repeat domain family member C |
| 2.00 | NEK3 | NIMA-related kinase 3 |
| 2.00 | IFRD1 | interferon-related developmental regulator 1 |
| 2.00 | TBPL1 | TBP-like 1 |
| 0.50 | ROR1 | receptor tyrosine kinase-like orphan receptor 1 |
| 0.50 | FAM111B | family with sequence similarity 111, member B |
| 0.50 | CDCA2 | cell division cycle-associated 2 |
| 0.50 | DIO1 | deiodinase, iodothyronine, type I |
| 0.50 | HIST1H1B | histone cluster 1, H1b |
| 0.50 | HOOK2 | hook microtubule-tethering protein 2 |
| 0.50 | CYP7A1 | cytochrome P450, family 7, subfamily A, polypeptide 1 |
| 0.50 | CYP4F2 | cytochrome P450, family 4, subfamily F, polypeptide 2 |
| 0.50 | PNPLA3 | patatin-like phospholipase domain-containing 3 |
| 0.49 | TSACC | TSSK6-activating co-chaperone |
| 0.49 | HIST1H4A | histone cluster 1, H4a |
| 0.49 | ANGPTL3 | angiopoietin-like 3 |
| 0.49 | ADHFE1; C8orf46 | alcohol dehydrogenase, iron-containing 1; chromosome 8 open reading frame 46 |
| 0.49 | GINS2 | GINS complex subunit 2 (Psf2 homolog) |
| 0.49 | ARHGEF39 | Rho guanine nucleotide exchange factor 39 |
| 0.49 | POTEF | POTE ankyrin domain family, member F |
| 0.49 | HIST1H2AG | histone cluster 1, H2ag |
| 0.49 | ZNF341 | zinc finger protein 341 |
| 0.49 | NAT6 | N-acetyltransferase 6 (GCN5-related) |
| 0.48 | FRY | FRY microtubule-binding protein |
| 0.48 | SOX6; MIR6073 | SRY box 6; microRNA 6073 |
| 0.48 | SGOL2 | shugoshin-like 2 (S. pombe) |
| 0.48 | MTHFR | methylenetetrahydrofolate reductase (NAD(P)H) |
| 0.48 | CD24 | CD24 molecule |
| 0.47 | ITIH1 | inter-alpha-trypsin inhibitor heavy-chain 1 |
| 0.47 | SCARA3 | scavenger receptor class A, member 3 |
| 0.47 | TMEM143 | transmembrane protein 143 |
| 0.47 | CDC25C | cell division cycle 25C |
| 0.46 | LEAP2 | liver-expressed antimicrobial peptide 2 |
| 0.46 | ZNF565 | zinc finger protein 565 |
| 0.46 | E2F8 | E2F transcription factor 8 |
| 0.46 | CYP3A7; CYP3A7-CYP3A51P | cytochrome P450, family 3, subfamily A, polypeptide 7; CYP3A7-CYP3A51P readthrough |
| 0.45 | ESCO2 | establishment of sister chromatid cohesion N-acetyltransferase 2 |
| 0.45 | ODAM | odontogenic, ameloblast-associated |
| 0.45 | C1orf116 | chromosome 1 open reading frame 116 |
| 0.44 | HIST2H3A | histone cluster 2, H3a |
| 0.44 | CHGB | chromogranin B |
| 0.44 | PRLR | prolactin receptor |
| 0.44 | HIST2H3A; HIST2H3C | histone cluster 2, H3a; histone cluster 2, H3c |
| 0.44 | CYP3A5 | cytochrome P450, family 3, subfamily A, polypeptide 5 |
| 0.43 | HIST1H2BM | histone cluster 1, H2bm |
| 0.43 | SNAI2 | snail family zinc finger 2 |
| 0.43 | PRRG2 | proline-rich Gla (G-carboxyglutamic acid) 2 |
| 0.42 | SGOL1 | shugoshin-like 1 (S. pombe) |
| 0.42 | SLC22A7 | solute carrier family 22 (organic anion transporter), member 7 |
| 0.41 | CYP3A43 | cytochrome P450, family 3, subfamily A, polypeptide 43 |
| 0.41 | LINC00612 | long intergenic non-protein coding RNA 612 |
| 0.41 | IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 |
| 0.40 | HMMR | hyaluronan-mediated motility receptor (RHAMM) |
| 0.39 | CDH1 | cadherin 1, type 1 |
| 0.39 | HIST1H3B | histone cluster 1, H3b |
| 0.38 | AFM | afamin |
| 0.36 | DEPDC4 | DEP domain-containing 4 |
| 0.36 | YPEL2 | yippee like 2 |
| 0.32 | G6PC | glucose-6-phosphatase, catalytic subunit |
| 0.30 | ANGPTL8 | angiopoietin like 8 |
| 0.29 | CENPI | centromere protein I |
| 0.20 | ALDOB | aldolase B, fructose-bisphosphate |
References
