Porcine Respirovirus 1 Suppresses Host Type I Interferon Production and the JAK-STAT Signaling Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Reagents
2.2. Immunofluorescence Assay (IFA)
2.3. Quantitative RT-PCR
2.4. Dual-Luciferase Reporter Assay
2.5. Immunoprecipitation Assay (IP)
2.6. GST Pull-Down Assay
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
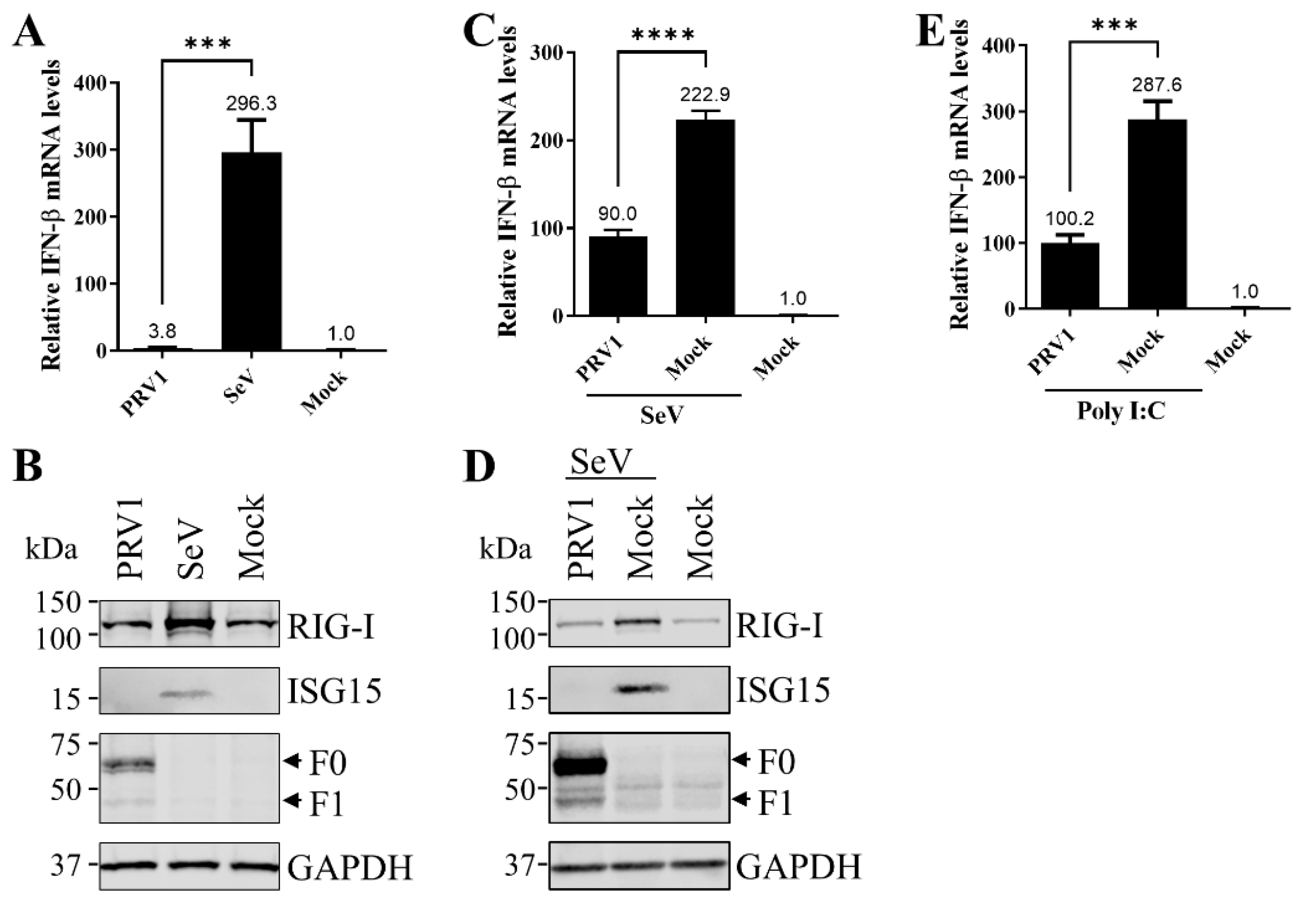
3.1. PRV1 Infection Antagonizes Type I IFN Production and Signaling Pathway
