Abstract
miRNAs, small non-coding RNAs that regulate gene expression, are involved in various pathological processes, including viral infections. Virus infections may interfere with the miRNA pathway through the inhibition of genes involved in miRNA biogenesis. A reduction in the number and the levels of miRNAs expressed in nasopharyngeal swabs of patients with severe COVID-19 was lately observed by us, pointing towards the potential of miRNAs as possible diagnostic or prognostic biomarkers for predicting outcomes among patients with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. The objective of the present study was to investigate whether SARS-CoV-2 infection influences the expression levels of messenger RNAs (mRNAs) of key genes involved in miRNA biogenesis. mRNA levels of AGO2, DICER1, DGCR8, DROSHA, and Exportin-5 (XPO5) were measured by quantitative reverse-transcription polymerase chain reaction (RT-qPCR) in nasopharyngeal swab specimens from patients with COVID-19 and controls, as well as in cells infected with SARS-CoV-2 in vitro. Our data showed that the mRNA expression levels of AGO2, DICER1, DGCR8, DROSHA, and XPO5 were not significantly different in patients with severe COVID-19 when compared to patients with non-severe COVID-19 and controls. Similarly, the mRNA expression of these genes was not affected by SARS-CoV-2 infection in NHBE and Calu-3 cells. However, in Vero E6 cells, AGO2, DICER1, DGCR8, and XPO5 mRNA levels were slightly upregulated 24 h after infection with SARS-CoV-2. In conclusion, we did not find evidence for downregulation of mRNA levels of miRNA biogenesis genes during SARS-CoV-2 infection, neither ex vivo nor in vitro.
1. Background
A novel coronavirus, named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) emerged in 2019 [1]. It rapidly spread around the globe and caused a pandemic with more than 670 million cases and more than 6.8 million deaths as of 10 March 2023 [2]. The end of COVID-19 as a public health emergency of international concern was recently declared by the head of the World Health Organization (WHO), however, stating that this does not mean that the disease is no longer a global threat [3]. The pandemic has had profound impacts on society and economy due to containment measures, including travel restrictions, school closures, and even complete lock-downs [4]. It also has shed light on the importance of having access to rapid and accurate diagnostics; however, massive diagnostic testing led to shortages in several reagents and plasticware needed for performing RT-PCR for SARS-CoV-2 detection [5,6].
SARS-CoV-2, the etiological agent of coronavirus disease 2019 (COVID-19) [7], belongs to the genus Betacoronavirus of the Coronaviridae family [8] and is an enveloped virus with a positive-sense single-stranded RNA genome of approximately 30 kb [9]. Severe forms of this disease are characterized by a state of hyperinflammation called ‘cytokine storm’ and are associated with severe lung disease with acute respiratory distress syndrome (ARDS) [10,11]. Several symptoms can be present, such as fatigue, fever, cough, dyspnea, headache, conjunctivitis, sore throat, dysgeusia, hyposmia, and gastrointestinal symptoms such as diarrhea, nausea, and vomiting [12,13]. Despite the development of vaccines and the availability of some treatment options, the understanding of the pathophysiology of the infection still remains a priority, especially due to observed symptoms of post-acute sequelae of SARS-CoV-2 infection (PASC), also known as “long COVID”, possibly related to a persistence of SARS-CoV-2 and reactivation of the latent pathogen [14]. Regarding the pathophysiology, several studies have shown that miRNAs are involved in infections caused by other coronaviruses [15,16,17,18,19,20]. microRNAs have also been shown to be involved in the pathophysiology of SARS-CoV-2 infection [21,22,23,24].
RNA silencing is a fundamental cellular mechanism of gene regulation and includes the small interfering RNA (siRNA) and microRNA (miRNA) pathways [25,26]. The miRNA pathway is evolutionarily conserved in metazoans [27]. miRNAs are small non-coding RNAs of approximately 22 nucleotides that regulate gene expression at the post-transcriptional level. miRNAs exert their function by binding most commonly to the 3′ untranslated regions (UTRs) or alternatively to 5′ UTRs or in the open reading frames (ORFs) of their target mRNAs [27,28,29]. miRNA biogenesis begins with the transcription from genes encoding the miRNA in the nucleus, resulting in a primary transcript with a hairpin structure, named pri-miRNA [30]. Then, the loop end of the pri-miRNA is cleaved by the microprocessor complex formed by the ribonuclease III Drosha and the co-factor DGCR8 (DiGeorge syndrome critical region gene 8) [31]. The cleavage results in the generation of the precursor miRNA, named pre-miRNA, which has a hairpin structure of about 70 nucleotides [32]. Next, the pre-miRNA is exported into the cytoplasm through Exportin-5 [31]. In the cytoplasm, the pre-miRNA is cleaved by Dicer, a cytoplasmic RNase endonuclease, to form the mature miRNA duplex of around 22 nucleotides [33]. This mature duplex miRNA associates with one argonaute protein (AGO), and following the expulsion of one of the miRNA strands, termed the passenger strand, the remaining single-stranded guide miRNA becomes part of the miRNA-induced silencing complex (miRISC) [34]. This complex allows the post-transcriptional regulation of gene expression through two mechanisms: translational repression or mRNA degradation [34,35,36].
The total number of human miRNAs is estimated to be 2300 [37]. miRNAs regulate the expression of most human genes and are involved in several physiological and pathological processes, such as development, cancers, viral infections, and antiviral immune responses [27]. Viruses interact with the miRNA machinery of their hosts and this interaction can either result in an increase or a repression of the expression of specific miRNAs. Thus virus infections profoundly affect cellular miRNA expression profiles [38,39,40,41,42]. Furthermore, several viruses encode their own viral miRNAs [38,39,41]. On the other hand, viruses can also globally interfere with the miRNA pathway. In fact, in plants and insects, RNA silencing pathways function as antiviral defense mechanisms [43,44]. Recent findings suggest they may also have antiviral functions in mammalian cells [44,45]. To escape this antiviral defense, plant and insect viruses possess virus-encoded suppressors of RNA interference that modulate the activity of central components of the RNA silencing pathways in order to favor viral replication or to inhibit host antiviral defense mechanisms [39,44,46]. There is some evidence that viruses also interfere with the miRNA pathway in mammals [39,44,47,48].
