Abstract
Integrase inhibitors (INIs) are a potent option for HIV treatment. Limited data exist on INI resistance in West Africa, particularly in children living with HIV/AIDS. We determined the prevalence of integrase gene polymorphisms and the frequency of naturally occurring amino acid (aa) substitutions at positions associated with INI resistance. Dried blood spot (DBS) samples were obtained from one hundred and seven (107) HIV-1-infected children aged less than 15 years old in two West African countries, Benin and Mali. All children were naïve to INI treatment, 56 were naïve to anti-retroviral therapy (ART), and 51 had received ART. Genetic sequencing of HIV integrase was successful in 75 samples. The aa changes at integrase positions associated with INI resistance were examined according to the Stanford HIV Genotypic Resistance database. The median ages were 2.6 and 10 years for ART-naïve and -treated children, respectively. The most common subtypes observed were CRF02_AG (74.7%) followed by CRF06_cpx (20%). No major INI-resistance mutations at positions 66, 92, 121, 143, 147, 148, 155, and 263 were detected. The most prevalent INI accessory resistance mutations were: L74I/M (14/75, 18.6%) followed by E157Q (8/75, 10.6%), G163E/N/T/Q (5/75, 6.6%), Q95A/H/P (2/75, 2.6%), and T97A (4/75, 5.3%). Other substitutions observed were M50I/L/P, H51E/P/S/Q, I72V, T112V, V201I, and T206S. Polymorphisms at positions which may influence the genetic barrier and/or drive the selection of specific INI-resistance pathways were detected. However, no transmitted drug resistance (TDR) to INI was detected among samples of INI-naïve patients. These findings support the use of this treatment class for children with HIV-1, particularly in West Africa.
1. Introduction
Major efforts have been directed towards the development of drugs targeting HIV-1 integrase, a crucial enzyme involved in HIV-1 replication [1]. Integrase-inhibitor (INI)-based drugs block the integrase strand transfer, that is, the binding of the INI to the integrase enzyme, thereby preventing the integration of the viral genome into the host DNA [2]. First-generation INIs, raltegravir (RAL) and elvitegravir (EVG), were approved by the United States Food and Drug Administration (US FDA) for clinical use in 2007 and 2012, respectively, with the second-generation INIs dolutegravir (DTG), bictegravir (BIC), and cabotegravir (CAB) being approved in 2012, 2018, and 2021, respectively [3]. INI-based regimens are presently among the most commonly used and recommended first-line antiretroviral therapies (ARTs) for HIV-1 infection, in both treatment-experienced and ART-naïve patients, due to their high potency, effectiveness, safety, low toxicity, and tolerability [1,4,5,6]. The combined use of INI-based drugs with other early therapies, such as nucleoside reverse-transcriptase inhibitors (NRTIs), the non-nucleoside reverse-transcriptase inhibitor (NNRTI) Etravirine, and the protease inhibitor (PI) Darunavir, have resulted in high levels of viral-load suppression in HIV-infected patients [4,6]. However, the long-term efficacy of these drugs has been restricted by the emergence of resistance-associated mutations [7,8].
An estimated 1.8 million children aged 0–14 were living with HIV at the end of 2019, and 150.000 children were newly infected. The sub-Saharan African region remains the most affected. Benin and Mali, two West African countries, have experienced similar HIV-1 epidemics, with prevalence rates of around 1% (UNAIDS report, 2021); the most prominent strain circulating within these countries is CRF02_AG [9]. HIV-positive children should be started on antiretroviral drugs (ARVs) immediately, although in 2018 almost half of all children living with HIV were not taking ARVs.
A multi-country analysis of HIV drug resistance in sub-Saharan African countries showed that 53.0% and 8.8% of newly diagnosed infants had resistance to one or more non-nucleoside reverse-transcriptase inhibitors (NNRTIs) and nucleoside reverse-transcriptase inhibitors (NRTIs), respectively, before treatment initiation [10]. The World Health Organization WHO) recommended dolutegravir-based ART as the preferred first- and second-line treatment for adults and children with HIV-1 infection [11]. In Benin and Mali, DTG and RAL are the only two INIs accessible since 2018.
The ODYSSEY trial, a randomized study that included African countries, showed evidence of the superior efficacy of dolutegravir-based ART, compared with standard care in children and adolescents, as first-line and second-line ART [12].
Since the implementation of INI regimens, surveillance of INI-selected mutations has gained in importance [13]. Several INI-resistance-associated mutations (RAMs) have been reported [7]. Three groups (major, accessory, and other mutations) have been classified in the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu, accessed on 2 January 2023) based on their associations with reduced drug susceptibility: (1) major mutations represent non-polymorphic mutations that, by themselves, reduce susceptibility to one or more INIs; (2) accessory mutations represent polymorphic or non-polymorphic mutations that can reduce INI susceptibility in combination with major RAMs; (3) other mutations are polymorphic or non-polymorphic mutations that can be selected under INI-based therapy and which may or may not potentially reduce susceptibility [1].