- Sreevalsan, S.; Safe, S. Reactive oxygen species and colorectal cancer. Curr. Color. Cancer Rep. 2013, 9, 350–357. [Google Scholar] [CrossRef]
- Marinescu, S.; Anghel, R.; Gruia, M.I.; Beuran, M. Involvement of reactive oxygen species in the mechanisms associated with cervical cancer specific treatment. Chirurgia 2015, 109, 806–811. [Google Scholar]
- Thyagarajan-Sahu, A.; Sahu, R.P. Potential contributions of antioxidants to cancer therapy: Immunomodulation and radiosensitization. Integr. Cancer Ther. 2018, 17, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Nunes, R.; Das Neves, J.; Sarmento, B. Nanoparticles for the regulation of intestinal inflammation: Opportunities and challenges. Nanomedicine 2019, 14, 2631–2644. [Google Scholar] [CrossRef]
- Tapeinos, C.; Battaglini, M.; Ciofani, G. Advances in the design of solid lipid nanoparticles and nanostructured lipid carriers for targeting brain diseases. J. Control. Release 2017, 264, 306–332. [Google Scholar] [CrossRef]
- Machuca, A.; Garcia-Calvo, E.; Anunciação, D.S.; Garcia, J.L.L. Rhodium nanoparticles as a novel photosensitizing agent in photodynamic therapy against cancer. Chem. A Eur. J. 2020, 26, 7685–7691. [Google Scholar] [CrossRef]
- Montalvo-Quiros, S.; Aragoneses-Cazorla, G.; Garcia-Alcalde, L.; Vallet-Regí, M.; González, B.; Luque-Garcia, J.L. Cancer cell targeting and therapeutic delivery of silver nanoparticles by mesoporous silica nanocarriers: Insights into the action mechanisms using quantitative proteomics. Nanoscale 2019, 11, 4531–4545. [Google Scholar] [CrossRef] [PubMed]
- Oueslati, M.H.; Ben Tahar, L.; Harrath, A.H. Catalytic, antioxidant and anticancer activities of gold nanoparticles synthesized by kaempferol glucoside from Lotus leguminosae. Arab. J. Chem. 2020, 13, 3112–3122. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Del Giovane, C.; Dennert, G.; Zwahlen, M.; Brinkman, M.; Zeegers, M.P.; Horneber, M.; D’Amico, R.; Crespi, C.M. Selenium for preventing cancer. Cochrane Database Syst. Rev. 2018, 1, CD005195. [Google Scholar] [CrossRef] [PubMed]
- Aboul-Fadl, T. Selenium derivatives as cancer preventive agents. Curr. Med. Chem. Agents 2005, 5, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Geoffrion, L.D.; Hesabizadeh, T.; Medina-Cruz, D.; Kusper, M.; Taylor, P.; Vernet-Crua, A.; Chen, J.; Ajo, A.; Webster, T.J.; Guisbiers, G. Naked selenium nanoparticles for antibacterial and anticancer treatments. ACS Omega 2020, 5, 2660–2669. [Google Scholar] [CrossRef]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloids Surf. B Biointerfaces 2014, 122, 184–193. [Google Scholar] [CrossRef]
- Malekifard, F.; Tavassoli, M.; Vaziri, K. In vitro assessment antiparasitic effect of selenium and copper nanoparticles on Giardia deodenalis cyst. Iran. J. Parasitol. 2020, 15, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Liu, L.; Tao, W.; Wang, G.; Sun, W.; Pei, X.; Xiao, Z.; Jin, Y.; Wang, M. Prooxidation and cytotoxicity of selenium nanoparticles at nonlethal level in Sprague-Dawley rats and Buffalo rat liver cells. Oxidative Med. Cell. Longev. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Toubhans, B.; Gazze, S.A.; Bissardon, C.; Bohic, S.; Gourlan, A.T.; Gonzalez, D.; Charlet, L.; Conlan, R.S.; Francis, L.W. Selenium nanoparticles trigger alterations in ovarian cancer cell biomechanics. Nanomed. Nanotechnol. Biol. Med. 2020, 29, 102258. [Google Scholar] [CrossRef]
