Abstract
Congenital cytomegalovirus (CMV) is a major cause of neonatal morbidity, particularly sensorineural hearing loss, yet its early detection remains challenging. While urinary PCR is the current diagnostic gold standard, its implementation in neonatal settings is often limited by feasibility issues. Salivary PCR presents a more practical alternative, but its diagnostic accuracy has remained uncertain. This systematic review and meta-analysis aimed to evaluate the performance of salivary PCR compared to urinary PCR in detecting congenital CMV in neonates. Following PRISMA guidelines, 15 observational studies involving 29,617 neonates were analyzed using a random-effects model. Pooled sensitivity and specificity were 0.99 and 1.00, respectively, with a negative predictive value (NPV) of 1.00 and a positive predictive value (PPV) of 0.91, despite moderate heterogeneity. Subgroup analysis showed high diagnostic performance across general neonates, infants of seropositive mothers and high-risk neonates (referring to neonates that are small for their gestational age (SGA), neonates who failed hearing screening, and neonates with CMV-related congenital abnormalities). The general group had the highest specificity (0.999), while high-risk neonates showed the highest sensitivity (0.981). Across all groups, NPV remained consistently above 0.994, with PPV ranging from 0.848 to 0.981. These findings confirm that salivary PCR is a highly accurate and feasible tool for congenital CMV diagnosis.
1. Introduction
Cytomegalovirus (CMV) is a double-stranded DNA virus and a member of the Herpesviridae family [1]. The human cytomegalovirus virion consists of an icosahedral capsid with attached proteins, an outer layer of proteins called the tegument, and a surrounding lipid envelope containing viral glycoproteins [1]. This pathogen infects individuals of all ages and targets a wide range of tissues including blood, brain, and heart [1]. It has high prevalence worldwide, affecting half the population in developed countries and almost 100% in developing countries, particularly in Asia and Africa [2,3].
CMV spreads through various transmission routes, primarily via direct contact with infected body fluids such as urine, saliva, and blood [4]. Additionally, a notable transmission route is during pregnancy, in which the virus can pass through the placenta, causing congenital CMV in the neonate [4]. Once a person is infected, CMV establishes lifelong latency by persisting in myeloid cells [4]. While the virus often remains dormant in healthy individuals, it has the potential to reactivate when the immune system is weakened. During reactivation, CMV remains largely cell-associated in the bloodstream [5]. Dissemination occurs primarily through circulating infected leukocytes (e.g., monocytes, polymorphonuclear cells) and endothelial cells [5]. This cell-associated viremia facilitates viral spread to multiple organ systems [5]. This ability to remain latent and reactivate under certain conditions makes CMV a major concern, particularly for immunocompromised individuals and newborns.
According to the CDC, approximately 1 in 200 babies are born with congenital CMV, and a staggering 1 in 5 suffer from birth defects or long-term ailments [6]. Around 10 to 15% of asymptomatic newborns lose their hearing later in life [7]. Hearing loss can worsen from mild to severe within the first two years of life, which is a crucial period for language development; over time, it affects the child’s ability to develop communication, language, and social skills, which causes cognitive impairment [8]. Infected patients could also develop vestibular disorders, leading to balance and motor deficits, among other severe complications, including microcephaly, intracranial calcifications, and cerebral hypoplasia [9]. Therefore, timely diagnosis and intervention for congenital CMV in neonates are crucial to avoid such complications.
The cost of illness for congenital CMV was estimated at $4 billion in the USA (2001) and £732 million in the UK (2016), highlighting an example of its substantial economic burden [10,11].
Traditionally, urine polymerase chain reaction (PCR) has been considered the gold-standard method of diagnosis for congenital CMV, but it faces challenges in time and collection [12,13]. A prospective study described it as “prohibitive”, energy-intensive, requires frequent bag changes, is prone to contamination by meconium and leakage, all of which led to sample loss and delay in testing. This can prevent discrimination between acquired and congenital CMV [4,6,13,14].
Given these challenges, saliva PCR has emerged as a viable alternative due to its feasibility. Infants with congenital CMV shed significant quantities of the virus in their saliva, making it a useful screening technique [15]. Saliva specimens are collected through cheek swabs from neonates and sent for PCR [15]. Delays in sampling can subject this mode to false positive congenital CMV, as CMV may be present in the breastmilk of seropositive mothers [16]. However, compared to urine samples, saliva remains are less prone to contamination and more easily collected [17].
Despite the promise of salivary PCR, the existing literature is limited in depth and scope. Therefore, the aim of this study is to assess the diagnostic test accuracy of salivary PCR as an alternative to urinary PCR in effectively detecting congenital CMV. This study aims to thoroughly explore diagnostic metrics, including specificity (SP), sensitivity (SN), Positive Predictive value (PPV), and Negative Predictive Value (NPV). Given the surge in scientific research, an updated meta-analysis is warranted to capture new global data. Also, this study will focus exclusively on the neonatal population, which is often underrepresented in similar reviews. Finally, reducing heterogeneity to strengthen the validity of conclusions will be a key priority.
2. Materials and Methods
2.1. Protocol and Registration
The protocol for this systematic review and meta-analysis was developed in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA) 2020 guidelines to ensure methodological transparency and rigor [18]. The protocol was prospectively registered in the PROSPERO database under the registration number CRD42024603785 [19].
2.2. Search Protocol
A literature search was comprehensively performed across eight electronic databases: PubMed, Scopus, Web of Science, ScienceDirect, ProQuest, Cochrane Library, CINAHL, and Wiley. The search strategy was developed using Medical Subject Headings (MeSH) terms and relevant keywords (Appendix A).
2.3. Study Selection
Study selection was performed in three phases using Covidence, a systematic review management tool. First was the removal of duplicates, which was conducted mainly by Covidence automatically. Secondly, for title and abstract screening, two independent reviewers screened all studies based on the following specified inclusion and exclusion criteria in Table 1, and conflicts were resolved by another two independent reviewers. Subsequently, the same process was followed for full-text screening, but by the opposite set of reviewers.

Table 1.
PICO Inclusion and Exclusion Criteria for Primary and Secondary Outcomes.
2.4. Quality Assessment
Risk of Bias was assessed using the QUDAS-2 tool (Quality Assessment of Diagnostic Accuracy Studies) as recommended by the Cochrane Collaboration [20]. QUADAS-2 assesses four key domains: patient selection, index test, reference standard, and flow and timing.
Studies were categorized as having low, high, or unclear risk of bias or applicability concern in each domain, based on standardized questions (Appendix B, Table A1). The assessment was performed independently by two reviewers, with conflicts resolved through group discussion. This ensured a transparent and unbiased quality assessment to determine the overall strength of the evidence.
2.5. Data Extraction
For each study, relevant data were extracted, including the total number of participants, sample collection method, and population type, among other variables. When diagnostic accuracy metrics such as SN, SP, PPV, and NPV were reported, they were directly extracted. In cases where these metrics were not available, raw values for true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) were extracted to enable calculation. All data were systematically recorded and managed using a standardized template within Covidence, as presented in Appendix C, Figure A1.
2.6. Missing Data Calculations
In this study, missing proportions were calculated from extracted raw data using the Proportions were calculated using the Binomial Proportion equation:
where p^ is the estimated proportion (e.g., sensitivity, specificity, PPV, or NPV), x is the number of successes (e.g., true positives for sensitivity), and n is the relevant sample size. This equation forms the basis for all diagnostic performance metrics presented in Equation (2).
p^ = x/n,
Diagnostic accuracy measures were calculated as follows:
where TP = true positives, TN = true negatives, FP = false positives, and FN = false negatives.
SP = TN/(TN + FP), SN = TP/(TP + FN), PPV = TP/(TP + FP), NPV = TN/(TN + FN),
The standard error for each proportion was calculated using the Delta Method (Equation (3)) to account for variability in Wilson-adjusted proportions [21]:
where z is the z-score for the desired confidence level (z = 1.96 for 95% confidence), and n is the sample size.
SEWilson = √[p^(1 − p^)/n + z2/(4n2)],
The 95% confidence interval (CI) was calculated using the Wilson Score Interval without Continuity Correction:
which provides more accurate interval estimates in small or extreme samples [21]. This approach improves the robustness of our estimates and aligns with best practices in diagnostic accuracy studies.
CI = (p^ + z2/(2n) ± z × √[p^(1 − p^)/n + z2/(4n2)])/(1 + z2/n),
2.7. Data Analysis
The meta-analysis was conducted using R software (version 4.4.3) with the ‘meta’ [22] and ‘metafor’ [23] packages to evaluate the diagnostic performance of salivary PCR compared to urinary PCR in detecting congenital CMV. A random-effects model employing the DerSimonian and Laird method was used to calculate pooled estimates of diagnostic accuracy, including SN, SP, PPV, and NPV; this model accounts for anticipated heterogeneity among studies [24].
Expanded diagnostic metrics were also calculated using the same method, which include Diagnostic Odds Ratio (DOR), Area Under the Curve (AUC), Positive Likelihood Ratio (PLR), and Negative Likelihood Ratio (NLR).
A Summary Receiver Operating Characteristic (SROC) Curve was made to visually represent the overall diagnostic performance of Salivary PCR. A funnel plot was then constructed to evaluate small-study effects and potential publication bias, which was further assessed using Egger’s test.
Then, a subgroup analysis was conducted across three clinically relevant neonatal populations: All neonates, neonates of seropositive mothers, and, lastly, high-risk neonates, which generally refers to the following three characteristics: neonates that are SGA, neonates who failed hearing screening, and neonates with CMV-related congenital abnormalities. Pooled diagnostic metrics were calculated and presented separately for each subgroup using random-effects forest plots. Additionally, the same expanded diagnostic metrics were calculated for each group. This allowed for comparative assessment of salivary PCR performance across different population contexts
3. Results
3.1. Screening Process
A total of 922 identified studies were explored from the following databases: 303 from PubMed, 282 from Scopus, 217 from Web of Science, 68 from ScienceDirect, 27 from ProQuest, 17 from Cochrane, 5 from CINAHL, and 3 from Wiley. However, only 408 studies were eligible for screening since 514 duplicates were excluded. A total of 369 studies were excluded during the abstract screening process, leaving 39 full-text papers for detailed evaluation. Among these, 9 were excluded for wrong outcomes, 6 for wrong interventions, 4 for wrong study design, 3 for wrong patient population, 1 for wrong comparator and finally 1 for wrong language, all of which amounted to 23 studies. A final 15 papers were selected for further data extraction. Refer to Figure 1 for more details.