Several studies have shown a difference in miRNA expression levels between COVID-19 patients and uninfected individuals [49,50,51,52,53,54,55,56,57,58,59,60,61,62]. In our previous study [55], a lower total number of miRNAs expressed in nasopharyngeal swabs of patients with severe COVID-19 was noted. Furthermore, most differentially regulated miRNAs were downregulated in severe COVID-19 patients [55]. Among the hypotheses put forward to explain this observation was that SARS-CoV-2 infection inhibited cellular miRNA biogenesis. In the current study, we therefore investigated the impact of SARS-CoV-2 infection on the expression of miRNA biogenesis genes.
2. Materials and Methods
2.1. Patients and Specimens
Nasopharyngeal swab specimens of 60 patients were used in the study, including 19 patients with severe COVID-19, 21 patients with non-severe COVID-19 and 20 patients without COVID-19 (control group) [55]. Patients with severe COVID-19 needed intensive care unit admission and oxygen treatment and patients with non-severe COVID-19 required neither intensive care nor oxygen treatment. Nasopharyngeal swab specimens were obtained for routine diagnostic purposes by using flocked swabs that were placed in universal transport medium. Nasopharyngeal swab specimens were stored at −80°C. The study was approved by the French Institutional Authority for Personal Data Protection (Commission Nationale de l’Informatique et des Libertés DR-2020-178, 22 October 2020) and the ethics committee (Comité de Protection des Personnes Nord Ouest IV, ECH20/09, 7 September 2020).
2.2. Cells
The Vero E6 cell line (ATCC, CRL-1586) is a clone of strain 76, isolated from the kidney of an African green monkey with the morphology of epithelial cells. The Calu-3 cell line (Merck #SCC438, Sigma Aldrich, Saint-Quentin-Fallavier, France) is an epithelial cell line derived from human lung adenocarcinoma. The Vero E6 and Calu-3 cell lines were cultivated with DMEM medium, 4.5 g/L glucose, 10% fetal calf serum, 1% L-glutamine, and 1% penicillin and streptomycin (Gibco #15070-063, Thermofisher Scientific, Courtaboeuf, France) at 37 °C in 95% air/5% CO2 atmosphere.
Normal Human Bronchial Epithelial (NHBE) (Lonza, Switzerland) are human epithelial cells isolated from the airway epithelium above the bifurcation of the lungs of healthy patients. They were cultured with PneumaCultTM-Ex Plus Medium (StemCell Technologies, Saint Égrève, France) at 37 °C in 95% air/5% CO2 atmosphere.
2.3. Virus
SARS-CoV-2, variant B.1.617.2 (delta variant, GISAID ID EPI_ISL_2143633) was isolated from a clinical specimen and cultivated on Vero E6 cells in a biosafety 3 (BSL3) facility.
2.4. Infection of Cells
For Vero E6 cells, 104 cells per well were plated in 96-well plates two days before infection. Four replicates were used. For Calu-3 cells, 105 cells per well were plated in 48-well plates one day before infection. Four replicates were used. For NHBE cells, 5 × 105 cells per well were plated in 6-well plates one day before infection. Four replicates were used. Cells were infected with SARS-CoV-2 with a multiplicity of infection (MOI) of 0.1 for 24 and 48 h, with incubation of cells with the virus for 1 h at 37 °C in 95% air/5% CO2 atmosphere, followed by three washes with DMEM culture medium.
2.5. RNA Extraction
RNA was extracted from nasopharyngeal swab specimens using the MagMAX mirVana Total RNA Isolation Kit (Thermofisher Scientific, Courtaboeuf, France) according to the manufacturer’s instructions.
RNA extraction from NHBE and Vero E6 cells was performed using miRNAeasy Tissue/Cells Advanced Micro Kit (QIAGEN, Courtaboeuf, France) according to the manufacturer’s instructions. RNA extraction from Calu-3 cells was performed using a QIAamp Viral RNA kit (QIAGEN, Courtaboeuf, France) according to the manufacturer’s instructions. RNA extracts were treated with DNAse (Jena Bioscience, Germany) for 10 min at 37 °C followed by 10 min at 65 °C. RNA extracts were stored at −80 °C.
2.6. RT-qPCR
The primer sequences for AGO2 (PrimerBank ID: 257467481c1), DICER1 (PrimerBank ID: 307133774c1), DGCR8 (PrimerBank ID: 298358603c1), DROSHA (PrimerBank ID: 155030235c1), and XPO5 (PrimerBank ID: 221136812c1) genes (Table 1) were retrieved from the PrimerBank of Harvard Medical School [63]. RT-qPCR was performed to quantify mRNA expression of the genes involved in miRNA biogenesis by using 5 ng of extracted RNA of cells and 5 microL of RNA extracts of patients’ specimens, respectively, and the Sybr green Luna Universal one-step RT-qPCR kit (New England BioLabs, Evry, France) according to the manufacturer’s instructions using a 7500 Real-Time PCR System (ThermoFisher Scientific, Courtaboeuf, France). The following thermal profile was used: 10 min at 55 °C followed by 1 min at 95 °C and 40 cycles of 10 s at 95 °C, 30 s at 53 °C and 60 s at 60 °C. The beta-actin (ACTB gene) was used for normalization. Results were analyzed by using the 7500 Software (v2.0.6, Life Technologies, Thermofisher Scientific, Courtaboeuf, France). Results were presented as delta Ct values = Ct of the gene of interest − Ct of beta-actin. Fold changes were calculated according to the 2 −delta delta Ct method, with delta delta Ct = delta Ct (infected cells)—delta Ct (uninfected cells) [64].

Table 1.
List and sequences of primers.