In developed counties, INI RAMs have been well studied but there are few data for Africa, particularly in children. The aim of this study was to characterize the prevalence of HIV-1 natural polymorphisms and RAMs in INI-naïve, HIV-1-infected children in two West African countries.
2. Materials and Methods
2.1. Patients and Sample Collection
Dried blood spots (DBSs) were obtained from 107 HIV-1-infected children aged less than 15 years old living in two West African countries, Mali and Benin. The DBSs were collected from 51 ART-treated children with virological failure, defined as having at least one viral load (VL) >1000 copies/mL, during a study conducted in Benin between 2015 and 2016 [14] and from 56 ART-naïve children, collected between 2018 and 2020 during early diagnosis testing and follow-up in Mali. All children were INI-naïve. Whatman 903 filter paper was used to collect the DBSs, with 50 uL of blood being dropped into each concentric circle. The DBSs were dried at ambient temperature and then sent to the Virology Laboratory at the Saint-Antoine Hospital (Paris, France) for sequencing.
2.2. Genotyping and Analysis of Resistance-Associated Mutations
HIV-1 RNA was extracted from the 107 DBSs using EasyMag technology as per the manufacturer’s instructions. Target integrase regions of the HIV-1 were subsequently amplified by PCR after reverse-transcription production of complementary DNA (cDNA) and sequenced using the Sanger method. Genotypic-resistance testing was conducted on the integrase gene’s target regions of HIV-1 using the ANRS ((French) National Agency for Research on AIDS and Viral Hepatitis) method (http://www.hivfrenchresistance.org/, accessed on 2 January 2023).
The primers used were: outer primers: INPS1: 5′-TAG TAG CCA GCT GTG ATA AAT GTC-3′ and INPR8: 5′-TTC CAT GTT CTA ATC CTC ATC CTG-3′; inner and sequencing primers: INPS3: 5′-GAA GCC ATG CAT GGA CAA G-3′ and INPR9: 5′-ATC CTC ATC CTG TCT ACT TGC C-3′. The PCR conditions were as follows: RT-PCR step, using the RT-PCR kit Access (PROMEGA): 45′ at 45 °C, 2′ at 95 °C, then: (30″ at 95 °C; 30″ at 50 °C; 1′ at 68 °C) × 40 cycles. Nested PCR step, using Ampli Taq Polymerase with Gene Amp (Roche): 5′ at 94 °C, then (30″ at 94 °C; 30″ at 55 °C (50 °C); 1′ at 72 °C) × 45 cycles 7′ at 72 °C.
Seventy-five (75) of the 107 samples were successfully sequenced, assembled, and aligned using Sescap and SmartGene HIV Software (Innovation Park, Lausanne, Switzerland). The other 32 samples could not be amplified. The INI sequences, successfully analyzed from 75 DBS samples, were from 31 ART-treated children with virological failure (20 from Benin and 11 from Mali) and 44 ART-naïve HIV-1-infected Malian children. Positions associated with INI resistance were analyzed, and the interpretation of INI resistance was performed following the Stanford HIV resistance algorithm, version 7.0. The HIV-1 subtypes and CRFs were determined according to the Stanford University HIV Drug Resistance Database HIVdb Program, latest version (https://hivdb.stanford.edu/hivdb, accessed on 2 January 2023), with a single integrase gene amplification. Regarding INI mutations, all mutations reported in the IAS and Stanford lists were considered; in particular, the major INI RAMs from the latest version of Stanford’s list, including T66A/I/K, E92Q, G118R, E138K/A/T, G140S/A/C/R, Y143R/C/H, S147G, Q148H/R/K, N155H, and R263K (https://hivdb.stanford.edu/, accessed on 2 January 2023). Some uncommon nonpolymorphic accessory mutations, such as M50I/L/P, H51Y, and F121Y, alone, do not reduce INI susceptibility [15]. We also analyzed 19 natural polymorphisms (I72V, L74I/M, T97A, T112I, A128T, E138K, Q148H, V151I, S153Y, S153A, M154I, N155H, K156N, E157Q, G163R, V165I, V201I, I203M, and T206S) [16]. HIV-1 strains were defined as resistant if they carried at least one major transmitted drug-resistance (TDR) mutation or natural polymorphism. The overall prevalence was defined as the percentage of HIV-1-infected patients with any strain with a TDR mutation or natural polymorphism.
All sequences were submitted to GenBank and registered under accession numbers OQ435656-OQ435729.