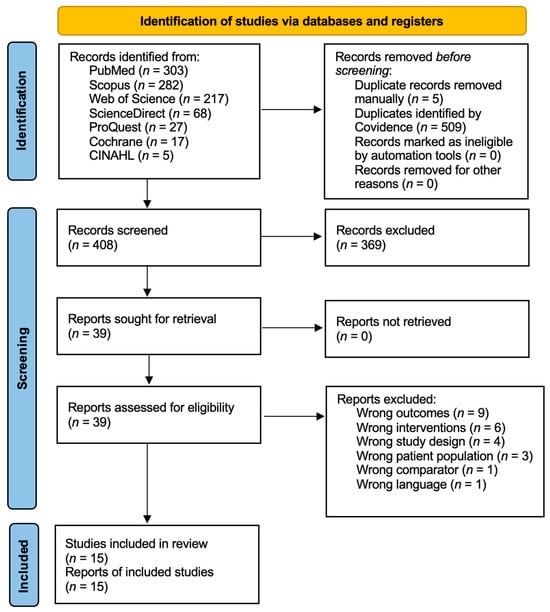
Figure 1.
PRIMA flow diagram.
3.2. Quality Assessment Results
Figure 2 presents the quality assessment of the included studies using the QUADAS-2 tool. Overall, studies showed low risk of bias and low applicability concern, supported by clearly described methods. However, some studies had high risk of bias due to discriminatory patient selection: 8 studies tested patients that are high risk, suspected or confirmed to have CMV. Studies marked as unclear lacked sufficient information for a definitive judgment.
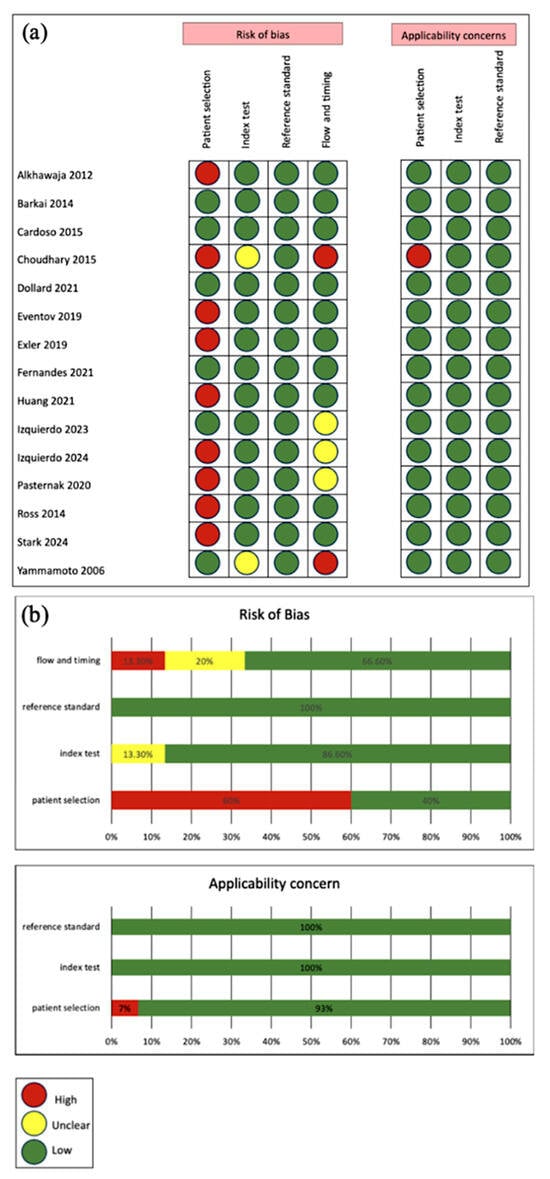
Figure 2.
(a) Study-level QUADAS-2 risk of bias and applicability assessments across domains [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38]; (b) Aggregated domain-level percentages for risk of bias and applicability concern.
3.3. Study Characteristics
The raw data tables are present in Appendix D, Table A2. From the 15 included studies [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38] we extracted 3 additional datasets; one from Izquierdo (2024) and two from Pasternak (2020); giving a total of 18 datasets. Specificity ranged from [0.904–1.000], sensitivity [0.245–1.000], PPV [0.456–1.000], and NPV [0.976 to 1.000]. Two anomalous studies were manually identified, calling for a Leave-One-Out (LOO) analysis to assess their impact. Most studies were prospective cohort studies conducted in the Americas WHO region. Study characteristics are detailed in Appendix E, Table A3. Three populations were represented: high-risk neonates, infants of seropositive mothers, and all neonates, with high-risk neonates being the most common.
3.4. Exclusion of Outliers
A Leave One Out (LOO) analysis was performed, identifying two studies contributing significantly to heterogeneity (Appendix F, Table A4, Table A5, Table A6 and Table A7).Huang 2021 was excluded in NPV and SN analysis as the estimates were notably higher, 0.99998 and 0.9875, respectively, after its exclusion [33]. Additionally, when excluded, the Tau2 and I2 values dropped to zero, confirming their role in heterogeneity.
Eventov-Friedman 2019 was excluded in PPV and SP analysis as it also gave higher estimates, 0.9078 and 0.99908, respectively, when excluded [29]. Moderate heterogeneity in PPV and SP (I2 = 51.49%, 57.23%) was significantly reduced upon exclusion, indicating its disproportionate influence on pooled PPV and SP results.
Table 2 and Table 3 are sample LOO tables showing an example of the significant changes with the exclusion of the previously mentioned studies in comparison to Dollard 2021 [17] as a control exclusion.

Table 2.
Eventov-Friedman 2019 [29] LOO Sample Table.

Table 3.
Huang 2021 [33] LOO Sample Table.
3.5. Meta-Analysis of Diagnostic Test Accuracy
3.5.1. Random Effects Forest Plots for All Studies
Using a Random Effects Model, the review assessed all four diagnostic metrics shown in Figure 3 and Figure 4. Sensitivity pooled at 0.99 (95% CI: [0.97, 1.00]), showing excellent detection of true positives. Most studies reported sensitivity above 0.85, and heterogeneity was low (I2 = 0%) suggesting low variability among studies. Specificity pooled at 1.00 (95% CI: [1.00, 1.00]), indicating a very low false-positive rate. Although most studies reported values near 1.00, moderate heterogeneity (I2 = 57.2%) suggests some study-level variation.
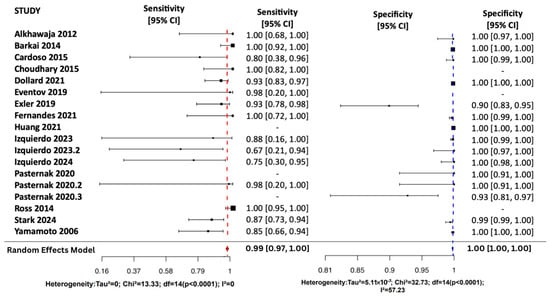
Figure 3.
Random Effects Forest Plots for SN and SP [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38].
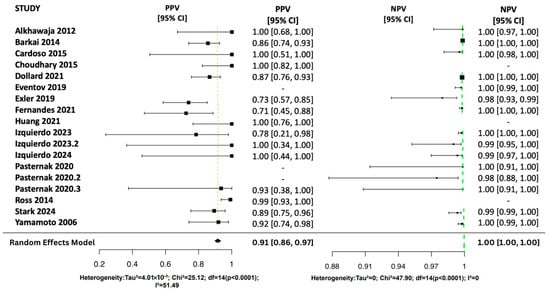
Figure 4.
Random Effects Forest Plots for PPV and NPV [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38].
The pooled NPV was 1.00 (95% CI: [1.00, 1.00]), indicating a strong ability to rule out disease with low variability (I2 = 0%). Individual NPVs ranged from 0.98 to 1.00 with narrow confidence intervals. The pooled PPV was 0.91 (95% CI: [0.86, 0.97]), reflecting a high likelihood of correctly identifying disease. However, PPV varied across studies (0.71–1.00), with moderate heterogeneity (I2 = 51.5%).
3.5.2. Expanded Diagnostic Metrics for All Studies
Table 4 displays the summary meta-analysis results for all the studies. The DOR of 2041.14 (95% CI: 778.995–5348.226) reflects a very high level of overall test accuracy, indicating strong separation between those with and without the disease. The AUC of 0.99 (95% CI: 0.97–1.00) further supports this, showing that salivary PCR has excellent discriminatory ability, with near-perfect sensitivity and specificity.

Table 4.
Summary Meta-analysis Results.
A high PLR of 109.87 [33.78–357.39] suggests that a positive result strongly confirms the presence of the disease. Meanwhile, the low NLR of 0.0885 [0.0409–0.1916] indicates that a negative result effectively rules it out, highlighting the test’s strong diagnostic accuracy.
3.5.3. The Summary Receiver Operating Characteristic (SROC) Curve
SROC curve assesses overall diagnostic accuracy by plotting sensitivity against specificity (False-positive rate) as shown in Figure 5. All 16 datasets were included. The sensitivity was 0.956 (95% CI: 0.924–0.988), and the specificity was 0.988 (95% CI: 0.900–0.999). The AUC was 0.994 (0.975–0.998), reflecting excellent discriminatory power with minimal misclassification. These findings show that the diagnostic test demonstrated high performance.
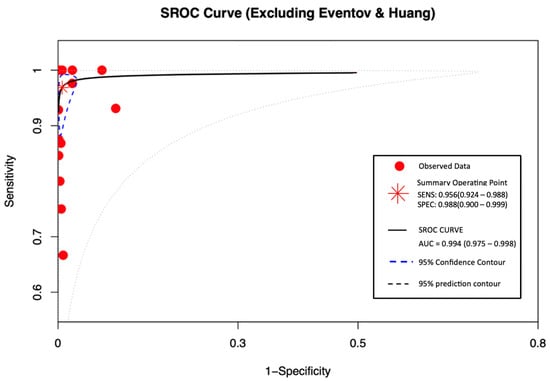
Figure 5.
SROC curve excluding Eventov and Huang.
3.5.4. Publication Bias
This meta-analysis confirms that the diagnostic test exhibits high sensitivity, specificity, and predictive accuracy across different populations. However, publication bias and study-level influences were evaluated using Egger’s test and the funnel plot to ensure the robustness of these findings as shown in Figure 6 and Table 5.