- Zhao, G.; Dong, R.; Teng, J.; Yang, L.; Liu, T.; Wu, X.; He, Y.; Wang, Z.; Pu, H.; Wang, Y. N-acetyl-l-cysteine enhances the effect of selenium nanoparticles on cancer cytotoxicity by increasing the production of selenium-induced reactive oxygen species. ACS Omega 2020, 5, 11710–11720. [Google Scholar] [CrossRef]
- Hussein, H.-A.A.; Darwesh, O.M.; Mekki, B.B.; El-Hallouty, S.M. Evaluation of cytotoxicity, biochemical profile and yield components of groundnut plants treated with nano-selenium. Biotechnol. Rep. 2019, 24, e00377. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Heras, I.; Sanchez-Diaz, R.; Anunciacao, D.S.; Madrid, Y.; Luque-Garcia, J.L.; Camara, C. Effect of chitosan-stabilized selenium nanoparticles on cell cycle arrest and invasiveness in hepatocarcinoma cells revealed by quantitative proteomics. J. Nanomed. Nanotechnol. 2014, 5. [Google Scholar] [CrossRef]
- Neagu, M.; Boda, D. Transcriptomics in cancer—Stages toward patents in biomarkers? Recent Pat. Biomark. 2012, 2, 75–82. [Google Scholar] [CrossRef]
- Alkhateeb, A.; Rezaeian, I.; Singireddy, S.; Cavallo-Medved, D.; Porter, L.A.; Rueda, L. transcriptomics signature from next-generation sequencing data reveals new transcriptomic biomarkers related to prostate cancer. Cancer Inform. 2019, 18. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, Y.; Zhou, Y.; Li, W.; Zheng, W. Modification and modulation of saccharides on elemental selenium nanoparticles in liquid phase. Mater. Lett. 2008, 62, 2311–2314. [Google Scholar] [CrossRef]
- Mees, D.R.; Pysto, W.; Tarcha, P.J. Formation of selenium colloids using sodium ascorbate as the reducing agent. J. Colloid Interface Sci. 1995, 170, 254–260. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Zhang, L. Creation of highly stable selenium nanoparticles capped with hyperbranched polysaccharide in water. Langmuir 2010, 26, 17617–17623. [Google Scholar] [CrossRef]
- Zhang, S.; Luo, Y.; Zeng, H.; Wang, Q.; Tian, F.; Song, J.; Cheng, W.-H. Encapsulation of selenium in chitosan nanoparticles improves selenium availability and protects cells from selenium-induced DNA damage response. J. Nutr. Biochem. 2011, 22, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Zhang, C.; Zhao, G.; Stoll, S.; Ren, F.; Leng, X. Antioxidant capacities of the selenium nanoparticles stabilized by chitosan. J. Nanobiotechnol. 2017, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Estevez, H.; Palacios, A.; Gil, D.; Anguita, J.; Vallet-Regi, M.; González, B.; Prados-Rosales, R.; Luque-Garcia, J.L. Antimycobacterial effect of selenium nanoparticles on mycobacterium tuberculosis. Front. Microbiol. 2020, 11, 800. [Google Scholar] [CrossRef] [PubMed]
- Charrier-Savournin, F.B.; Château, M.-T.; Gire, V.; Sedivy, J.; Piette, J.; Dulić, V. p21-mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol. Biol. Cell 2004, 15, 3965–3976. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Eichbaum, Q.; Silberman, E.D.; Mayer, B.J.; Dutta, A. p21CIP1 and Cdc25A: Competition between an inhibitor and an activator of cyclin-dependent kinases. Mol. Cell. Biol. 1997, 17, 4338–4345. [Google Scholar] [CrossRef]
- Feng, Y.; Qian, W.; Zhang, Y.; Peng, W.; Li, J.; Gu, Q.; Ji, D.; Zhang, Z.; Wang, Q.; Zhang, D.; et al. CDCA2 promotes the proliferation of colorectal cancer cells by activating the AKT/CCND1 pathway in vitro and in vivo. BMC Cancer 2019, 19, 576. [Google Scholar] [CrossRef]
- Uchida, F.; Uzawa, K.; Kasamatsu, A.; Takatori, H.; Sakamoto, Y.; Ogawara, K.; Shiiba, M.; Bukawa, H.; Tanzawa, H. Overexpression of CDCA2 in human squamous cell carcinoma: Correlation with prevention of G1 phase arrest and apoptosis. PLoS ONE 2013, 8, e56381. [Google Scholar] [CrossRef]