2.3. Data Analysis
The participant characteristics and the proportions of resistance or polymorphism mutations were compared between the naïve and treated patients. Fisher’s test was used to compare categorical variables, and the Wilcoxon test was used to compare continuous variables. All statistical analyses were computed using STATA, version 11 (Stata Corporation, College Station, TX, USA). All statistical tests were two-tailed, with α = 0.05.
3. Results
3.1. Characteristics of Study Subjects
The characteristics of the patients providing the 75 samples that were successfully sequenced are shown in Table 1. At enrollment, 90.3% of the treated children were receiving non-nucleoside-reverse-transcriptase-inhibitor (NNRTI)-based regimens, and only 9.7% received both nucleoside-reverse-transcriptase inhibitor (NRTI)- and PI-based regimens. INI-resistance mutations in sequenced samples are shown in Table 2.

Table 1.
Characteristics of Patients Providing the Sequenced Samples.

Table 2.
INI-Resistance Mutations in Sequenced Samples.
3.2. INI-Resistance-Associated Mutations and Natural Polymorphism Patterns
No residue substitutions of major INI RAMs at codons 66, 92, 121, 143, 147, 148, 155, or 263 were observed in the 75 sequences. Of the 75 sequences, 25/75, 33%, harbored at least one INI accessory polymorphism mutation (L74I/M, Q95A/H/P, T97A, G118V, E138Q, S153F, E157Q, G163E/N/T/Q, or S230N) and 48 (64%) harbored at least one polymorphism mutation, described as having no impact on INI sensitivity [15,16].
Among the detected INI accessory mutations, L74I/M was the most prevalent (14/75, 18%), followed by E157Q (8/75, 10.6%), G163EN/T/Q (5/75, 6.6%), Q95A/H/P (2/75, 2.6%), and T97A (4/75, 5.3%). The lowest prevalence of residue substitutions was found at codons G118V, S153F, and S230N, all with the same prevalence (1.3%), representing only 1 of the 75 samples. The prevalence of E138Q was 2.6% (2/75) (Figure 1). Some natural polymorphism patterns (no known impact on INI sensitivity) were also observed (Figure 2). The highest prevalence in this group was observed at codon T206S (74, 7%), followed by T112V (62.6%), V201I (61/75, 81.3%), and V72I (26/75, 34.6%). Other mutations, H51E/P/S/Q and M50I/L/P, were found to have prevalences of 28% and 26%, respectively. The prevalence of INI accessory polymorphism mutations at the described positions was higher in samples with ART than those without ART, particularly the E157Q, E138Q, T97A, and Q95A/H/P mutations (Figure 1), whereas, the presence of polymorphisms described as having no known impact on INI sensitivity was higher in ART-naïve patient samples (Figure 2).
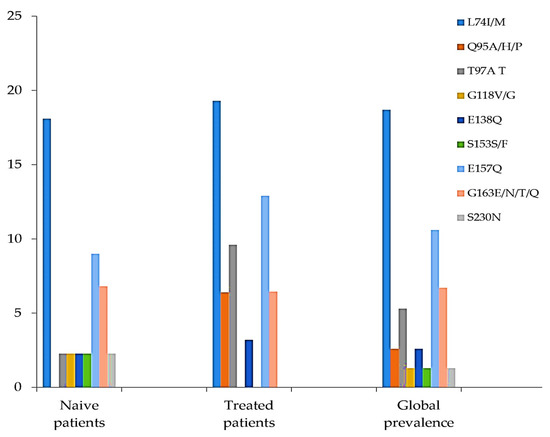
Figure 1.
Prevalence of INI accessory polymorphism mutations at described positions.
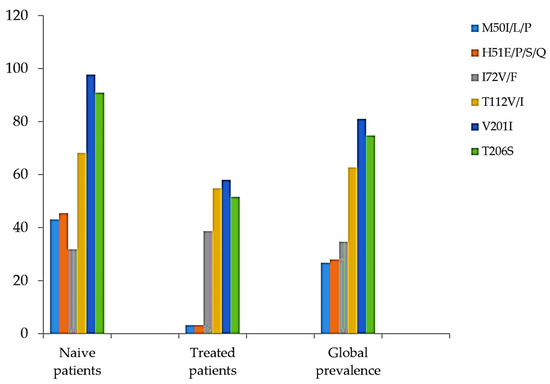
Figure 2.
Presence of polymorphism mutations described as having no known impact on INI sensitivity (Adapted from [17]).