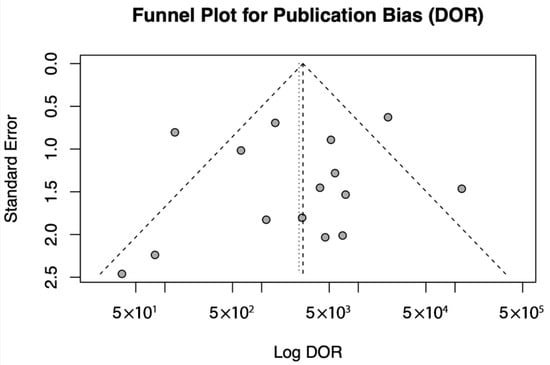
Figure 6.
Funnel Plot for Publication Bias.

Table 5.
Egger’s Test.
A funnel plot of log DOR against standard error was generated to assess publication bias. Individual studies are shown as points, and the pooled effect is represented by a vertical dotted line. The dashed lines correspond to the expected 95% confidence region in the absence of bias.
In the assessment of publication bias, a funnel plot was generated for DOR, and Egger’s test was performed to statistically evaluate asymmetry. The funnel plot exhibited some visual asymmetry; however, Egger’s test did not provide significant evidence of publication bias (t = −0.4149, df = 13, p = 0.685). The estimated effect size when the standard error approached zero was 8.3678 (95% CI: 5.6490–11.0866), indicating a stable overall effect. These findings suggest that any observed asymmetry is likely due to random variation or small-study effects rather than systematic publication bias.
3.6. Meta-Analysis of Diagnostic Test Acuuracy of Subgroups
3.6.1. SN and SP Random Effects Forest Plots for Subgroups
Figure 7, Figure 8 and Figure 9 display the forest plots of sensitivity and specificity for salivary PCR in each subgroup. Specificity was nearly perfect in all neonate subgroups at 0.9990 (95% CI: 0.9986–0.9994) and high-risk populations at 0.9956 (95% CI: 0.9909–1.00), but notably lower in the seropositive mother subgroup at 0.9566 (95% CI: 0.8629–1.00). Heterogeneity in specificity was only significant in the seropositive group (I2 = 89.46%).
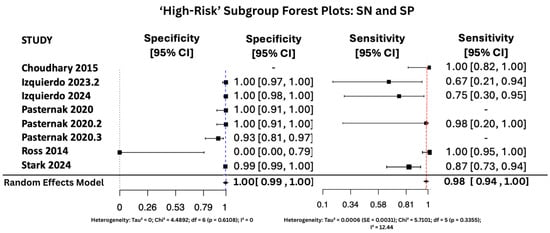
Figure 7.
SP and SN Forest Plots for ‘High-Risk’ Subgroup [15,28,34,35,36,37]. The red dashed vertical line corresponds to the pooled sensitivity estimate, while the blue dashed vertical line denotes the pooled specificity estimate.
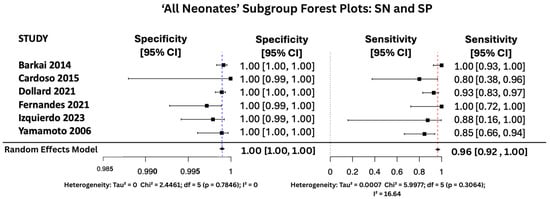
Figure 8.
SP and SN Forest Plots for ‘All Neonates’ Subgroup [17,26,27,31,34,38].
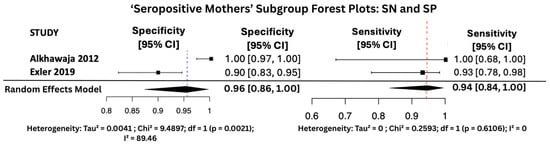
Figure 9.
SP and SN Forest Plots for ‘Seropositive Mothers’ Subgroup [25,30].
Sensitivity was also consistently high, with the highest observed in the high-risk group at 0.9810 (95% CI: 0.9353–1.00), followed by 0.9648 (95% CI: 0.9163–1.00) in All neonates group, and 0.9436 (95% CI: 0.8410–1.00) in seropositive mothers. Heterogeneity for sensitivity was low across all groups, with I2 values ranging from 0.00% to 16.64%, indicating good consistency between studies.
3.6.2. PPV and NPV Random Effects Forest Plots for Subgroups
Figure 10, Figure 11 and Figure 12 display the forest plots of PPV and NPV for salivary PCR in each subgroup. In terms of predictive values, the Positive Predictive Value (PPV) was highest in the high-risk population (0.9817, 95% CI: 0.9507–1.00), moderate in All neonates group (0.8674, 95% CI: 0.8092–0.925), and lowest in the seropositive group (0.8484, 95% CI: 0.5857–1.00). PPV heterogeneity was moderate in the seropositive mothers (I2 = 71.15%) but negligible in the other two subgroups.
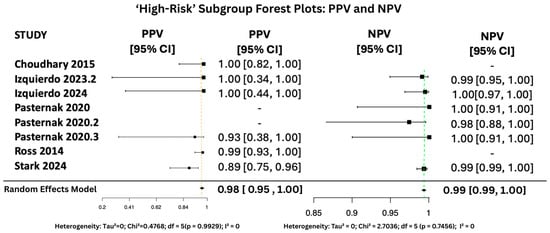
Figure 10.
PPV and NPV Forest Plots for ‘High-Risk’ Subgroup [15,28,34,35,36,37]. The yellow dashed vertical line corresponds to the pooled PPV estimate, while the green dashed vertical line denotes the pooled NPV estimate.
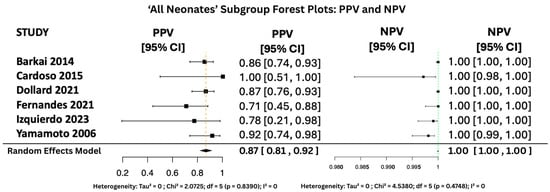
Figure 11.
PPV and NPV Forest Plots for ‘All Neonates’ Subgroup [17,26,27,31,34,38].
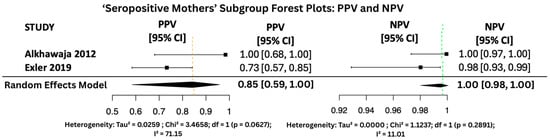
Figure 12.
PPV and NPV Forest Plots for ‘Seropositive Mothers’ Subgroup [25,30].
The Negative Predictive Value (NPV) was consistently very high across all groups, with values of 0.99999 (95% CI: 0.9998–1.00) in All neonates group, 0.9965 (95% CI: 0.9816–1.00) in seropositive mothers, and 0.9940 (95% CI: 0.9889–0.9991) in high-risk individuals. Heterogeneity for NPV was minimal to none (I2 = 0–11%).
3.6.3. Expanded Diagnostic Metrics for Subgroups
Table 6 displays the summary meta-analysis results for all the subgroups. The diagnostic performance was highest in the general neonate population, with a DOR of 8635.79 (95% CI: 3197.51–23,323.41), compared to 897.15 (95% CI: 351.84–2287.65) in high-risk populations and 437.03 (95% CI: 10.75–17,754.31) in seropositive mothers.

Table 6.
Expanded Diagnostic Metrics for Subgroups.
AUC was similarly high across all subgroups, reflecting excellent overall test performance: 0.9818 (95% CI: 0.9575–1.00) in all neonate groups, 0.9424 (95% CI: 0.8681–1.00) in seropositive mothers, and 0.9259 (95% CI: 0.8681–0.9836) in high-risk populations.
PLR was markedly highest in the All-neonate population at 833.01 (95% CI: 517.11–135.20), followed by 37.10 (95% CI: 1.416–971.59) in seropositive mothers, and 29.49 (95% CI: 6.43–135.20) in high-risk groups.
Finally, NLR was lowest in the seropositive mother group (0.0849, 95% CI: 0.0270–0.2667) and remained similarly low in the high-risk group (0.0914, 95% CI: 0.0382–0.2188). In the general neonatal group, the NLR was slightly higher at 0.1141 (95% CI: 0.0620–0.2099), yet still within a range suggestive of good diagnostic performance.
4. Discussion
4.1. All Studies Discussion
Findings from the 15 studies [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38] confirm excellent diagnostic performance, with high NPV (99.99%), SN (98.75%), and SP (99.91%), indicating low false-negative and false-positive rates. A meta-analysis by Zheng et al. (2022), conducted on all infants rather than neonates alone, supports these results, reporting 100% SP and 97% SN [39].
Additionally, authoritative sources from the CDC, Cleveland Clinic and Boston Children’s Hospital all recognize salivary PCR as a diagnostic method for congenital CMV further supporting the reliability of salivary PCR’s diagnostic ability [40,41,42].
The pooled PPV is 90.78% ([95% CI: 86.00–96.28]), with variability likely driven by differences in population prevalence (high-risk neonates, infants of seropositive mothers, and all neonate populations). Biologically, this is supported by the shedding of CMV in the breast milk of seropositive mothers, which may lead to transient viral presence in the infant’s saliva and potential false-positive results [40]. This reflects postnatal exposure rather than congenital infection, making this distinction essential for accurate interpretation of positive saliva PCR results.
To address PPV variability, likelihood ratios were assessed, with PLR > 10 (109.87 [95% CI: 33.77–357.39]) and NLR < 0.1 (0.088 [95% CI: 0.0409–0.1916]) indicating strong diagnostic performance of salivary PCR in confirming and excluding congenital CMV cases [43]. Similarly, Zheng et al. (2022) reported an exceptionally high PLR of 797.5 and an NLR of 0.09 [39].
This study’s SROC curve demonstrates a high TP rate of 0.956 (95% CI: 0.924–0.988) and low FP rate with a SP of 0.988 (95% CI: 0.900–0.999), along with an AUC of 0.994. This confirms near-perfect discriminatory ability for salivary PCR, supporting the CDC’s decision to use it as one of the diagnostic tools as mentioned above [6].
Given its high sensitivity, specificity, and ease of sample collection, salivary PCR also shows promise as a potential tool for early screening of congenital CMV, particularly in asymptomatic neonates. Its non-invasive nature and logistical feasibility make it a strong candidate for integration into large-scale newborn screening programs [44]. However, the clinical and economic implications of routine screening remain to be fully evaluated [45].
In conclusion, the findings of this systematic review and meta-analysis confirm that saliva PCR is a highly efficient diagnostic tool for congenital CMV due to its exceptional performance, supported by comparisons with Zheng et al. (2022) and authoritative sources [6,39,40,41,42]
4.2. Subgroup Discussion
This meta-analysis confirms the robust diagnostic performance of salivary PCR testing for congenital CMV detection across multiple neonatal subgroups. Overall, the test demonstrated exceptional accuracy, with pooled sensitivity and specificity approaching perfection, highlighting its utility as a non-invasive tool for early diagnosis of congenital CMV [34,43].