3.2. Multiple PRV1 Proteins Antagonize Type I IFN Production
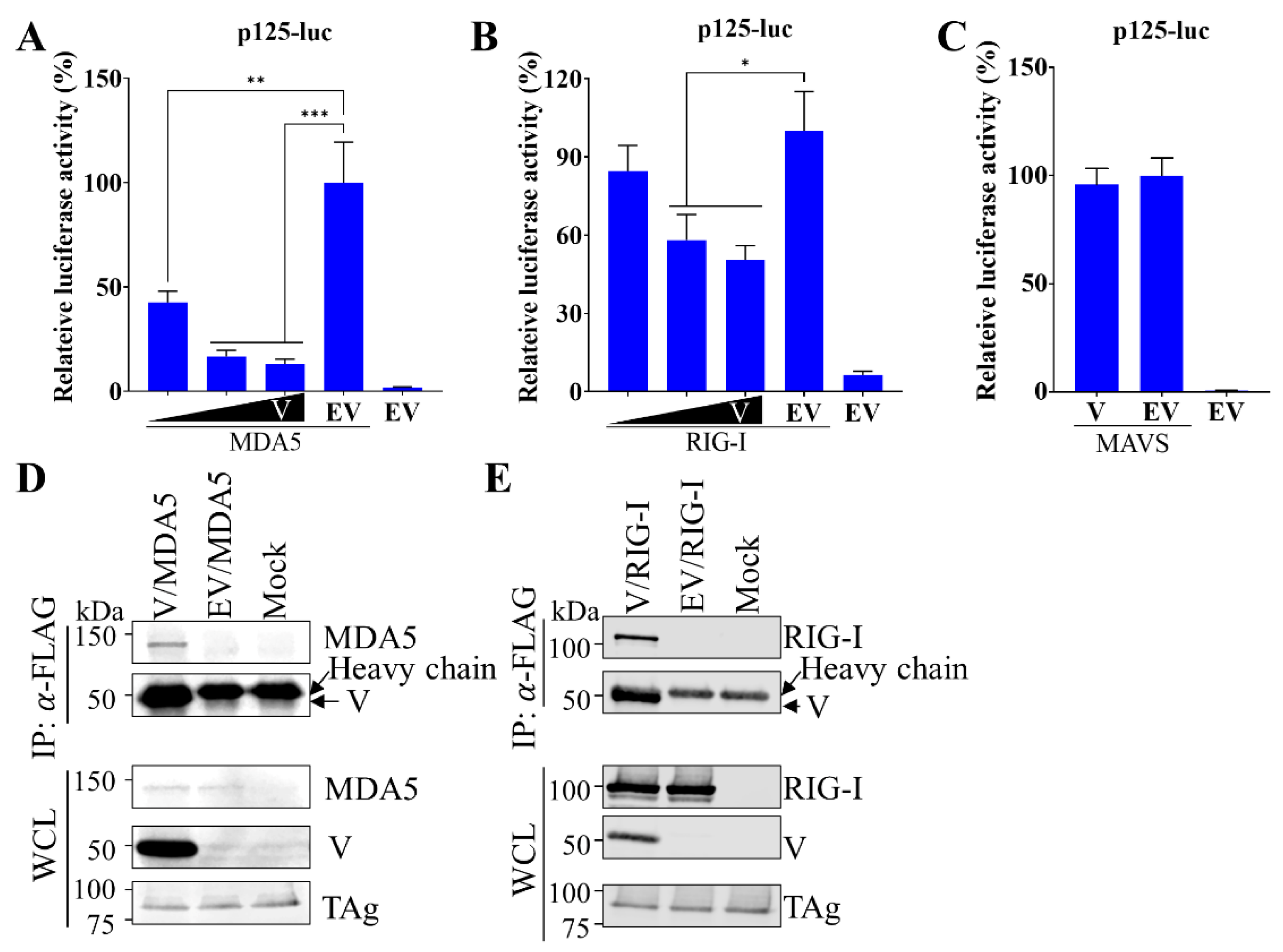
3.3. V Protein Suppresses IFNβ Production by Targeting MDA5 and RIG-I
3.4. V Protein Suppresses RIG-I Ubiquitination
3.5. PRV1 P Gene Products Inhibit Interferon-Stimulated Gene Expression from ISRE Promoter by Blocking the Nucleus Translocation of STAT1
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses (ICTV). Virus Taxonomy: 2021 Release. 2021. Available online: https://ictv.global/taxonomy (accessed on 1 January 2022).