2.7. Statistical Analysis
Statistical analysis was performed with Prism 9 for Windows (Version 9.5.1) using nonparametric and unpaired tests; Kruskal–Wallis test and Mann–Whitney U test. The results were considered significant when the p-value was below 0.05.
3. Results
3.1. Expression of miRNA Biogenesis Genes in COVID-19 Patients’ Specimens
In our previous study, the number of miRNAs detected in nasopharyngeal swabs of severe COVID-19 patients was lower than in non-severe COVID-19 patients and controls. Furthermore, most differentially expressed miRNAs were downregulated in severe COVID-19 patients compared to patients with non-severe COVID-19 and controls [55]. In order to determine whether this could be due to an inhibition of miRNA biogenesis during severe COVID-19, we compared mRNA expression of genes involved in miRNA biogenesis, namely AGO2, DICER1, DGCR8, DROSHA, and XPO5, in nasopharyngeal swabs of severe and non-severe COVID-19 patients and controls. As shown in Figure 1, there were no statistically significant differences in mRNA levels of these genes between the three different patient groups.
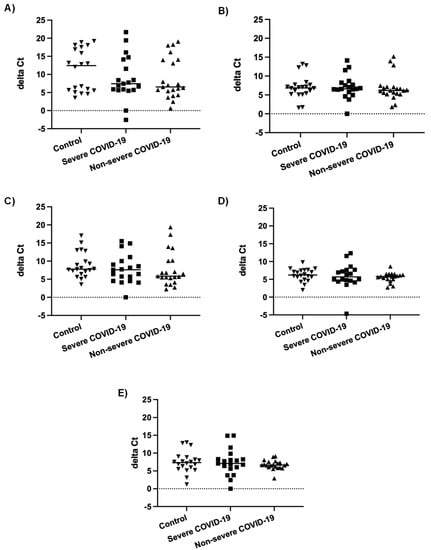
Figure 1.
mRNA expression levels of genes involved in miRNA biogenesis. Normalized mRNA expression (delta Ct values) of AGO2 (A), DICER1 (B), DGCR8 (C), DROSHA (D), and XPO5 (E) genes in nasopharyngeal swabs of patients with severe COVID-19 (n = 19), patients with non-severe COVID-19 (n = 21) and controls (n = 20). Each data point represents one nasopharyngeal swab specimen. The bar indicates the median value.
3.2. Expression of miRNA Biogenesis Genes in SARS-CoV-2 Infected Cells
We next investigated whether SARS-CoV-2 infection impacted the expression of genes involved in miRNA biogenesis in vitro. To this end, primary cultures of human bronchial epithelial (NHBE) cells were used as they represent a cellular model close to the human respiratory tract. In addition, two epithelial cell lines that are commonly used for in vitro studies concerning SARS-CoV-2, namely Calu-3 and Vero E6 cell lines, were also included in this study. The three cell types were infected with the SARS-CoV-2 delta variant at an MOI of 0.1, and mRNA expression levels of AGO2, DICER1, DGCR8, DROSHA, and XPO5 were measured 24 h and 48 h after infection. In parallel, we confirmed that SARS-CoV2 effectively infected these cells by measuring the viral RNA production (data not shown). Again, as shown in Figure 2 and Figure 3, no significant differences in mRNA levels of these genes were observed after infection of NHBE and Calu-3 cells with SARS-CoV-2 at 24 h and 48 h post infection. However, as shown in Figure 4, mRNA levels of miRNA biogenesis genes were impacted by SARS-CoV-2 infection in Vero E6 cells at 24 h post infection. A significant increase in mRNA levels of AGO2, DICER1, DGCR8, and XPO (all p-values= 0.03) was found 24 h after infection (Figure 4A). In contrast, 48 h after infection, no significant differences in mRNA levels of these genes were observed (Figure 4B). We next calculated the fold change expression of mRNA levels in infected Vero E6 cells. Indeed, mRNA levels were higher in infected Vero E6 cells as compared to uninfected cells (Figure 5). mRNA level changes of DICER1, DGCR8, and XPO were approximately 2-fold, and of AGO2 approximately 4.6-fold, 24 h after infection. Thus, we observed a slight and transient overexpression of mRNA levels of key miRNA biogenesis genes after SARS-CoV-2 infection in Vero E6 cells at 24 h post infection but not at 48 h post infection.
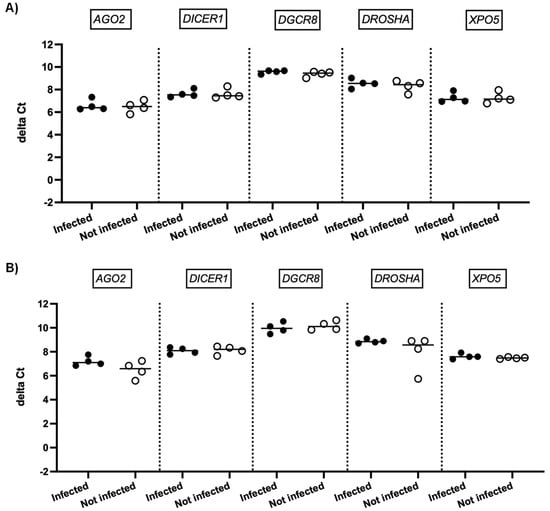
Figure 2.
mRNA expression of genes involved in miRNA biogenesis in NHBE cells. Normalized mRNA levels (delta Ct values) of AGO2, DICER1, DGCR8, DROSHA, and XPO5 genes were measured in NHBE cells infected with SARS-CoV-2 (black circles) or in non-infected cells (empty circles), 24 h (A) and 48 h (B) post infection. Four independent infections are shown, each data point represents one replicate. The bar indicates the median.