In the general neonate population, salivary PCR showed outstanding performance, with the highest DOR of 8635.78 and an AUC of 0.98, indicating near-perfect diagnostic accuracy. Both sensitivity (96.5%) and specificity (99.9%) were high, with minimal heterogeneity, reflecting excellent consistency across studies and affirming the test’s reliability in diagnosing congenital CMV.
In the high-risk population subgroup, comprising infants with elevated baseline risk due to clinical symptoms of congenital CMV, diagnostic performance remained highly favorable. Sensitivity reached 98.1%, with specificity of 99.6%, and heterogeneity was low across all diagnostic parameters. These findings suggest that salivary PCR is particularly well-suited to high-risk settings, where early and accurate detection is critical for timely intervention and prevention of long-term complications [46].
The seropositive mothers’ subgroup, while also showing promising results (sensitivity 94.4%, specificity 95.7%), included only two datasets. This limited sample size likely explains the observed heterogeneity in specificity (I2 = 89.46%) and PPV (I2 = 71.15%). Although the results remain encouraging, the small number of studies reduces the statistical power and generalizability of findings within this subgroup.
As such, caution should be exercised in interpreting these results, and further research is needed to validate test performance in seropositive populations, especially given the potential biological variability and differing baseline prevalence of congenital CMV [47].
Across all subgroups, the negative predictive value (NPV) consistently exceeded 99%, and negative likelihood ratios remained exceptionally low (0.08–0.11), indicating that a negative salivary PCR result is highly reliable for excluding congenital CMV infection. Meanwhile, positive likelihood ratios were extremely high, particularly in the general neonate group (PLR = 833.01), underscoring the strong diagnostic confirmation offered by a positive result.
Together, these findings strongly support the use of salivary PCR testing. Its ease of collection [34], high diagnostic performance, and consistency across both general and high-risk populations, make it an ideal candidate for early detection of congenital CMV. Future studies should aim to expand the evidence base in seropositive maternal cohorts to strengthen confidence in targeted diagnostic strategies.
4.3. Strengths
During the study scoping, all the search strategies included title and abstract searches for the key words, increasing the cohesion between the different search strategies for each electronic database.
The use of Wilson Score Interval in this study to estimate missing confidence intervals guaranteed greater accuracy than the traditional Wald method, particularly for small/extreme proportions, which were often present in specificities of 0 or 1 [21].
Additionally, the performance of the LOO for all 4 proportions resulted in exclusion of 2 studies from the meta-analysis, which decreased heterogeneity and increased the results’ accuracy and robustness [48].
The statistical power of this meta-analysis of 15 studies, incorporating a total population of 29,617 neonates, is extremely high (1.0 or 100%), indicating a strong ability to detect a true effect. This is primarily driven by the large sample size, with 480 PCR-positive cases and 29,137 PCR-negative cases [49].
The inclusion of an SROC curve in the analysis provided a comprehensive evaluation of diagnostic performance across studies. The resulting AUC of 0.994 demonstrates near-perfect discriminative ability of the salivary PCR test, reinforcing the robustness and clinical relevance of the pooled findings.
The use of funnel plot analysis and Egger’s test strengthened the validity of the meta-analysis by demonstrating no significant publication bias, thereby increasing confidence in the reliability of the pooled estimates.
The variability in data, indicated by Tau2 and I2, is moderate or low, which makes the effect sizes more distinct and reliable. Another strength of this review is the targeted focus on the neonatal population, addressing a gap in existing literature where this specific group has been underrepresented [48].
Lastly, these findings demonstrate this meta-analysis provides a highly reliable and statistically robust evaluation of salivary PCR as a diagnostic tool for congenital CMV.
4.4. Limitations
Although Alkhawaja (2012) included both congenital and perinatal CMV cases, it was retained despite potential inaccuracies in the congenital CMV population due to its small sample size and limited impact on pooled estimates [25].
Baraki (2014), Fernandez (2021), and Izquierdo (2023) retested only a subset of negative salivary PCR results to evaluate screening protocols [26,31,34]. They were included based on subgroup retesting and shared assumptions, though the possibility of undetected false negatives in untested cases remains.
The exclusion of Huang and Eventov-Friedman may have contributed to overestimating salivary PCR performance, as shown in the LOO analysis in Appendix F, Table A4, Table A5, Table A6 and Table A7 [29,33]. Notably, Eventov-Friedman was the only study to collect neonatal samples within the first 10 days of life, which differs from CDC guidelines recommending collection at 2–3 weeks [4,29].
Both studies also collected and tested saliva and urine samples concurrently, unlike most others. These design differences, along with broader population and epidemiological factors, likely contributed to the observed heterogeneity beyond methodological variation alone.
Moderate heterogeneity was observed in PPV (I2 = 51.49%) and specificity (I2 = 57.23%), likely due to varying prevalence across studies, directly affecting PPV. Therefore, confirmatory testing may be relevant, as recommended by the CDC [40].
Finally, as previously noted, the inclusion of only two studies within the seropositive mother subgroup significantly limits the strength and generalizability of its findings. The small sample size reduces the statistical power and increases the potential for bias, making it difficult to draw definitive conclusions about diagnostic accuracy in this specific population. Additional studies focusing on seropositive cohorts are needed to validate and strengthen the evidence in this subgroup.
4.5. Future Directions
Salivary PCR testing offers a promising avenue for enhancing early detection of congenital CMV. Future research should prioritize prospective cohort studies that evaluate not only their cost-effectiveness but also long-term clinical outcomes following early diagnosis, as well as their feasibility across diverse healthcare settings. The development of standardized protocols, particularly for the optimal timing of sample collection while accounting for potential postnatal exposures, will be essential to maximize diagnostic accuracy.
5. Conclusions
This systematic review supports the use of salivary PCR as a reliable diagnostic tool for congenital CMV in neonates, demonstrated by its high sensitivity, specificity, and negative predictive value. The strong diagnostic accuracy, coupled with the practicality and non-invasiveness of saliva collection compared to urine, highlights its potential for widespread use in large-scale screening programs. Subgroup analyses suggest consistent effectiveness across various neonatal populations; however, further research is warranted in seropositive mothers to clarify performance in this subgroup. Future studies should also aim to standardize salivary PCR protocols to enhance consistency and global applicability.
Author Contributions
Conceptualization, S.M.A.R.; Methodology, S.M.A.R., M.I.A., M.A. and S.D.; Writing—original draft preparation, S.M.A.R., S.S., M.A. and M.I.A.; Writing—reviewing and editing, S.M.A.R., M.I.A., S.S. and M.A.; Visualization, S.M.A.R., M.I.A. and S.S.; Supervision, J.K.S. and S.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study has received an APC grant, funded by the Royal College of Surgeons in Ireland-Bahrain (RCSI-Bahrain), P.O. Box 15503, Adliya, Bahrain.
Data Availability Statement
The raw data table has been provided in Appendix D.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CMV | Cytomegalovirus |
| SGA | Small for Gestational Age |
| PCR | Polymerase Chain Reaction |
| SN | Sensitivity |
| SP | Specificity |
| PPV | Positive Predicted Value |
| NPV | Negative Predicted Value |
| DOR | Diagnostic Odds Ratio |
| AUC | Area Under the Curve |
| PLR | Positive Likelihood Ratio |
| NLR | Negative Likelihood Ratio |
| SROC | Summary Receiver Operating Characteristic |
Appendix A. Search Strategies
PUBMED:
(newborns[MeSH Terms] OR newborn*[Title/Abstract] OR neonate*[Title/Abstract] OR infant*[Title/Abstract] OR babies[Title/Abstract] OR baby[Title/Abstract]) AND (cytomegalovirus[MeSH Terms] OR cytomegalovirus[Title/Abstract] OR “CMV”[Title/Abstract] OR “HCMV”[Title/Abstract] OR “Human Herpesvirus 5”[Title/Abstract] OR “HHV-5”[Title/Abstract]) AND (saliva[MeSH Terms] OR saliva*[Title/Abstract])
SCOPUS:
TITLE-ABS ((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))
Web Of Science:
TI = ((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*)) OR AB = ((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))
ProQuest:
abstract(((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))) AND title(((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*)))
Science direct:
Title, abstract, keywords ((Newborn OR neonate OR infant OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus”) AND (saliva OR salivary))
Cochrane library:
((Newborn OR neonate OR infant OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus”) AND (saliva OR salivary))
CINAHL:
TI (((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))) AND AB (((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*)))
Wiley’s online library:
“((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))” in Title and “((newborn* OR neonate* OR infant* OR babies OR baby) AND (cytomegalovirus OR “CMV” OR “HCMV” OR “Human Herpesvirus 5” OR “HHV-5”) AND (saliva OR saliva*))” in Abstract
Appendix B. Quality Assessment Template

Table A1.
QUDAS-2 Survey.
Table A1.
QUDAS-2 Survey.
| QUDAS-2 Survey |
|---|
| Patient Selection |
| Was a case–control design avoided? |
| Were inappropriate exclusions avoided? |
| Risk of Bias: Was patient selection conducted in a way that avoids bias? |
| Applicability Concern: Do the included patients match the review question? |
| Index Test |
| Was the index test (test being evaluated) result interpreted without knowledge of the reference standard? (Blinded interpretation) |
| Was the test threshold pre-specified (rather than determined post hoc)? |
| Risk of Bias: Was the index test conducted in a way that avoids bias? |
| Applicability Concern: Does the index test match the review question? |
| Reference Standard |
| Is the reference standard likely to correctly classify the target condition? |
| Was the reference standard interpreted without knowledge of the index test? (Blinded interpretation) |
| Risk of Bias: Was the reference standard appropriate? |
| Applicability Concern: Does the reference standard match the review question? |
| Flow and Timing |
| Was there an appropriate time interval between the index test and reference standard? |
| Did all patients receive the same reference standard? |
| Were all patients included in the final analysis? |
| Risk of Bias: Was patient flow and timing handled appropriately? |
Appendix C. Data Extraction Template

Figure A1.
Data Extract Template on Covidence.
Appendix D. Raw Data Table

Table A2.
Raw Data Table for meta-analysis [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38].
Table A2.