- Lamb, R.; Parks, G. Paramyxoviridae: The viruses and their 542 replication. In Fields Virology; Lippincott-Raven Press: Philadelphia, PA, USA, 2007; Volume 543, pp. 1449–1496. [Google Scholar]
- Zhao, J.; Sun, J.; Li, X.; Xing, G.; Zhang, Y.; Lai, A.; Baele, G.; Ji, X.; Su, S. Divergent Viruses Discovered in Swine Alter the Understanding of Evolutionary History and Genetic Diversity of the Respirovirus Genus and Related Porcine Parainfluenza Viruses. Microbiol. Spectr. 2022, 10, e0024222. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.; Selleck, P.; Hooper, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Westbury, H.; Hiley, L.; Selvey, L.; Rodwell, B.; et al. A morbillivirus that caused fatal disease in horses and humans. Science 1995, 268, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Chadha, M.S.; Comer, J.A.; Lowe, L.; Rota, P.A.; Rollin, P.E.; Bellini, W.J.; Ksiazek, T.G.; Mishra, A. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 2006, 12, 235–240. [Google Scholar] [CrossRef] [PubMed]
- McCormack, J.G. Hendra and Nipah viruses: New zoonotically-acquired human pathogens. Respir. Care Clin. N. Am. 2005, 11, 59–66. [Google Scholar] [CrossRef]
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Tan, P.S.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; Tan, C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354, 1257–1259. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Wu, Y.; Wong, A.Y.; Wong, B.H.; Lau, C.C.; Fan, R.Y.; Cai, J.P.; Tsoi, H.W.; Chan, K.H.; et al. Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs. J. Gen.Virol. 2013, 94, 2184–2190. [Google Scholar] [CrossRef]
- Palinski, R.M.; Chen, Z.; Henningson, J.N.; Lang, Y.; Rowland, R.R.; Fang, Y.; Prickett, J.; Gauger, P.C.; Hause, B.M. Widespread detection and characterization of porcine parainfluenza virus 1 in pigs in the USA. J. Gen. Virol. 2016, 97, 281–286. [Google Scholar] [CrossRef]
- Park, J.; Kim, H.R.; Kim, J.M.; Lee, K.K.; Kim, W.I.; Lyoo, Y.S.; Kwon, O.D.; Park, C.K.; Park, S.C. First report of Porcine respirovirus 1 in South Korea. Transbound. Emerg. Dis. 2022, 69, 4041–4047. [Google Scholar] [CrossRef]
- Stadejek, T.; Cybulski, P.; Gauger, P.C.; Wozniak, A. European and American Strains of Porcine Parainfluenza Virus 1 (PPIV-1) Belong to Two Distinct Genetic Lineages. Pathogens 2022, 11, 375. [Google Scholar] [CrossRef]
- Wozniak, A.; Cybulski, P.; Denes, L.; Balka, G.; Stadejek, T. Detection of Porcine Respirovirus 1 (PRV1) in Poland: Incidence of Co-Infections with Influenza A Virus (IAV) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) in Herds with a Respiratory Disease. Viruses 2022, 14, 148. [Google Scholar] [CrossRef]
- Schuele, L.; Lizarazo-Forero, E.; Cassidy, H.; Strutzberg-Minder, K.; Boehmer, J.; Schuetze, S.; Loebert, S.; Lambrecht, C.; Harlizius, J.; Friedrich, A.W.; et al. First detection of porcine respirovirus 1 in Germany and the Netherlands. Transbound. Emerg. Dis. 2021, 68, 3120–3125. [Google Scholar] [CrossRef] [PubMed]
- Denes, L.; Csagola, A.; Schonhardt, K.; Halas, M.; Solymosi, N.; Balka, G. First report of porcine parainfluenza virus 1 (species Porcine respirovirus 1) in Europe. Transbound. Emerg. Dis. 2021, 68, 1731–1735. [Google Scholar] [CrossRef]
- Aguero, B.; Mena, J.; Berrios, F.; Tapia, R.; Salinas, C.; Dutta, J.; van Bakel, H.; Mor, S.K.; Brito, B.; Medina, R.A.; et al. First report of porcine respirovirus 1 in South America. Vet. Microbiol. 2020, 246, 108726. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Welch, M.W.; Harmon, K.M.; Zhang, J.; Pineyro, P.E.; Li, G.; Hause, B.M.; Gauger, P.C. Detection, isolation, and in vitro characterization of porcine parainfluenza virus type 1 isolated from respiratory diagnostic specimens in swine. Vet. Microbiol. 2019, 228, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, F.; Yan, X.; Matta, T.; Cino-Ozuna, G.A.; Fang, Y. Characterization of an emerging porcine respirovirus 1 isolate in the US: A novel viral vector for expression of foreign antigens. Virology 2022, 570, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.; Park, J.; Harmon, K.; Zhang, J.; Pineyro, P.; Gimenez-Lirola, L.; Zhang, M.; Wang, C.; Patterson, A.; Gauger, P.C. Pathogenesis of a novel porcine parainfluenza virus type 1 isolate in conventional and colostrum deprived/caesarean derived pigs. Virology 2021, 563, 88–97. [Google Scholar] [CrossRef]