- Wiese, K.E.; Walz, S.; Von Eyss, B.; Wolf, E.; Athineos, D.; Sansom, O.; Eilers, M. The role of MIZ-1 in MYC-dependent tumorigenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a014290. [Google Scholar] [CrossRef]
- Koyama, T.; Ogawara, K.; Kasamatsu, A.; Okamoto, A.; Kasama, H.; Minakawa, Y.; Shimada, K.; Yokoe, H.; Shiiba, M.; Tanzawa, H.; et al. ANGPTL3 is a novel biomarker as it activates ERK/MAPK pathway in oral cancer. Cancer Med. 2015, 4, 759–769. [Google Scholar] [CrossRef]
- Sun, H.; Liu, K.; Huang, J.; Sun, Q.; Shao, C.; Luo, J.; Xu, L.; Shen, Y.; Ren, B. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. OncoTargets Ther. 2019, 12, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Regulski, M.J. Cellular senescence: What, why, and how. Wounds 2017, 29, 168–174. [Google Scholar] [PubMed]
- Cruickshanks, H.A.; McBryan, T.; Nelson, D.M.; VanderKraats, N.D.; Shah, P.P.; Van Tuyn, J.; Rai, T.S.; Brock, C.; Donahue, G.; Dunican, D.S.; et al. Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 2013, 15, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.W.; Lee, S.K.; Lee, Y.W.; Lee, J.O.; Kim, N.; Lee, H.J.; Seo, J.S.; Kim, J.; Kim, H.S.; Park, S.-H. Alcohol induces cell proliferation via hypermethylation of ADHFE1 in colorectal cancer cells. BMC Cancer 2014, 14, 377. [Google Scholar] [CrossRef]
- Murakami-Tonami, Y.; Ikeda, H.; Yamagishi, R.; Inayoshi, M.; Inagaki, S.; Kishida, S.; Komata, Y.; Koster, J.; Takeuchi, I.; Kondo, Y.; et al. SGO1 is involved in the DNA damage response in MYCN-amplified neuroblastoma cells. Sci. Rep. 2016, 6, 31615. [Google Scholar] [CrossRef]
- Hernandez, M.X.; Jiang, S.; Cole, T.A.; Chu, S.-H.; Fonseca, M.I.; Fang, M.J.; Hohsfield, L.A.; Torres, M.D.; Green, K.N.; Wetsel, R.A.; et al. Prevention of C5aR1 signaling delays microglial inflammatory polarization, favors clearance pathways and suppresses cognitive loss. Mol. Neurodegener. 2017, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Castellano, G.; Franzin, R.; Sallustio, F.; Stasi, A.; Banelli, B.; Romani, M.; De Palma, G.; Lucarelli, G.; Divella, C.; Battaglia, M.; et al. Complement component C5a induces aberrant epigenetic modifications in renal tubular epithelial cells accelerating senescence by Wnt4/βcatenin signaling after ischemia/reperfusion injury. Aging 2019, 11, 4382–4406. [Google Scholar] [CrossRef]
- Gao, S.; Jin, L.; Liu, G.; Wang, P.; Sun, Z.; Cao, Y.; Shi, H.; Liu, X.; Shi, Q.; Zhou, X.; et al. Overexpression of RASD1 inhibits glioma cell migration/invasion and inactivates the AKT/mTOR signaling pathway. Sci. Rep. 2017, 7, 3202. [Google Scholar] [CrossRef]
- Krones-Herzig, A.; Mittal, S.; Yule, K.; Liang, H.; English, C.; Urcis, R.; Soni, T.; Adamson, E.D.; Mercola, D. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 2005, 65, 5133–5143. [Google Scholar] [CrossRef]
- Ruoslahti, E. Fibronectin and its integrin receptors in cancer. Adv. Cancer Res. 1999, 76, 1–20. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, Y.; Li, Y.; Grün, K.; Berndt, A.; Zhou, Z.; Petersen, I. Cystatin A suppresses tumor cell growth through inhibiting epithelial to mesenchymal transition in human lung cancer. Oncotarget 2017, 9, 14084–14098. [Google Scholar] [CrossRef]
- Kidger, A.M.; Keyse, S.M. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs). Semin. Cell Dev. Biol. 2016, 50, 125–132. [Google Scholar] [CrossRef]