Raw Data Table for meta-analysis [17,25,26,27,28,29,30,31,32,33,34,35,36,37,38].
| Study ID | Total Number of Participants | TP | FP | TN | FN | SP | Standard Error | Lower CI | Upper CI | SN | Standard Error 2 | Lower CI 2 | Upper CI 2 | PPV | Standard Error 3 | Lower CI 3 | Upper CI 3 | NPV | Standard Error 4 | Lower CI 4 | Upper CI 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 150 | 8 | 0 | 142 | 0 | 1 | 0.006901408 | 0.973659093 | 1 | 1 | 0.1225 | 0.67558438 | 1 | 1 | 0.1225 | 0.67558438 | 1 | 1 | 0.006901408 | 0.973659093 | 1 |
| Barkai 2014 [26] | 9845 | 48 | 8 | 9789 | 0 | 0.9992 | 0.000302652 | 0.998411364 | 0.999597297 | 1 | 0.020416667 | 0.925897349 | 1 | 0.8571 | 0.049933812 | 0.74258818 | 0.925762933 | 1 | 0.000100112 | 0.999607713 | 1 |
| Cardoso 2015 [27] | 333 | 4 | 0 | 328 | 1 | 1 | 0.002987805 | 0.988423392 | 1 | 0.8 | 0.265360133 | 0.375528264 | 0.963776839 | 1 | 0.245 | 0.51009998 | 1 | 0.997 | 0.004238393 | 0.983052342 | 0.999475082 |
| Choudhary 2015 [28] | 18 | 18 | 0 | 0 | 0 | - | - | - | - | 1 | 0.054444444 | 0.824115449 | 1 | 1 | 0.054444444 | 0.824115449 | 1 | - | - | - | - |
| Dollard 2021 [17] | 12,554 | 52 | 8 | 12,490 | 4 | 0.999 | 0.000293396 | 0.998271786 | 0.999421546 | 0.929 | 0.038523879 | 0.830800296 | 0.972119412 | 0.867 | 0.046782772 | 0.758739536 | 0.931092799 | 1 | 7.84377 E-05 | 0.999692619 | 1 |
| Eventov-Friedman 2019 [29] | 856 | 57 | 68 | 730 | 1 | 0.915 | 0.009948395 | 0.893606311 | 0.932417183 | 0.983 | 0.988489251 | 0.199595396 | 0.999925424 | 0.456 | 0.062094275 | 0.343155986 | 0.573549656 | 0.999 | 0.001778738 | 0.992923231 | 0.999859431 |
| Exler 2019 [30] | 133 | 27 | 10 | 94 | 2 | 0.904 | 0.030385127 | 0.832175114 | 0.947041811 | 0.931 | 0.057940588 | 0.780304765 | 0.980863997 | 0.73 | 0.077643808 | 0.570498487 | 0.846233472 | 0.98 | 0.017560659 | 0.928436508 | 0.994625623 |
| Fernandes 2021 [31] | 1492 | 10 | 4 | 1478 | 0 | 0.9973 | 0.001501406 | 0.993079098 | 0.998949392 | 1 | 0.098 | 0.722459831 | 1 | 0.7143 | 0.139559386 | 0.453518208 | 0.882796921 | 1 | 0.000663058 | 0.99740755 | 1 |
| Huang 2021 [33] | 6234 | 12 | 0 | 6185 | 37 | 1 | 0.000158448 | 0.99937927 | 1 | 0.245 | 0.06461424 | 0.146101715 | 0.380975436 | 1 | 0.081666667 | 0.757499243 | 1 | 0.994 | 0.000991636 | 0.991752774 | 0.995637589 |
| Izquierdo 2023 [34] | 1651 | 7 | 2 | 1641 | 1 | 0.998 | 0.001253247 | 0.994387678 | 0.999288945 | 0.875 | 1.03429928 | 0.158743682 | 0.996163786 | 0.777 | 0.57160782 | 0.211260159 | 0.978413903 | 0.999 | 0.000982148 | 0.995914755 | 0.999755788 |
| Izquierdo 2023.2 [34] | 119 | 2 | 0 | 116 | 1 | 1 | 0.008448276 | 0.967944353 | 1 | 0.666 | 0.425275336 | 0.207287919 | 0.938292062 | 1 | 0.49 | 0.342371953 | 1 | 0.9914 | 0.011959543 | 0.953082694 | 0.998473723 |
| Izquierdo 2024 [34] | 190 | 3 | 0 | 186 | 1 | 1 | 0.005268817 | 0.979764182 | 1 | 0.75 | 0.326955654 | 0.300636052 | 0.954413937 | 1 | 0.326666667 | 0.43849392 | 1 | 0.995 | 0.007353136 | 0.970913725 | 0.99915779 |
| Pasternak 2020 [35] | 83 | 42 | 0 | 41 | 0 | 1 | 0.023902439 | 0.914329551 | 1 | 1 | - | - | - | 1 | - | - | - | 1 | 0.023902439 | 0.914329551 | 1 |
| Pasternak 2020.2 [35] | 83 | 41 | 0 | 41 | 1 | 1 | 0.023902439 | 0.914329551 | 1 | 0.976 | 0.991879025 | 0.196777328 | 0.999851885 | 1 | - | - | - | 0.976 | 0.033198776 | 0.876493769 | 0.995727096 |
| Pasternak 2020.3 [35] | 83 | 42 | 3 | 38 | 0 | 0.927 | 0.047136385 | 0.805946267 | 0.97489117 | 1 | - | - | - | 0.933 | 0.357138784 | 0.382925624 | 0.99681011 | 1 | 0.025789474 | 0.908187067 | 1 |
| Ross 2014 [36] | 80 | 79 | 1 | 0 | 0 | 0 | 0.98 | 0 | 0.793456709 | 1 | 0.012405063 | 0.953627163 | 1 | 0.988 | 0.017270278 | 0.93334121 | 0.997938727 | - | - | - | - |
| Stark 2024 [37] | 880 | 33 | 4 | 838 | 5 | 0.994 | 0.002904787 | 0.986088851 | 0.997423901 | 0.868 | 0.060665104 | 0.726226125 | 0.942199556 | 0.892 | 0.057490953 | 0.753044839 | 0.95721137 | 0.994 | 0.002902787 | 0.986095372 | 0.997422679 |
| Yamamoto 2006 [38] | 1923 | 22 | 2 | 1895 | 4 | 0.999 | 0.000890787 | 0.996249107 | 0.999733936 | 0.846 | 0.080197463 | 0.664506503 | 0.938410231 | 0.917 | 0.069560425 | 0.741935722 | 0.976988478 | 0.998 | 0.00114778 | 0.994749493 | 0.999239707 |
Appendix E. Study Characteristics Table

Table A3.
Study Characteristics Table.
Table A3.
Study Characteristics Table.
| Study | Year of Publication | Country in Which the Study Conducted | Total Population | Temporal Classification | Study Design | Type of Test Conducted for Saliva | Type of Test Conducted for Urine | Saliva Sample | Urine Sample | Population Type | Age of Children | SP | SN | P PV | NPV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 2012 | Bahrain | 150 | prospective | cohort | nested PCR | nested PCR | Liquid | unclear | seropositive mothers | <6 wks | 1.000 | 1.000 | 1.000 | 1.000 |
| Barkai 2014 [26] | 2014 | Israel | 9845 | prospective | cross-sectional | RT-PCR | RT-PCR | unclear | unclear | All | <4 wks | 0.999 | 1.000 | 0.857 | 1.000 |
| Cardoso 2015 [27] | 2015 | Brazil | 333 | prospective | cross-sectional | nested PCR | nested PCR | Liquid | Bag | All | <3 wks | 1.000 | 0.800 | 1.000 | 0.997 |
| Choudhary 2015 [28] | 2015 | India | 18 | prospective | cohort | PCR | PCR | Liquid | Sterile tube | High-risk | <10 months | - | 1.000 | 1.000 | - |
| Dollard 2021 [17] | 2021 | United States | 12,554 | prospective | cohort | PCR | PCR | Dry | unclear | All | <3 wks | 0.999 | 0.929 | 0.867 | 1.000 |
| Eventov-Friedman 2019 [29] | 2019 | Israel | 856 | prospective | cohort | PCR | PCR | Liquid | Bag | High-risk | <10 days | 0.915 | 0.983 | 0.456 | 0.999 |
| Exler 2019 [30] | 2019 | Germany | 133 | retrospective | cohort | PCR | PCR | Liquid | unclear | seropositive mothers | <2 wks | 0.904 | 0.931 | 0.730 | 0.980 |
| Fernandes 2021 [31] | 2021 | Portugal | 1492 | prospective | cross-sectional | qPCR | qPCR | Liquid | Bag | All | <2 wks | 0.997 | 1.000 | 0.714 | 1.000 |
| Huang 2021 [33] | 2021 | China | 6234 | prospective | cohort | qPCR | qPCR | Liquid | Bag | seropositive mothers | <3 wks | 1.000 | 0.245 | 1.000 | 0.994 |
| Izquierdo 2023 [34] | 2023 | Chile | 1651 | prospective | cross-sectional | RT-PCR | RT-PCR | Dry | Bag | All | <3 wks | 0.998 | 0.875 | 0.777 | 0.999 |
| Izquierdo 2023.2 [34] | 2023 | Chile | 119 | prospective | cross-sectional | RT-PCR | RT-PCR | Dry | Bag | High-risk | <3 wks | 1.000 | 0.666 | 1.000 | 0.991 |
| Izquierdo 2024 [34] | 2024 | Chile | 190 | prospective | cross-sectional | RT-PCR | RT-PCR | Dry | Bag | High-risk | <3 wks | 1.000 | 0.750 | 1.000 | 0.995 |
| Pasternak 2020 [35] | 2020 | Israel | 83 | prospective | case–control | qPCR | qPCR | Liquid | Bag | High-risk | <4 wks | 1.000 | 1.000 | 1.000 | 1.000 |
| Pasternak 2020.2 [35] | 2020 | Israel | 83 | prospective | case–control | qPCR | qPCR | Wet | Bag | High-risk | <4 wks | 1.000 | 0.976 | 1.000 | 0.976 |
| Pasternak 2020.3 [35] | 2020 | Israel | 83 | prospective | case–control | qPCR | qPCR | Dry | Bag | High-risk | <4 wks | 0.927 | 1.000 | 0.933 | 1.000 |
| Ross 2014 [36] | 2014 | United States | 80 | prospective | cohort | qPCR | qPCR | Liquid | Bag | High-risk | <3 wks | 0.000 | 1.000 | 0.988 | - |
| Stark 2024 [37] | 2024 | United States | 880 | retrospective | cohort | qPCR | PCR | unclear | unclear | High-risk | <3 wks | 0.994 | 0.868 | 0.892 | 0.994 |
| Yamamoto 2006 [38] | 2006 | Brazil | 1923 | prospective | cross-sectional | PCR | PCR | Liquid | Sterile tube | All | <3 wks | 0.999 | 0.846 | 0.917 | 0.998 |
Appendix F. Full LOO Tables

Table A4.