- Pisanelli, G.; Pagnini, U.; Iovane, G.; Garcia-Sastre, A. Type I and Type II Interferon Antagonism Strategies Used by Paramyxoviridae: Previous and New Discoveries, in Comparison. Viruses 2022, 14, 1107. [Google Scholar] [CrossRef]
- Teijaro, J.R. Type I interferons in viral control and immune regulation. Curr. Opin. Virol. 2016, 16, 31–40. [Google Scholar] [CrossRef]
- Thanos, D.; Maniatis, T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 1995, 83, 1091–1100. [Google Scholar] [CrossRef]
- Lee, A.J.; Ashkar, A.A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr.; Akira, S.; et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Takeuchi, K.; Gotoh, B. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect. 2007, 9, 954–962. [Google Scholar] [CrossRef]
- Treier, M.; Staszewski, L.M.; Bohmann, D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 1994, 78, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shyu, D.L.; Shang, P.; Bai, J.; Ouyang, K.; Dhakal, S.; Hiremath, J.; Binjawadagi, B.; Renukaradhya, G.J.; Fang, Y. Mutations in a Highly Conserved Motif of nsp1beta Protein Attenuate the Innate Immune Suppression Function of Porcine Reproductive and Respiratory Syndrome Virus. J. Virol. 2016, 90, 3584–3599. [Google Scholar] [CrossRef]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- Andrejeva, J.; Childs, K.S.; Young, D.F.; Carlos, T.S.; Stock, N.; Goodbourn, S.; Randall, R.E. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 2004, 101, 17264–17269. [Google Scholar] [CrossRef]
- Childs, K.; Stock, N.; Ross, C.; Andrejeva, J.; Hilton, L.; Skinner, M.; Randall, R.; Goodbourn, S. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 2007, 359, 190–200. [Google Scholar] [CrossRef]
- Parisien, J.P.; Bamming, D.; Komuro, A.; Ramachandran, A.; Rodriguez, J.J.; Barber, G.; Wojahn, R.D.; Horvath, C.M. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 2009, 83, 7252–7260. [Google Scholar] [CrossRef]
- Childs, K.S.; Andrejeva, J.; Randall, R.E.; Goodbourn, S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J. Virol. 2009, 83, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Horvath, C.M. Dissociation of paramyxovirus interferon evasion activities: Universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 2010, 84, 11152–11163. [Google Scholar] [CrossRef] [PubMed]
- Motz, C.; Schuhmann, K.M.; Kirchhofer, A.; Moldt, M.; Witte, G.; Conzelmann, K.K.; Hopfner, K.P. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science 2013, 339, 690–693. [Google Scholar] [CrossRef]
- Davis, M.E.; Wang, M.K.; Rennick, L.J.; Full, F.; Gableske, S.; Mesman, A.W.; Gringhuis, S.I.; Geijtenbeek, T.B.; Duprex, W.P.; Gack, M.U. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe 2014, 16, 19–30. [Google Scholar] [CrossRef]
- Sanchez-Aparicio, M.T.; Feinman, L.J.; Garcia-Sastre, A.; Shaw, M.L. Paramyxovirus V Proteins Interact with the RIG-I/TRIM25 Regulatory Complex and Inhibit RIG-I Signaling. J. Virol. 2018, 92, e01960-17. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Kowalinski, E.; Lunardi, T.; McCarthy, A.A.; Louber, J.; Brunel, J.; Grigorov, B.; Gerlier, D.; Cusack, S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011, 147, 423–435. [Google Scholar] [CrossRef]