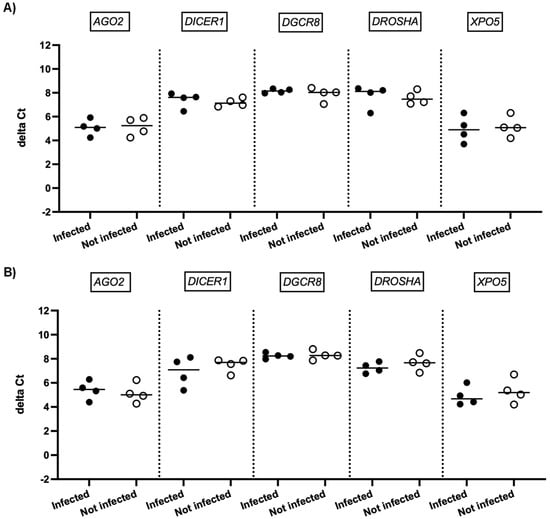
Figure 3.
mRNA expression of genes involved in miRNA biogenesis in Calu-3 cells. Normalized mRNA levels (delta Ct values) of AGO2, DICER1, DGCR8, DROSHA, and XPO5 genes were measured in Calu-3 cells infected with SARS-CoV-2 (black circles) or in non-infected cells (empty circles) 24 h (A) and 48 h (B) post infection. Four independent infections are shown. Each data point represents one replicate. The bar indicates the median.
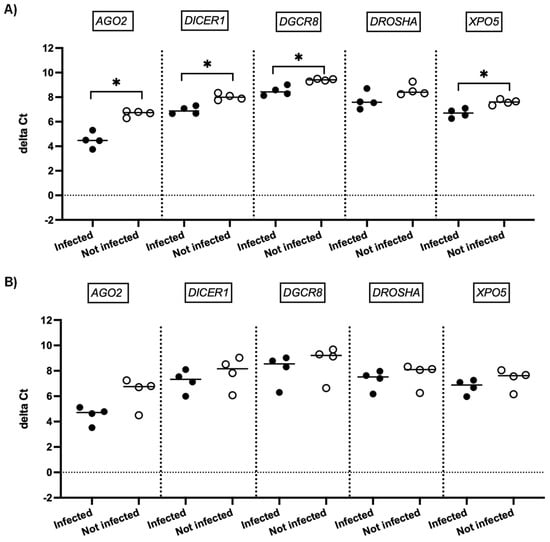
Figure 4.
mRNA expression of genes involved in miRNA biogenesis in Vero E6 cells. Normalized mRNA levels (delta Ct values) of AGO2, DICER1, DGCR8, DROSHA, and XPO5 genes were measured in Vero E6 cells infected with SARS-CoV-2 (black circles) or in non-infected cells (empty circles), 24 h (A) and 48 h (B) post infection. Four independent infections are shown. Each data point represents one replicate. The bar indicates the median. * p < 0.05.
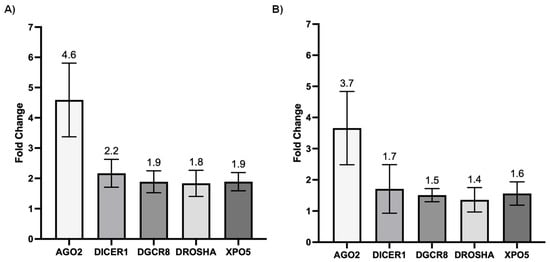
Figure 5.
Fold changes of mRNA expression of genes involved in miRNA biogenesis in infected Vero E6 cells. Fold changes of mRNA expression of AGO2, DICER1, DGCR8, DROSHA, and XPO5 genes in SARS-CoV-2 infected compared to uninfected Vero E6 cells at 24 h (A) and 48 h (B) post infection. Means and standard deviations of four independent infections are shown.
4. Discussion
While a reduced number of miRNAs was expressed in nasopharyngeal swabs specimens of patients with severe COVID-19 compared to non-severe COVID-19 patients and controls, and most miRNAs were downregulated in severe COVID-19 patients [55], the reasons for these observations were not clear. The inhibition of miRNA biogenesis during SARS-CoV-2 infection was considered to be the most likely underlying reason, as supported by a study by Mousavi et al., who noted that AGO2, DICER, and DROSHA were downregulated in COVID-19 patients compared to controls [65] and suggested that viruses may interact with the miRNA biogenesis pathway [44,46].
A dysregulation of the expression of genes involved in miRNA biogenesis had actually been found in several other viral infections: Dengue virus infection led to a decrease of mRNA levels of DICER, DROSHA, AGO1, and AGO2 in Huh-7 cells and this was associated with increased viral replication [66]. In another study, infection of A549 cells with dengue virus 4 resulted in reduced mRNA levels of DICER, DROSHA, and DGCR8 [67]. Vaccinia virus infection led to a general decrease of miRNA expression in infected cells and was associated with a decrease of DICER expression at the mRNA and protein levels [68]. Influenza virus A infection also led to a decrease of DICER mRNA and protein levels in infected A549 and Vero cells [69]. Interestingly, DROSHA mRNA levels were not impacted by vaccinia virus infection. In contrast, infection with herpes simplex type 1 and type 2, influenza A virus, and human respiratory syncytial virus had no effect on DICER expression [68], suggesting that the impact of virus infection on the expression of genes involved in miRNA biogenesis differs between viruses. One study even observed a different impact of yellow fever virus genotype I versus genotype II on mRNA levels of miRNA biogenesis components in infected cells [70].
Furthermore, most studies investigated the effect of virus infection on miRNA biogenesis genes by using in vitro infected cells. To date, only a few studies have investigated this effect in clinical samples. Apart from the above-mentioned study [65], one study investigated miRNA expression and expression of miRNA biogenesis genes in HTLV-1 infected patients [71]. Interestingly, the authors measured DROSHA, DGCR8, XPO5, DICER1, AGO2, and AGO3 mRNA expression, and only DICER1 mRNA was differently expressed in CD8+ T-cell–depleted PBMCs from HTLV-1 asymptomatic carriers when compared to patients with acute adult T cell Leukemia. This led to a reduction in the expression level of several miRNAs [71]. Another study found that patients with chronic hepatitis B who had high hepatitis B virus loads had reduced mRNA levels of DROSHA, DICER1, and AGO2 compared with patients with low virus loads [72]. Interestingly, reduced mRNA expression levels of DICER, DROSHA, and AGO2 were also observed in hepatitis B virus replicon-transfected HepG2 cells [72].