Leave-Out-Analysis 1-Specificity.
Table A4.
Leave-Out-Analysis 1-Specificity.
| Study | Estimate | SE | Z_Value | p_Value | CI_Lower | CI_Upper | Q | Qp | Tau2 | I2 | H2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 0.99835 | 0.00068 | 1473.69292 | <0.0001 | 0.99702 | 0.99968 | 105.06197 | 5.04392E-16 | 2.48461E-06 | 86.67453 | 7.50443 |
| Barkai 2014 [26] | 0.99790 | 0.00088 | 1128.29218 | <0.0001 | 0.99616 | 0.99963 | 102.93035 | 1.29820E-15 | 3.99736E-06 | 86.39857 | 7.35217 |
| Cardoso 2015 [27] | 0.99830 | 0.00069 | 1448.59914 | <0.0001 | 0.99695 | 0.99965 | 105.04763 | 5.07616E-16 | 2.49311E-06 | 86.67271 | 7.50340 |
| Dollard 2021 [17] | 0.99793 | 0.00089 | 1126.86722 | <0.0001 | 0.99620 | 0.99967 | 99.91582 | 4.92191E-15 | 4.01000E-06 | 85.98820 | 7.13684 |
| Eventov-Friedman 2019 [29] | 0.99908 | 0.00038 | 2662.53506 | <0.0001 | 0.99835 | 0.99982 | 32.73220 | 3.14837E-03 | 5.10888E-07 | 57.22866 | 2.33801 |
| Exler 2019 [30] | 0.99847 | 0.00065 | 1547.15384 | <0.0001 | 0.99720 | 0.99973 | 95.16536 | 3.97717E-14 | 2.21286E-06 | 85.28876 | 6.79753 |
| Fernandes 2021 [31] | 0.99849 | 0.00071 | 1413.18368 | <0.0001 | 0.99710 | 0.99987 | 102.69548 | 1.44050E-15 | 2.46067E-06 | 86.36746 | 7.33539 |
| Huang 2021 [33] | 0.99759 | 0.00096 | 1037.24469 | <0.0001 | 0.99570 | 0.99947 | 88.89298 | 6.13968E-13 | 4.98788E-06 | 84.25072 | 6.34950 |
| Izquierdo 2023 [34] | 0.99841 | 0.00072 | 1391.73049 | <0.0001 | 0.99700 | 0.99982 | 103.41226 | 1.04860E-15 | 2.49966E-06 | 86.46195 | 7.38659 |
| Izquierdo 2023.2 [34] | 0.99836 | 0.00068 | 1476.01648 | <0.0001 | 0.99703 | 0.99968 | 105.06307 | 5.04146E-16 | 2.48396E-06 | 86.67467 | 7.50451 |
| Izquierdo 2024 [34] | 0.99834 | 0.00068 | 1468.91767 | <0.0001 | 0.99701 | 0.99967 | 105.05961 | 5.04923E-16 | 2.48601E-06 | 86.67423 | 7.50426 |
| Pasternak 2020 [35] | 0.99837 | 0.00067 | 1480.26758 | <0.0001 | 0.99704 | 0.99969 | 105.06500 | 5.03715E-16 | 2.48282E-06 | 86.67492 | 7.50464 |
| Pasternak 2020.2 [35] | 0.99837 | 0.00067 | 1480.26758 | <0.0001 | 0.99704 | 0.99969 | 105.06500 | 5.03715E-16 | 2.48282E-06 | 86.67492 | 7.50464 |
| Pasternak 2020.3 [35] | 0.99839 | 0.00067 | 1495.79361 | <0.0001 | 0.99709 | 0.99970 | 102.69279 | 1.44222E-15 | 2.41803E-06 | 86.36711 | 7.33520 |
| Stark 2024 [37] | 0.99857 | 0.00068 | 1471.41615 | <0.0001 | 0.99724 | 0.99990 | 101.33737 | 2.62705E-15 | 2.39212E-06 | 86.18476 | 7.23838 |
| Yamamoto 2006 [38] | 0.99824 | 0.00074 | 1350.65444 | <0.0001 | 0.99679 | 0.99969 | 104.59739 | 6.19929E-16 | 2.59703E-06 | 86.61534 | 7.47124 |

Table A5.
Leave-Out-Analysis 2-Sensitivity.
Table A5.
Leave-Out-Analysis 2-Sensitivity.
| Study | Estimate | SE | Z_Value | p_Value | CI_Lower | CI_Upper | Q | Qp | Tau2 | I2 | H2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 0.86748 | 0.04870 | 17.81205 | <0.0001 | 0.77202 | 0.96293 | 142.47142 | 2.28664E-23 | 0.01954 | 90.17347 | 10.17653 |
| Barkai 2014 [26] | 0.85824 | 0.06241 | 13.75202 | <0.0001 | 0.73592 | 0.98056 | 140.02984 | 6.99861E-23 | 0.03393 | 90.00213 | 10.00213 |
| Cardoso 2015 [27] | 0.87799 | 0.04743 | 18.51029 | <0.0001 | 0.78502 | 0.97096 | 142.10809 | 2.70109E-23 | 0.01927 | 90.14834 | 10.15058 |
| Choudhary 2015 [28] | 0.86208 | 0.05076 | 16.98425 | <0.0001 | 0.76260 | 0.96156 | 142.24243 | 2.53976E-23 | 0.02067 | 90.15765 | 10.16017 |
| Dollard 2021 [17] | 0.86898 | 0.05233 | 16.60448 | <0.0001 | 0.76640 | 0.97155 | 141.23326 | 4.03325E-23 | 0.02211 | 90.08732 | 10.08809 |
| Eventov-Friedman 2019 [29] | 0.87584 | 0.04691 | 18.67088 | <0.0001 | 0.78390 | 0.96778 | 142.52594 | 2.23019E-23 | 0.01928 | 90.17723 | 10.18042 |
| Exler 2019 [30] | 0.86978 | 0.05050 | 17.22255 | <0.0001 | 0.77080 | 0.96876 | 142.02524 | 2.80564E-23 | 0.02046 | 90.14260 | 10.14466 |
| Fernandes 2021 [31] | 0.86571 | 0.04917 | 17.60662 | <0.0001 | 0.76934 | 0.96208 | 142.44039 | 2.31941E-23 | 0.01969 | 90.17133 | 10.17431 |
| Huang 2021 [33] | 0.98751 | 0.00961 | 102.79747 | <0.0001 | 0.96868 | 1.00633 | 13.33019 | 5.00705E-01 | 0.00000 | 0.00000 | 1.00000 |
| Izquierdo 2023 [34] | 0.87608 | 0.04690 | 18.67837 | <0.0001 | 0.78415 | 0.96801 | 142.51738 | 2.23896E-23 | 0.01927 | 90.17664 | 10.17981 |
| Izquierdo 2023.2 [34] | 0.87842 | 0.04706 | 18.66679 | <0.0001 | 0.78619 | 0.97066 | 142.00995 | 2.82538E-23 | 0.01922 | 90.14154 | 10.14357 |
| Izquierdo 2024 [34] | 0.87832 | 0.04724 | 18.59445 | <0.0001 | 0.78574 | 0.97090 | 142.06694 | 2.75252E-23 | 0.01924 | 90.14549 | 10.14764 |
| Pasternak 2020.2 [35] | 0.87586 | 0.04691 | 18.67139 | <0.0001 | 0.78392 | 0.96780 | 142.52605 | 2.23007E-23 | 0.01928 | 90.17723 | 10.18043 |
| Ross 2014 [36] | 0.85778 | 0.06461 | 13.27719 | <0.0001 | 0.73116 | 0.98440 | 129.70910 | 7.75839E-21 | 0.03686 | 89.20662 | 9.26494 |
| Stark 2024 [37] | 0.87669 | 0.04996 | 17.54918 | <0.0001 | 0.77878 | 0.97460 | 139.54507 | 8.73733E-23 | 0.01995 | 89.96740 | 9.96751 |
| Yamamoto 2006 [38] | 0.87880 | 0.04926 | 17.84000 | <0.0001 | 0.78225 | 0.97535 | 140.04435 | 6.95228E-23 | 0.01953 | 90.00317 | 10.00317 |

Table A6.
Leave-Out-Analysis 3-PPV.
Table A6.
Leave-Out-Analysis 3-PPV.