- Johnston, M.D. The characteristics required for a Sendai virus preparation to induce high levels of interferon in human lymphoblastoid cells. J. Gen. Virol. 1981, 56, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.H.; Kew, C.; Lui, P.Y.; Chan, C.P.; Satoh, T.; Akira, S.; Jin, D.Y.; Kok, K.H. PACT- and RIG-I-Dependent Activation of Type I Interferon Production by a Defective Interfering RNA Derived from Measles Virus Vaccine. J. Virol. 2016, 90, 1557–1568. [Google Scholar] [CrossRef]
- Goodbourn, S.; Randall, R.E. The regulation of type I interferon production by paramyxoviruses. J. Interf. Cytokine Res. 2009, 29, 539–547. [Google Scholar] [CrossRef]
- Fontana, J.M.; Bankamp, B.; Rota, P.A. Inhibition of interferon induction and signaling by paramyxoviruses. Immunol. Rev. 2008, 225, 46–67. [Google Scholar] [CrossRef] [PubMed]
- Bharaj, P.; Wang, Y.E.; Dawes, B.E.; Yun, T.E.; Park, A.; Yen, B.; Basler, C.F.; Freiberg, A.N.; Lee, B.; Rajsbaum, R. The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKepsilon Kinase-Mediated Type-I IFN Antiviral Response. PLoS Pathog. 2016, 12, e1005880. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Zhang, L.; Li, Z.; Zhong, Y.; Tang, Q.; Qin, Y.; Chen, M. The Matrix Protein of Human Parainfluenza Virus Type 3 Induces Mitophagy that Suppresses Interferon Responses. Cell Host Microbe 2017, 21, 538–547.e534. [Google Scholar] [CrossRef] [PubMed]
- Takayama, I.; Sato, H.; Watanabe, A.; Omi-Furutani, M.; Sugai, A.; Kanki, K.; Yoneda, M.; Kai, C. The nucleocapsid protein of measles virus blocks host interferon response. Virology 2012, 424, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Sugai, A.; Sato, H.; Takayama, I.; Yoneda, M.; Kai, C. Nipah and Hendra Virus Nucleoproteins Inhibit Nuclear Accumulation of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT2 by Interfering with Their Complex Formation. J. Virol. 2017, 91, e01136-17. [Google Scholar] [CrossRef]



| Name | Sequence (5′ to 3′) | Usage |
|---|---|---|
| 3FLAG-HindIII-N-F | aaAAGCTTATGGCAGGGTTATTAAGTGT | The expression plasmid for N |
| BamHI-N-R | aaGGATCCTTATATTCCTCCTAGTGCATTCAT | |
| 3FLAG-HindIII-P-F | aaAAGCTTATGGATCAGGAcGCCCTC | The expression plasmid for P |
| SalI-P-R | ggGTCGACTTATTCATTACTTGATTCTATATCTTCCTC | |
| 3FLAG-HindIII-C-F | ggAAGCTTATGCCCTCTTTTCTGAAGAA | The expression plasmid for C |
| SalI-C-R | ttGTCGACCTACTCTTGGATTATGTGTGC | |
| PRV1-F-SacI-F | aaGAGCTCgccaccATGCAAATCATCATCCTCAGACC | The expression plasmid for F |
| PRV1-F-FLAG-R | aaCTCGAGCTACTTGTCGTCATCGTCTTTGTAGTCTCCCATGAAATTAGTAGGC | |
| PRV1-M-SacI-F | aaGAGCTCgccaccATGGCCGAGATCTACAAGT | The expression plasmid for M |
| PRV1-M-FLAG-R | aCTCGAGCTACTTGTCGTCATCGTCTTTGTAGTCAACTTTTATTTTCCCAATATTTTTTG | |
| PRV1-HN-SacI-F | aaGAGCTCgccaccATGGAAGAGACCAAAGTTAAG | The expression plasmid for HN |
| PRV1-HN-FLAG-R | aaCTCGAGTTACTTGTCGTCATCGTCTTTGTAGTCTAAATTGCTTATCCTGCAA | |
| V-F | GATTGGTAAAAAGGGGCACAGAAGAGAATAC | The expression plasmid for V |
| V-R | GTATTCTCTTCTGTGCCCCTTTTTACCAATC | |
| W-F | GATTGGTAAAAAGGGGGCACAGAAGAGAATAC | The expression plasmid for W |
| W-R | GTATTCTCTTCTGTGCCCCCTTTTTACCAATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Li, C. Porcine Respirovirus 1 Suppresses Host Type I Interferon Production and the JAK-STAT Signaling Pathway. Viruses 2023, 15, 1176. https://doi.org/10.3390/v15051176
Li Y, Li C. Porcine Respirovirus 1 Suppresses Host Type I Interferon Production and the JAK-STAT Signaling Pathway. Viruses. 2023; 15(5):1176. https://doi.org/10.3390/v15051176
Chicago/Turabian StyleLi, Yanhua, and Chenxi Li. 2023. "Porcine Respirovirus 1 Suppresses Host Type I Interferon Production and the JAK-STAT Signaling Pathway" Viruses 15, no. 5: 1176. https://doi.org/10.3390/v15051176
APA StyleLi, Y., & Li, C. (2023). Porcine Respirovirus 1 Suppresses Host Type I Interferon Production and the JAK-STAT Signaling Pathway. Viruses, 15(5), 1176. https://doi.org/10.3390/v15051176