In the present study, we investigated the expression levels of genes implicated in miRNA biogenesis both in vitro and ex vivo. Measured mRNA levels of AGO2, DICER1, DGCR8, DROSHA, and XPO5 were not significantly different in nasopharyngeal swab specimens of severe COVID-19 patients compared to non-severe COVID-19 patients or controls (Figure 1). No impact of SARS-CoV-2 infection on mRNA expression levels of key genes involved in miRNA biogenesis was observed, in contrast to the study by Mousavi et al., who found that mRNA levels of AGO2, DICER1, and DROSHA, but not DGCR8, differed between COVID-19 patients and controls [65]. There are several differences between our study and the one from Mousavi et al. First, in the present study, we used nasopharyngeal swab specimens, whereas Mousavi et al. used whole blood specimens. Second, there were differences in the experimental protocols: Mousavi et al. used RT followed by qPCR, whereas we used one-step RT qPCR. The primer sequences used in the two studies were not the same. The gene used for normalization was GAPDH in the study by Mousavi et al., whereas we used beta-actin. While all of these differences may have an impact on the results, the most likely explanation for the discrepancy in the findings of the two studies is, in our opinion, the fact that we measured mRNA expression in very different specimen types [65]. When investigating mRNA expression in nasopharyngeal swabs, we studied the local effect of SARS-CoV-2 infection directly on the targeted respiratory epithelium and confirmed that in vitro SARS-CoV2 infection of airway epithelial cells did not affect mRNA expression of these genes. In contrast, measuring mRNA expression in the blood reflected a systemic effect of infection in leucocytes, which may or may not have been caused by SARS-CoV-2 infection directly. Of note, it was not mentioned whether SARS-CoV-2 viruses had been detected in the patients’ blood [65].
The impact of SARS-CoV-2 infection on the expression of genes involved in miRNA biogenesis in in vitro models was tested to validate the clinical observations. The results showed that infection of NHBE and Calu-3 cells with SARS-CoV-2 did not impact mRNA expression levels of AGO2, DICER1, DGCR8, DROSHA, and XPO5 at 24 and 48 h post infection (Figure 2 and Figure 3), in good concordance with our results on patient samples. On the other hand, infection of Vero E6 cells with SARS-CoV-2 impacted mRNA levels of AGO2, DICER1, DGCR8, and XPO5 at 24 h (Figure 4A). However, rather than being downregulated as we had hypothesized, mRNA levels were slightly and transiently increased in SARS-CoV-2 infected cells at 24 h post infection. mRNA expression changes were around two-fold in most cases (Figure 5), and even if differences were statistically significant, it is uncertain whether these slight changes have a biological impact. Furthermore, at 48 h post infection, mRNA levels were not significantly different between SARS-CoV-2 infected and uninfected cells (Figure 4B). Indeed, increased mRNA expression of genes involved in miRNA biogenesis has been observed in other virus infections. For example, increased DICER1 and DROSHA mRNA expression were found in some human papillomavirus (HPV) positive cervical cancer cell lines [73]. Expression of the HPV E6 and E7 oncoproteins in primary human foreskin keratinocytes resulted in the upregulation of DICER1 mRNA and DROSHA mRNA expression [73], suggesting that these viral proteins are responsible for the induction of gene expression. Another example is that infection with Kaposi’s sarcoma-associated herpesvirus (KSHV) resulted in increased expression of DICER1 mRNA in primary human umbilical vein endothelial cells, whereas mRNA expression levels of DGCR8, DROSHA, and XPO5 did not change significantly [74]. The authors hypothesized that KSHV induced upregulation of DICER1 expression in order to counteract the miRNA biogenesis inhibition caused by the human MCP-1-induced protein-1 [74].
Taken together, we found no evidence that SARS-CoV-2 infection inhibited miRNA biogenesis by downregulation of mRNA levels of key miRNA biogenesis genes. However, the fact that the mRNA levels of genes involved in miRNA biogenesis were not downregulated after SARS-CoV-2 infection does not necessarily mean that the miRNA pathway is not affected by SARS-CoV-2 infection. There are several alternative mechanisms by which SARS-CoV-2 infection may interfere with the miRNA pathway [39]. For example, there could be a direct interaction of viral factors with central components of the miRNA pathway, leading to their inhibition. Indeed it was shown that human adenovirus virus-associated RNAs inhibited DICER activity [47]. Similarly, the insect flock house virus B2 protein interacted with DICER and thereby inhibited siRNA biogenesis [75]. Furthermore, the Zika virus capsid interacted with DICER and inhibited miRNA biogenesis [76]. Interestingly, and in parallel to our observation of reduced miRNA expression in severe COVID-19 [55], Zika virus infection of neural stem cells resulted in a reduction of total miRNA reads, and 138 miRNAs were significantly downregulated; in contrast, only two miRNAs were upregulated [76]. HSV-1 used a different mechanism to interfere with miRNA biogenesis, namely by blocking pre-miRNA nuclear export [77]. Reduced expression of mature miRNAs can also be explained by cleavage of miRNA precursors. Indeed, human MCP-1-induced protein-1 cleaved the terminal loops of pre-miRNAs leading to the destabilization of pre-miRNAs and resulting in their degradation [74,78]. Increased turnover of mature miRNAs could also underlie a reduced miRNA expression [39,74,79]. For example, poxvirus-encoded poly(A)-polymerase mediated poly-adenylation of cellular miRNAs, resulting in their degradation. This phenomenon was observed in insect and mammalian cells. Interestingly, restoring miRNA function resulted in reduced virus replication suggesting that the virus-induced degradation of host miRNAs favored virus replication [80].