- Goeppert, B.; Schmezer, P.; Dutruel, C.; Oakes, C.; Renner, M.; Breinig, M.; Warth, A.; Vogel, M.N.; Mittelbronn, M.; Mehrabi, A.; et al. Down-regulation of tumor suppressor a kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology 2010, 52, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Xu, X.; Wu, M.; Guan, Z.; Su, X.; Chen, S.; Wang, H.; Teng, L. GPRC5A: An emerging biomarker in human cancer. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Cetin, M.; Odabas, G.; Douglas, L.R.; Duriez, P.J.; Balcik-Ercin, P.; Yalim-Camci, I.; Sayan, A.E.; Yagci, T. ROR1 expression and its functional significance in hepatocellular carcinoma cells. Cells 2019, 8, 210. [Google Scholar] [CrossRef] [PubMed]
- Asad, A.S.; Candia, A.J.N.; Gonzalez, N.; Zuccato, C.F.; Abt, A.; Orrillo, S.J.; Lastra, Y.; De Simone, E.; Boutillon, F.; Goffin, V.; et al. Prolactin and its receptor as therapeutic targets in glioblastoma multiforme. Sci. Rep. 2019, 9, 19578. [Google Scholar] [CrossRef]
- Tilghman, J.; Wu, H.; Sang, Y.; Shi, X.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Eberhart, C.G.; Laterra, J.; Ying, M. HMMR maintains the stemness and tumorigenicity of glioblastoma stem-like cells. Cancer Res. 2014, 74, 3168–3179. [Google Scholar] [CrossRef]
- Thangavelu, P.U.; Lin, C.-Y.; Vaidyanathan, S.; Nguyen, T.H.; Dray, E.; Duijf, P.H. Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer. Oncotarget 2017, 8, 62167–62182. [Google Scholar] [CrossRef]
- Bu, P.; Chen, K.-Y.; Xiang, K.; Johnson, C.; Crown, S.B.; Rakhilin, N.; Ai, Y.; Wang, L.; Xi, R.; Astapova, I.; et al. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab. 2018, 27, 1249–1262.e4. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 5, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Paardekooper, L.M.; Vos, W.; Van Der Bogaart, G. Oxygen in the tumor microenvironment: Effects on dendritic cell function. Oncotarget 2019, 10, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Holmannova, D.; Borska, L.; Andrys, C.; Borsky, P.; Kremlacek, J.; Hamakova, K.; Rehacek, V.; Malkova, A.; Svadlakova, T.; Palicka, V.; et al. The impact of psoriasis and metabolic syndrome on the systemic inflammation and oxidative damage to nucleic acids. J. Immunol. Res. 2020, 2020, 7352637. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, J.; Biard, D.; Paget, V.; Goislard, M.; Morel-Altmeyer, S.; Konge, J.; Chevillard, S.; Lebeau, J. Forced extinction of CD24 stem-like breast cancer marker alone promotes radiation resistance through the control of oxidative stress. Mol. Carcinog. 2015, 55, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Hrycay, E.G.; Bandiera, S.M. Involvement of cytochrome P450 in reactive oxygen species formation and cancer. Adv. Pharmacol. 2015, 74, 35–84. [Google Scholar] [PubMed]
- Veith, A.; Moorthy, B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species. Curr. Opin. Toxicol. 2018, 7, 44–51. [Google Scholar] [CrossRef]
- Mishra, P.; Tang, W.; Ambs, S. ADHFE1 is a MYC-linked oncogene that induces metabolic reprogramming and cellular de-differentiation in breast cancer. Mol. Cell. Oncol. 2018, 5, e1432260. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, A.; Ohmura, S.; Orth, M.F.; Knott, M.M.L.; Colombo, M.V.; Arrigoni, C.; Bardinet, V.; Saucier, D.; Wehweck, F.S.; Li, J.; et al. Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma. Nat. Commun. 2020, 11, 2423. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).