| Study | Estimate | SE | Z_Value | p_Value | CI_Lower | CI_Upper | Q | Qp | Tau2 | I2 | H2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 0.84579 | 0.04740 | 17.84378 | <0.0001 | 0.75289 | 0.93869 | 93.34868 | 8.81038E-14 | 0.01986 | 85.00247 | 6.66776 |
| Barkai 2014 [26] | 0.85437 | 0.04999 | 17.08923 | <0.0001 | 0.75639 | 0.95236 | 90.07972 | 3.66575E-13 | 0.02176 | 84.45821 | 6.43427 |
| Cardoso 2015 [27] | 0.85655 | 0.04610 | 18.57963 | <0.0001 | 0.76619 | 0.94691 | 93.22106 | 9.31590E-14 | 0.01944 | 84.98194 | 6.65865 |
| Choudhary 2015 [28] | 0.83993 | 0.05006 | 16.77826 | <0.0001 | 0.74182 | 0.93805 | 92.57471 | 1.23559E-13 | 0.02189 | 84.87708 | 6.61248 |
| Dollard 2021 [17] | 0.85318 | 0.05056 | 16.87489 | <0.0001 | 0.75408 | 0.95227 | 90.39569 | 3.19487E-13 | 0.02237 | 84.51254 | 6.45684 |
| Eventov-Friedman 2019 [29] | 0.90788 | 0.02801 | 32.41077 | <0.0001 | 0.85298 | 0.96279 | 28.86187 | 1.09092E-02 | 0.00401 | 51.49310 | 2.06156 |
| Exler 2019 [30] | 0.86662 | 0.04674 | 18.54056 | <0.0001 | 0.77501 | 0.95824 | 85.50974 | 2.65604E-12 | 0.01859 | 83.62760 | 6.10784 |
| Fernandes 2021 [31] | 0.86325 | 0.04650 | 18.56453 | <0.0001 | 0.77211 | 0.95439 | 90.71594 | 2.77911E-13 | 0.01909 | 84.56721 | 6.47971 |
| Huang 2021 [33] | 0.84242 | 0.04843 | 17.39460 | <0.0001 | 0.74750 | 0.93734 | 93.11580 | 9.75457E-14 | 0.02044 | 84.96496 | 6.65113 |
| Izquierdo 2023 [34] | 0.85572 | 0.04568 | 18.73342 | <0.0001 | 0.76619 | 0.94525 | 93.44245 | 8.45646E-14 | 0.01941 | 85.01752 | 6.67446 |
| Izquierdo 2023.2 [34] | 0.85725 | 0.04572 | 18.75095 | <0.0001 | 0.76764 | 0.94685 | 93.09243 | 9.85470E-14 | 0.01934 | 84.96118 | 6.64946 |
| Izquierdo 2024 [34] | 0.85702 | 0.04590 | 18.67118 | <0.0001 | 0.76705 | 0.94698 | 93.16475 | 9.54805E-14 | 0.01939 | 84.97286 | 6.65463 |
| Pasternak 2020.3 [35] | 0.85412 | 0.04592 | 18.60090 | <0.0001 | 0.76412 | 0.94412 | 93.53004 | 8.13867E-14 | 0.01947 | 85.03155 | 6.68072 |
| Ross 2014 [36] | 0.83938 | 0.04929 | 17.02881 | <0.0001 | 0.74277 | 0.93598 | 58.00863 | 2.61361E-07 | 0.02065 | 75.86566 | 4.14347 |
| Stark 2024 [37] | 0.85096 | 0.04975 | 17.10584 | <0.0001 | 0.75346 | 0.94846 | 92.55589 | 1.24578E-13 | 0.02157 | 84.87400 | 6.61114 |
| Yamamoto 2006 [38] | 0.84903 | 0.04905 | 17.30819 | <0.0001 | 0.75289 | 0.94518 | 93.33104 | 8.87861E-14 | 0.02096 | 84.99963 | 6.66650 |

Table A7.
Leave-Out-Analysis 4-NPV.
Table A7.
Leave-Out-Analysis 4-NPV.
| Study | Estimate | SE | Z_Value | p_Value | CI_Lower | CI_Upper | Q | Qp | Tau2 | I2 | H2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkhawaja 2012 [25] | 0.99929 | 2.926E-04 | 3415.01153 | <0.0001 | 0.99872 | 0.99986 | 48.19143 | 1.22202E-05 | 2.58765E-07 | 70.94919 | 3.44224 |
| Barkai 2014 [26] | 0.99807 | 7.727E-04 | 1291.64409 | <0.0001 | 0.99655 | 0.99958 | 47.96598 | 1.33172E-05 | 3.08023E-06 | 70.81265 | 3.42614 |
| Cardoso 2015 [27] | 0.99931 | 2.916E-04 | 3427.09513 | <0.0001 | 0.99873 | 0.99988 | 47.70285 | 1.47204E-05 | 2.55168E-07 | 70.65165 | 3.40735 |
| Dollard 2021 [17] | 0.99807 | 7.734E-04 | 1290.47523 | <0.0001 | 0.99655 | 0.99958 | 47.60547 | 1.52758E-05 | 3.08688E-06 | 70.59162 | 3.40039 |
| Eventov-Friedman 2019 [29] | 0.99930 | 2.955E-04 | 3381.54984 | <0.0001 | 0.99872 | 0.99988 | 47.89841 | 1.36645E-05 | 2.57413E-07 | 70.77147 | 3.42131 |
| Exler 2019 [30] | 0.99931 | 2.884E-04 | 3464.84240 | <0.0001 | 0.99874 | 0.99988 | 46.89921 | 1.99691E-05 | 2.48935E-07 | 70.14875 | 3.34994 |
| Fernandes 2021 [31] | 0.99918 | 3.151E-04 | 3170.71180 | <0.0001 | 0.99856 | 0.99980 | 48.18820 | 1.22353E-05 | 2.65545E-07 | 70.94725 | 3.44201 |
| Huang 2021 [33] | 0.99999 | 6.121E-05 | 16337.66443 | <0.0001 | 0.99987 | 1.00011 | 11.90201 | 6.14172E-01 | 0.00000 | 0.00000 | 1.00000 |
| Izquierdo 2023 [34] | 0.99932 | 3.012E-04 | 3317.47315 | <0.0001 | 0.99873 | 0.99991 | 47.22766 | 1.76324E-05 | 2.54414E-07 | 70.35635 | 3.37340 |
| Izquierdo 2023.2 [34] | 0.99930 | 2.909E-04 | 3435.58932 | <0.0001 | 0.99873 | 0.99987 | 47.67887 | 1.48553E-05 | 2.54845E-07 | 70.63689 | 3.40563 |
| Izquierdo 2024 [34] | 0.99930 | 2.912E-04 | 3431.71177 | <0.0001 | 0.99873 | 0.99987 | 47.73599 | 1.45360E-05 | 2.55311E-07 | 70.67202 | 3.40971 |
| Pasternak 2020 [35] | 0.99929 | 2.924E-04 | 3418.07605 | <0.0001 | 0.99872 | 0.99986 | 48.19146 | 1.22201E-05 | 2.58709E-07 | 70.94921 | 3.44225 |
| Pasternak 2020.2 [35] | 0.99930 | 2.908E-04 | 3436.84941 | <0.0001 | 0.99873 | 0.99987 | 47.67048 | 1.49028E-05 | 2.54764E-07 | 70.63172 | 3.40503 |
| Pasternak 2020.3 [35] | 0.99929 | 2.924E-04 | 3418.11551 | <0.0001 | 0.99872 | 0.99986 | 48.19146 | 1.22201E-05 | 2.58708E-07 | 70.94921 | 3.44225 |
| Stark 2024 [37] | 0.99939 | 2.804E-04 | 3564.54656 | <0.0001 | 0.99884 | 0.99994 | 43.97065 | 5.98460E-05 | 2.27075E-07 | 68.16058 | 3.14076 |
| Yamamoto 2006 [38] | 0.99939 | 2.917E-04 | 3425.77611 | <0.0001 | 0.99882 | 0.99996 | 45.26014 | 3.70113E-05 | 2.38587E-07 | 69.06770 | 3.23287 |
References
- Varnum, S.M.; Streblow, D.N.; Monroe, M.E.; Smith, P.; Auberry, K.J.; Paša-Tolić, L.; Wang, D.; Camp, D.G., 2nd; Rodland, K.; Wiley, S.; et al. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). About Cytomegalovirus. Cytomegalovirus (CMV) and Congenital CMV Infection. 2024. Available online: https://www.cdc.gov/cytomegalovirus/about/index.html (accessed on 15 June 2025).
- Al Mana, H.; Yassine, H.M.; Younes, N.N.; Al-Mohannadi, A.; Al-Sadeq, D.W.; Alhababi, D.; Nasser, E.A.; Nasrallah, G.K. The status of cytomegalovirus (CMV) prevalence in the MENA region: A systematic review. Pathogens 2019, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Clinical Overview of CMV and Congenital CMV. Cytomegalovirus (CMV) and Congenital CMV Infection. 2024. Available online: https://www.cdc.gov/cytomegalovirus/hcp/clinical-overview/index.html (accessed on 15 June 2025).
- Sinzger, C.; Digel, M.; Jahn, G. Cytomegalovirus cell tropism. Med. Microbiol. Immunol. 2021, 210, 325–339. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). CMV in Newborns. Cytomegalovirus (CMV) and Congenital CMV Infection. 2024. Available online: https://www.cdc.gov/cytomegalovirus/congenital-infection/index.html (accessed on 15 June 2025).
- National Deaf Children’s Society. Cytomegalovirus Childhood Deafness|Deafness Caused by CMV. Available online: https://www.ndcs.org.uk/information-and-support/childhood-deafness/causes-of-deafness/cytomegalovirus-cmv/ (accessed on 15 June 2025).
- Grosse, S.D. Congenital cytomegalovirus (CMV) infection as a cause of permanent bilateral hearing loss: A quantitative assessment. J. Clin. Virol. 2007, 41, 57–62. [Google Scholar] [CrossRef]
- Aldè, M.; Fancello, V.; Di Mauro, P.; Canelli, R.; Zaouche, S.; Falanga, C. Audiological and vestibular follow-up for children with congenital cytomegalovirus infection: From current limitations to future directions. Children 2024, 11, 1211. [Google Scholar] [CrossRef]
- Grosse, S.D. Economic assessments of the burden of congenital cytomegalovirus infection and the cost-effectiveness of prevention strategies. Expert Rev. Pharmacoecon Outcomes Res. 2021, 21, 271–283. [Google Scholar] [CrossRef]
- Retzler, J.; Hex, N.; Bartlett, C.; Webb, A.; Wood, S.; Star, C.; Griffiths, P.; Jones, C.E. Economic cost of congenital CMV in the UK. Arch. Dis. Child. 2018, 104, 559–563. [Google Scholar] [CrossRef]
- de Vries, J.J.; van der Eijk, A.A.; Wolthers, K.C.; Rusman, L.G.; Pas, S.D.; Molenkamp, R.; Claas, E.C.; Kroes, A.C.; Vossen, A.C. Real-time PCR versus viral culture on urine as a gold standard in the diagnosis of congenital cytomegalovirus infection. J. Clin. Virol. 2012, 53, 167–170. [Google Scholar] [CrossRef]
- Ross, S.A.; Ahmed, A.; Palmer, A.L.; Michaels, M.G.; Sánchez, P.J.; Stewart, A.; Bernstein, D.I.; Feja, K.; Novak, Z.; Fowler, K.B.; et al. Urine collection method for the diagnosis of congenital cytomegalovirus infection. Pediatr. Infect. Dis. J. 2015, 34, 903–905. [Google Scholar] [CrossRef]
- Waters, A.; Jennings, K.; Fitzpatrick, E.; Coughlan, S.; Molloy, E.J.; De Gascun, C.F.; Hall, W.W.; Knowles, S.J. Incidence of congenital cytomegalovirus infection in Ireland: Implications for screening and diagnosis. J. Clin. Virol. 2014, 59, 156–160. [Google Scholar] [CrossRef]
- Izquierdo, G.; Guerra, C.; Reyes, R.; Araya, L.; Sepulveda, B.; Cabrera, C.; Medina, P.; Mardones, E.; Villavicencio, L.; Montecinos, L.; et al. Universal and expanded screening strategy for congenital cytomegalovirus infection: Is pool testing by a rapid molecular test in saliva a new choice in developing countries? Viruses 2024, 16, 772. [Google Scholar] [CrossRef]
- Ross, S.A.; Michaels, M.G.; Ahmed, A.; Palmer, A.L.; Sánchez, P.J.; Bernstein, D.I.; Feja, K.; Stewart, A.; Boppana, S.B.; Fowler, K.B. Contribution of breastfeeding to false-positive saliva polymerase chain reaction for newborn congenital cytomegalovirus screening. JAMA Pediatr. 2018, 172, 535–543. [Google Scholar] [CrossRef]
- Dollard, S.C.; Dreon, M.; Hernandez-Alvarado, N.; Amin, M.M.; Wong, P.; Lanzieri, T.M.; Osterholm, E.A.; Sidebottom, A.; Rosendahl, S.; McCann, M.T.; et al. Sensitivity of dried blood spot testing for detection of congenital cytomegalovirus infection. JAMA Pediatr. 2021, 175, e20544. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Shetty, J.K.; Rady, S.; Alameddine, M.; Abde, M.I.; Sabra, S. Evaluation of Salivary Sample in Diagnosis, Screening and Management of Cytomegalovirus (CMV) Infection in Newborns-Systematic Review and Metanalysis. 2024. Available online: https://www.crd.york.ac.uk/PROSPERO/view/CRD42024603785 (accessed on 20 April 2025).