Altogether, these findings show that many viruses interact with the miRNA pathway and that the mechanisms used are different. Concerning SARS-CoV-2 infection, the possible mechanisms need to be explored in future studies.
5. Conclusions
Taken together, our results suggest that there is no detectable downregulation of mRNA expression of genes involved in miRNA biogenesis during SARS-CoV-2 infection, neither ex vivo nor in vitro.
Author Contributions
Conceptualization, I.E.; formal analysis, N.G. and I.E.; funding acquisition, I.E., D.H. and S.S.; investigation, N.G., F.S. (Famara Sane), L.M. and I.E.; methodology, F.S. (Famara Sane), L.M. and F.S. (Fabrice Soncin), P.G. and I.E.; resources, I.E.; supervision, S.S. and I.E.; validation, S.S. and D.H.; visualization, N.G.; writing—original draft, N.G. and I.E.; writing—review and editing, F.S. (Famara Sane), L.M., F.S. (Fabrice Soncin), P.G., D.H. and S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by I-SITE ULNE, the Centre Hospitalier Universitaire de Lille and Université de Lille. N.G. wants to thank the Ecole Doctorale Biologie-Santé for a PhD fellowship.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the the French Institutional Authority for Personal Data Protection (Commission Nationale de l’Informatique et des Libertés DR-2020-178, 22 October 2020) and the ethics committee (Comité de Protection des Personnes Nord Ouest IV, ECH20/09, 7 September 2020).
Informed Consent Statement
Patient consent was waived by the ethics committee due to the retrospective nature of the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The excellent technical assistance of the technicians of the virology laboratory is acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Johns Hopkins Coronavirus Resource Center. COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 10 May 2023).
- Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (accessed on 10 May 2023).
- Carlsten, C.; Gulati, M.; Hines, S.; Rose, C.; Scott, K.; Tarlo, S.M.; Torén, K.; Sood, A.; de la Hoz, R.E. COVID-19 as an occupational disease. Am. J. Ind. Med. 2021, 64, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Ranney, M.L.; Valerie Griffeth, M.P.H.; Jha, A.K. Critical Supply Shortages—The Need for Ventilators and Personal Protective Equipment during the COVID-19 Pandemic. N. Engl. J. Med. 2020, 382, e41. [Google Scholar] [CrossRef]
- O’dowd, A. COVID-19: Government was too slow to respond to ventilator shortages, say MPs. BMJ 2020, 371, m4594. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Cabler, S.S.; French, A.R.; Orvedahl, A. A Cytokine Circus with a Viral Ringleader: SARS-CoV-2-Associated Cytokine Storm Syndromes. Trends Mol. Med. 2020, 26, 1078–1085. [Google Scholar] [CrossRef]
- Copaescu, A.; Smibert, O.; Gibson, A.; Phillips, E.J.; Trubiano, J.A. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020, 146, 518–534.e1. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Deng, Y.; Li, W. Coronavirus Disease 2019 (COVID-19): What we know? J. Med. Virol. 2020, 92, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Pascarella, G.; Strumia, A.; Piliego, C.; Bruno, F.; Del Buono, R.; Costa, F.; Scarlata, S.; Agrò, F.E. COVID-19 diagnosis and management: A comprehensive review. J. Intern. Med. 2020, 288, 192–206. [Google Scholar] [CrossRef]
- Chen, B.; Julg, B.; Mohandas, S.; Bradfute, S.B. Viral persistence, reactivation, and mechanisms of long COVID. eLife 2023, 12, e86015. [Google Scholar] [CrossRef] [PubMed]
- Lai, F.W.; Stephenson, K.B.; Mahony, J.; Lichty, B.D. Human Coronavirus OC43 Nucleocapsid Protein Binds MicroRNA 9 and Potentiates NF-κB Activation. J. Virol. 2014, 88, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, X.; Zhang, Z.-K.; Zou, W.-C.; Wang, H.-N. miR-146a-5p promotes replication of infectious bronchitis virus by targeting IRAK2 and TNFRSF18. Microb. Pathog. 2018, 120, 32–36. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, C.; Xue, M.; Fu, F.; Zhang, X.; Li, L.; Yin, L.; Xu, W.; Feng, L.; Liu, P. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J. Virol. 2018, 92, e00728-18. [Google Scholar] [CrossRef] [PubMed]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. MicroRNome Analysis Unravels the Molecular Basis of SARS Infection in Bronchoalveolar Stem Cells. PLoS ONE 2009, 4, e7837. [Google Scholar] [CrossRef]
- Peng, X.; Gralinski, L.; Ferris, M.T.; Frieman, M.B.; Thomas, M.J.; Proll, S.; Korth, M.J.; Tisoncik, J.R.; Heise, M.; Luo, S.; et al. Integrative Deep Sequencing of the Mouse Lung Transcriptome Reveals Differential Expression of Diverse Classes of Small RNAs in Response to Respiratory Virus Infection. mBio 2011, 2, e00198-11. [Google Scholar] [CrossRef]
- Zheng, H.; Xu, L.; Liu, Y.; Li, C.; Zhang, L.; Wang, T.; Zhao, D.; Xu, X.; Zhang, Y. MicroRNA-221-5p Inhibits Porcine Epidemic Diarrhea Virus Replication by Targeting Genomic Viral RNA and Activating the NF-κB Pathway. Int. J. Mol. Sci. 2018, 19, 3381. [Google Scholar] [CrossRef]
- Aslani, M.; Mortazavi-Jahromi, S.S.; Mirshafiey, A. Cytokine storm in the pathophysiology of COVID-19: Possible functional disturbances of miRNAs. Int. Immunopharmacol. 2021, 101 Pt A, 108172. [Google Scholar] [CrossRef]
- Meidert, A.S.; Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Billaud, J.-N.; Klein, M.; Lindemann, A.; Aue, E.; Schelling, G.; et al. Extracellular Vesicle Associated miRNAs Regulate Signaling Pathways Involved in COVID-19 Pneumonia and the Progression to Severe Acute Respiratory Corona Virus-2 Syndrome. Front. Immunol. 2021, 12, 784028. [Google Scholar] [CrossRef]
- Molinero, M.; Benítez, I.D.; González, J.; Gort-Paniello, C.; Moncusí-Moix, A.; Rodríguez-Jara, F.; García-Hidalgo, M.C.; Torres, G.; Vengoechea, J.J.; Gómez, S.; et al. Bronchial Aspirate-Based Profiling Identifies MicroRNA Signatures Associated With COVID-19 and Fatal Disease in Critically Ill Patients. Front. Med. 2022, 8, 756517. Available online: https://www.frontiersin.org/articles/10.3389/fmed.2021.756517 (accessed on 10 May 2023). [CrossRef]
- Rarani, F.Z.; Rashidi, B.; Abadi, M.H.J.N.; Hamblin, M.R.; Hashemian, S.M.R.; Mirzaei, H. Cytokines and microRNAs in SARS-CoV-2: What do we know? Mol. Ther.-Nucleic Acids 2022, 29, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.C.; Conte, D. Revealing the world of RNA interference. Nature 2004, 431, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Frédérick, P.; Simard, M.J. Regulation and different functions of the animal microRNA-induced silencing complex. Wiley Interdiscip. Rev. RNA 2021, 13, e1701. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Paturi, S.; Deshmukh, M.V. A Glimpse of “Dicer Biology” Through the Structural and Functional Perspective. Front. Mol. Biosci. 2021, 8, 643657. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a Link Between Genetic and Biochemical Analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef]
- Sand, M. The pathway of miRNA maturation. Methods Mol. Biol. 2014, 1095, 3–10. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Ghosh, Z.; Mallick, B.; Chakrabarti, J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2008, 37, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Libri, V.; Miesen, P.; Van Rij, R.P.; Buck, A.H. Regulation of microRNA biogenesis and turnover by animals and their viruses. Cell. Mol. Life Sci. 2013, 70, 3525–3544. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, I.; Alidjinou, E.K.; Bertin, A.; Sane, F.; Hober, D. miRNAs in enterovirus infection. Crit. Rev. Microbiol. 2018, 44, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Sskalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Ivens, A.; Gordon, K.; Craig, N.; Houzelle, A.; Roche, A.; Turnbull, N.; Beard, P.M. Quantitative Analysis of MicroRNAs in Vaccinia virus Infection Reveals Diversity in Their Susceptibility to Modification and Suppression. PLoS ONE 2015, 10, e0131787. [Google Scholar] [CrossRef]
- Ding, S.-W.; Lu, R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 2011, 1, 533–544. [Google Scholar] [CrossRef]
- Schütz, S.; Sarnow, P. Interaction of viruses with the mammalian RNA interference pathway. Virology 2006, 344, 151–157. [Google Scholar] [CrossRef]
- Takahashi, T.; Heaton, S.M.; Parrish, N.F. Mammalian antiviral systems directed by small RNA. PLOS Pathog. 2021, 17, e1010091. [Google Scholar] [CrossRef] [PubMed]
- Mockenhaupt, S.; Schürmann, N.; Grimm, D. When cellular networks run out of control: Global dysregulation of the RNAi machinery in human pathology and therapy. Prog. Mol. Biol. Transl. Sci. 2011, 102, 165–242. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.G.; Haasnoot, P.C.J.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjärvi, G. Suppression of RNA Interference by Adenovirus Virus-Associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar] [CrossRef]
- Lu, S.; Cullen, B.R. Adenovirus VA1 Noncoding RNA Can Inhibit Small Interfering RNA and MicroRNA Biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo-Calvo, D.; Benítez, I.D.; Pinilla, L.; Carratalá, A.; Moncusí-Moix, A.; Gort-Paniello, C.; Molinero, M.; González, J.; Torres, G.; Bernal, M.; et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021, 236, 147–159. [Google Scholar] [CrossRef]
- Donyavi, T.; Bokharaei-Salim, F.; Baghi, H.B.; Khanaliha, K.; Janat-Makan, M.A.; Karimi, B.; Nahand, J.S.; Mirzaei, H.; Khatami, A.; Garshasbi, S.; et al. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155–5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021, 97, 107641. [Google Scholar] [CrossRef]
- Duecker, R.P.; Adam, E.H.; Wirtz, S.; Gronau, L.; Khodamoradi, Y.; Eberhardt, F.J.; Donath, H.; Gutmann, D.; Vehreschild, M.J.G.T.; Zacharowski, K.; et al. The MiR-320 Family Is Strongly Downregulated in Patients with COVID-19 Induced Severe Respiratory Failure. Int. J. Mol. Sci. 2021, 22, 10351. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLOS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID -19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Garnier, N.; Pollet, K.; Fourcot, M.; Caplan, M.; Marot, G.; Goutay, J.; Labreuche, J.; Soncin, F.; Boukherroub, R.; Hober, D.; et al. Altered microRNA expression in severe COVID-19: Potential prognostic and pathophysiological role. Clin. Transl. Med. 2022, 12, e899. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef]
- Li, C.-X.; Chen, J.; Lv, S.-K.; Li, J.-H.; Li, L.-L.; Hu, X. Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients. Mediat. Inflamm. 2021, 2021, 6635925. [Google Scholar] [CrossRef]
- Parray, A.; Mir, F.A.; Doudin, A.; Iskandarani, A.; Danjuma, I.M.M.; Kuni, R.A.T.; Abdelmajid, A.; Abdelhafez, I.; Arif, R.; Mulhim, M.; et al. SnoRNAs and miRNAs Networks Underlying COVID-19 Disease Severity. Vaccines 2021, 9, 1056. [Google Scholar] [CrossRef] [PubMed]
- Pollet, K.; Garnier, N.; Szunerits, S.; Madder, A.; Hober, D.; Engelmann, I. Host miRNAs as Biomarkers of SARS-CoV-2 Infection: A Critical Review. 2022. Available online: https://www.semanticscholar.org/paper/Host-miRNAs-as-biomarkers-of-SARS-CoV-2-infection%3A-Pollet-Garnier/a3fe2285ad9831b5c8135e7541275e849e683c12 (accessed on 17 November 2022).