- Reitsma, J.B.; Rutjes, A.; Whiting, P.; Yang, B.; Leeflang, M.M.; Bossuyt, P.M.; Deeks, J.J. Chapter 8: Assessing risk of bias and applicability. In Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy, Version 2.0; Deeks, J.J., Bossuyt, P.M., Leeflang, M.M., Takwoingi, Y., Eds.; Cochrane: London, UK, 2023; Available online: https://training.cochrane.org/handbook-diagnostic-test-accuracy/current (accessed on 7 July 2025).
- Erdoğan, S.; Gülhan, O.T. Alternative Confidence Interval Methods Used in the Diagnostic Accuracy Studies. Comput. Math. Methods Med. 2016, 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, G. meta: An R package for meta-analysis. R. News 2007, 7, 40–45. [Google Scholar]
- Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- AlKhawaja, S.; Ismaeel, A.; Botta, G.; Senok, A.C. The prevalence of congenital and perinatal cytomegalovirus infections among newborns of seropositive mothers. J. Infect. Dev. Ctries. 2011, 6, 410–415. [Google Scholar] [CrossRef]
- Barkai, G.; Ari-Even Roth, D.; Barzilai, A.; Tepperberg-Oikawa, M.; Mendelson, E.; Hildesheimer, M.; Kuint, J. Universal neonatal cytomegalovirus screening using saliva—Report of clinical experience. J. Clin. Virol. 2014, 60, 361–366. [Google Scholar] [CrossRef]
- Cardoso, E.S.d.C.; Jesus, B.L.S.d.; da Gomes, L.G.S.; Sousa, S.M.B.; Gadelha, S.R.; Marin, L.J. The use of saliva as a practical and feasible alternative to urine in large-scale screening for congenital cytomegalovirus infection increases inclusion and detection rates. Rev. Soc. Bras. Med. Trop. 2015, 48, 206–207. [Google Scholar] [CrossRef][Green Version]
- Choudhary, A.; Pati, S.K.; Patro, R.K.; Deorari, A.K.; Dar, L. Comparison of conventional, immunological and molecular techniques for the diagnosis of symptomatic congenital human cytomegalovirus infection in neonates and infants. Indian J. Med. Microbiol. 2015, 33, 15–19. [Google Scholar] [CrossRef]
- Eventov-Friedman, S.; Manor, H.; Bar-Oz, B.; Averbuch, D.; Caplan, O.; Lifshitz, A.; Bdolah-Abram, T.; Wolf, D.G. Saliva Real-Time Polymerase Chain Reaction for Targeted Screening of Congenital Cytomegalovirus Infection. J. Infect. Dis. 2019, 220, 1790–1796. [Google Scholar] [CrossRef]
- Exler, S.; Daiminger, A.; Grothe, M.; Schalasta, G.; Enders, G.; Enders, M. Primary cytomegalovirus (CMV) infection in pregnancy: Diagnostic value of CMV PCR in saliva compared to urine at birth. J. Clin. Virol. 2019, 117, 33–36. [Google Scholar] [CrossRef]
- Fernandes, C.; Marques, A.; de Jesus Chasqueira, M.; Braz, M.C.; Ferreira, A.R.; Neto, A.S.; Mendes, C.; Lito, D.; Menezes, M.F.; Sousa, M.J.; et al. Saliva pools for screening of human cytomegalovirus using real-time PCR. Eur. J. Pediatr. 2020, 180, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Forner, G.; Saldan, A.; Mengoli, C.; Gussetti, N.; Palù, G.; Abate, D. Cytomegalovirus (CMV) Enzyme-Linked Immunosorbent Spot Assay but Not CMV QuantiFERON Assay Is a Novel Biomarker To Determine Risk of Congenital CMV Infection in Pregnant Women. J. Clin. Microbiol. 2016, 54, 2149–2154. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, H.; Li, T.; Tang, J.; Yu, H.; Guo, X.; Song, Q.; Wei, F.; Wang, J.; Liang, C.; et al. Comparison of detection strategies for screening and confirming congenital cytomegalovirus infection in newborns in a highly seroprevalent population: A mother-child cohort study. Lancet Reg. Health West. Pac. 2021, 12, 100182. Available online: https://www.thelancet.com/action/showPdf?pii=S2666-6065%2821%2900091-2 (accessed on 28 January 2022). [CrossRef] [PubMed]
- Izquierdo, G.; Farfan, M.J.; Villavicencio, L.; Montecinos, L.; Tarque, F.; Acevedo, W.; Reyes, R.; Guerra, C.; Araya, L.; Sepúlveda, B.; et al. Optimizing congenital cytomegalovirus detection by pool testing in saliva by a rapid molecular test. Eur. J. Pediatr. 2023, 182, 5131–5136. [Google Scholar] [CrossRef]
- Pasternak, Y.; Tepperberg Oikawa, M.; Mendelson, E.; Osovsky, M.; Klinger, G.; Bilavsky, E. Diagnosing congenital cytomegalovirus by saliva on Guthrie paper. J. Clin. Virol. 2020, 126, 104337. [Google Scholar] [CrossRef]
- Ross, S.A.; Ahmed, A.; Palmer, A.L.; Michaels, M.G.; Sánchez, P.J.; Bernstein, D.I.; Tolan, R.W., Jr.; Novak, Z.; Chowdhury, N.; Fowler, K.B.; et al. Detection of congenital cytomegalovirus infection by real-time polymerase chain reaction analysis of saliva or urine specimens. J. Infect. Dis. 2014, 210, 1415–1418. [Google Scholar] [CrossRef]
- Stark, A.; Greenberg, R.G.; Pittman, R.; Weimer, K.E.D. Concordance of cytomegalovirus saliva and urine testing in infants for the detection of congenital infection. Pediatr. Infect. Dis. J. 2024, 43, 1191–1193. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.Y.; Mussi-Pinhata, M.M.; Marin, L.J.; Brito, R.M.; Oliveira, P.F.C.; Coelho, T.B. Is saliva as reliable as urine for detection of cytomegalovirus DNA for neonatal screening of congenital CMV infection? J. Clin. Virol. 2006, 36, 228–230. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Wu, F.F.; Li, X.X.; Shen, R.; Zheng, Z.; Liu, H.Y. Diagnostic test accuracy of PCR by saliva specimen for cytomegalovirus infection in newborn: A protocol for systematic review and meta-analysis. Medicine 2022, 101, e31776. [Google Scholar] [CrossRef] [PubMed]
- CDC. Laboratory Testing for CMV and Congenital CMV. Cytomegalovirus (CMV) and Congenital CMV Infection. 2024. Available online: https://www.cdc.gov/cytomegalovirus/php/laboratories/index.html (accessed on 2 March 2025).
- Cytomegalovirus (CMV) Infection: Causes & Symptoms. Cleveland Clinic. 2022. Available online: https://my.clevelandclinic.org/health/diseases/21166-cytomegalovirus#diagnosis-and-tests (accessed on 15 March 2025).
- Boston Children’s Hospital. Childrenshospital.org. 2025. Available online: https://www.childrenshospital.org/conditions/congenital-cytomegalovirus (accessed on 23 March 2025).
- Chiereghin, A.; Pavia, C.; Turello, G.; Borgatti, E.C.; Pillastrini, F.B.; Gabrielli, L.; Gibertoni, D.; Marsico, C.; De Paschale, M.; Manco, M.T.; et al. Universal Newborn Screening for Congenital Cytomegalovirus Infection—From Infant to Maternal Infection: A Prospective Multicenter Study. Front. Pediatr. 2022, 10, 909646. [Google Scholar] [CrossRef]
- Manicklal, S.; Emery, V.C.; Lazzarotto, T.; Boppana, S.B.; Gupta, R.K. The “silent” global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef]
- LetamendiaRichard, E.; PérillaudDubois, C.; de La Guillonnière, L.; Thouard, I.; Cordier, A.G.; RoqueAfonso, A.M.; de Luca, D.; Benachi, A.; VauloupFellous, C. Feasibility, performances and predictive value of congenital CMV screening using saliva swabs. BJOG Int. J. Obstet. Gynaecol. 2022, 129, 291–299. [Google Scholar] [CrossRef]
- Pinninti, S.; Boppana, S. Congenital cytomegalovirus infection diagnostics and management. Curr. Opin. Infect. Dis. 2022, 35, 436–441. [Google Scholar] [CrossRef]
- Salomè, S.; Corrado, F.R.; Mazzarelli, L.L.; Maruotti, G.M.; Capasso, L.; Blazquez-Gamero, D.; Raimondi, F. Congenital cytomegalovirus infection: The state of the art and future perspectives. Front. Pediatr. 2023, 11, 1276912. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thomas, J.; Chandler, J.; Cumpston, M.; Li, T.; Page, M.J.; Welch, V.A. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions, Version 6.3; Cochrane: London, UK, 2022; Available online: https://training.cochrane.org/handbook (accessed on 15 March 2025).
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Lawrence Erlbaum Associates: Hillsdale, NJ, USA, 1988. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).