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Baroni, S.S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2020, 193, 111413. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- Wilson, J.C.; Kealy, D.; James, S.R.; Plowman, T.; Newling, K.; Jagger, C.; Filbey, K.; Mann, E.R.; Konkel, J.E.; Menon, M.; et al. Integrated miRNA/cytokine/chemokine profiling reveals severity-associated step changes and principal correlates of fatality in COVID-19. iScience 2022, 25, 103672. [Google Scholar] [CrossRef]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, D792–D799. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Mousavi, S.R.; Sajjadi, M.S.; Khosravian, F.; Feizbakhshan, S.; Salmanizadeh, S.; Esfahani, Z.T.; Beni, F.A.; Arab, A.; Kazemi, M.; Shahzamani, K.; et al. Dysregulation of RNA interference components in COVID-19 patients. BMC Res. Notes 2021, 14, 401. [Google Scholar] [CrossRef] [PubMed]
- Kakumani, P.K.; Ponia, S.S.; Rajgokul, K.S.; Sood, V.; Chinnappan, M.; Banerjea, A.C.; Medigeshi, G.; Malhotra, P.; Mukherjee, S.K.; Bhatnagar, R.K. Role of RNA Interference (RNAi) in Dengue Virus Replication and Identification of NS4B as an RNAi Suppressor. J. Virol. 2013, 87, 8870–8883. [Google Scholar] [CrossRef]
- Casseb, S.; Simith, D.; Melo, K.; Mendonça, M.; Santos, A.; Carvalho, V.; Cruz, A.; Vasconcelos, P. Drosha, DGCR8, and Dicer mRNAs are down-regulated in human cells infected with dengue virus 4, and play a role in viral pathogenesis. Genet. Mol. Res. 2016, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Grinberg, M.; Gilad, S.; Meiri, E.; Levy, A.; Isakov, O.; Ronen, R.; Shomron, N.; Bentwich, Z.; Shemer-Avni, Y. Vaccinia virus infection suppresses the cell microRNA machinery. Arch. Virol. 2012, 157, 1719–1727. [Google Scholar] [CrossRef]
- Matskevich, A.A.; Moelling, K. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 2007, 88, 2627–2635. [Google Scholar] [CrossRef]
- Holanda, G.M.; Casseb, S.M.M.; Mello, K.F.L.; Vasconcelos, P.F.C.; Cruz, A.C.R. Yellow Fever Virus Modulates the Expression of Key Proteins Related to the microRNA Pathway in the Human Hepatocarcinoma Cell Line HepG2. Viral Immunol. 2017, 30, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Gazon, H.; Belrose, G.; Terol, M.; Meniane, J.-C.; Mesnard, J.-M.; Césaire, R.; Jr, J.-M.P. Impaired expression of DICER and some microRNAs in HBZ expressing cells from acute adult T-cell leukemia patients. Oncotarget 2016, 7, 30258–30275. [Google Scholar] [CrossRef]
- Chinnappan, M.; Singh, A.K.; Kakumani, P.K.; Kumar, G.; Rooge, S.B.; Kumari, A.; Varshney, A.; Rastogi, A.; Singh, A.K.; Sarin, S.K.; et al. Key elements of the RNAi pathway are regulated by hepatitis B virus replication and HBx acts as a viral suppressor of RNA silencing. Biochem. J. 2014, 462, 347–358. [Google Scholar] [CrossRef]
- Harden, M.E.; Munger, K. Perturbation of DROSHA and DICER expression by human papillomavirus 16 oncoproteins. Virology 2017, 507, 192–198. [Google Scholar] [CrossRef]
- Happel, C.; Ramalingam, D.; Ziegelbauer, J.M. Virus-Mediated Alterations in miRNA Factors and Degradation of Viral miRNAs by MCPIP1. PLOS Biol. 2016, 14, e2000998. [Google Scholar] [CrossRef]
- Singh, G.; Popli, S.; Hari, Y.; Malhotra, P.; Mukherjee, S.; Bhatnagar, R.K. Suppression of RNA silencing by Flock house virus B2 protein is mediated through its interaction with the PAZ domain of Dicer. FASEB J. 2009, 23, 1845–1857. [Google Scholar] [CrossRef]
- Zeng, J.; Dong, S.; Luo, Z.; Xie, X.; Fu, B.; Li, P.; Liu, C.; Yang, X.; Chen, Y.; Wang, X.; et al. The Zika Virus Capsid Disrupts Corticogenesis by Suppressing Dicer Activity and miRNA Biogenesis. Cell Stem Cell 2020, 27, 618–632.e9. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Li, G.; Morris-Love, J.; Qi, S.; Feng, L.; Mertens, M.E.; Jurak, I.; Knipe, D.M.; Coen, D.M. Herpes Simplex Virus 1 Lytic Infection Blocks MicroRNA (miRNA) Biogenesis at the Stage of Nuclear Export of Pre-miRNAs. mBio 2019, 10, e02856-18. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Arase, M.; Matsuyama, H.; Choi, Y.L.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. MCPIP1 Ribonuclease Antagonizes Dicer and Terminates MicroRNA Biogenesis through Precursor MicroRNA Degradation. Mol. Cell 2011, 44, 424–436. [Google Scholar] [CrossRef]
- Chatterjee, S.; Großhans, H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 2009, 461, 546–549. [Google Scholar] [CrossRef]
- Backes, S.; Shapiro, J.S.; Sabin, L.R.; Pham, A.M.; Reyes, I.; Moss, B.; Cherry, S.; Tenoever, B.R. Degradation of Host MicroRNAs by Poxvirus Poly(A) Polymerase Reveals Terminal RNA Methylation as a Protective Antiviral Mechanism. Cell Host Microbe 2012, 12, 200–210. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).