4. Discussion
This first report from Benin and Mali found that no major INI RAMs were identified, consistent with prior studies [7,14,18], further supporting evidence that primary INI RAMs are rare. This finding is congruent with studies from the United States [19] and Europe [20], where INI drug-resistance mutations (DRMs) have rarely been identified in ART-naïve patients. The absence of major INI DRMs among INI-naïve patients in our study was also consistent with studies from other African countries, such as Zimbabwe, Uganda, Morocco, and Cameroon [21,22,23,24]. However, a multi-country study from SSA, including Kenya, South Africa, Uganda, and Zambia, found a 2% prevalence of INI RAMs using Illumina next-generation sequencing [25]. This difference can be explained by the fact that our analysis was based on the Sanger sequencing method, which might have underestimated the prevalence of INI RAMs. The Sanger method does not detect drug-resistance minority variants below 20% of the virus population. The most frequently reported major INI RAMs include codons T66A/I/K, E92G/Q, F121Y, Y143C/H/R/S, S147G, Q148H/R/K, N155H, and R263K [1,13,26], while accessory RAMs and other mutations have been largely reported in recent studies from SSA [21,22,23,24,25,27]. The prevalence of both major and accessory mutations in the ART-naïve population is very low (0.5%) and likely results from cases of transmitted INI resistance and the cumulative natural occurrence of mutations without selective drug pressure [4,26,28]. Nine accessory INI-resistance mutations were found in this study, among which L74I/M, T97A, and E157Q, were identified most frequently. These mutations were observed in Cameroon and Ethiopia [24,27], and L74M/I and M50I have been reported in the untreated population. Although many reports have shown that accessory INI-resistance mutations (including naturally occurring polymorphisms) do not compromise the effectiveness of INI-based regimens [29,30], other studies have revealed that several mutations can act synergistically or additively to decrease drug susceptibility [31,32]; for instance, the combination of L74M and G163R associated with T79A, has been reported by Abram et al. as further reducing susceptibility to RAL and/or EVG in the absence of primary INI-resistance mutations. Therefore, the accessory INI-resistance mutations found in this study should be closely monitored, as they may develop synergistic combinations. In addition, some accessory INI RAMs may also pre-exist in treatment-naïve patients. Although prevalence was low, a prior study reported that prevalence can increase over time and emerge in combination with nonpolymorphic INI-resistance mutations [26]. The subtypes found in our study were non-B subtypes, notably, CRF02_AG and CRF06_cpx, which are prevalent in West Africa. L74M tends to be more prevalent in non-B subtypes [1], explaining its high prevalence in our sequenced samples. The second most common mutation was E157Q in those sequences, a polymorphic mutation that was weakly selected in RAL-treated patients and in vitro with EVG. E157Q was the most common polymorphic accessory mutation detected in an Ethiopian analysis, and natural polymorphisms were present in 1–10% of untreated individuals, depending on the subtype [27]. It has been shown that E157Q decreases RAL susceptibility five-fold and EVG two-fold [33]. In addition, this mutation may increase DTG resistance because it may be a compensatory mutation that partially restores the enzymatic activity and infectivity lost with the R263K mutation [34]. However, no samples contained R263K in this study. This mutation was not detected in any non-B specimens in a Canadian study [1].
The mutation T97A is a naturally occurring and lowly prevalent integrase polymorphism that emerges more frequently among patients carrying non-B, as in our setting, compared to those carrying HIV subtype B [7,18,29,33,35,36]. The prevalence of T97A (5.6%) was high compared to prior studies [7,35,37]. T97A is associated with low-level decreased susceptibility to EVG and/or RAL [38,39] and has been detected in patients with virological failure and receiving INI-based therapy [7,40,41,42]. Therefore, the presence of T97A in this study suggests the need to evaluate the efficacy of INI-based treatment in HIV-infected patients. INI-associated polymorphic substitution of E157Q or L74I was identified during the IMPAACT P1093 study which evaluated the safety, tolerability, efficacy, and pharmacokinetics of dolutegravir in combination with an optimized background regimen for the treatment of HIV-1 in infants, children, and adolescents aged 4 weeks to 18 years in age-defined cohorts [43].
Some substitutions with no known effect on INI susceptibility were also identified, (V72I, T112V, V201I, T206S, Y227F, and S57N/G), with prevalences from 5% to 75%. Such frequencies can be attributed to natural genetic variability, frequently observed in the integrase gene [35,44]. The oscillation of the frequency of the unknown-effect polymorphism mutations may result from genetic drift [45] since HIV-1 is subject to genetic variability. Further investigations are required to elucidate the association of these unknown mutations with INI-based regimens.
We have reported a higher prevalence of some accessory INI-resistance mutations in ART-treated samples compared to untreated samples. Indeed, 15% of naïve patients and 11% of treated patients harbored at least one INI accessory polymorphism mutation at described positions. These prevalences were statically significant, with 38% and 17% (p = 0.0035) prevalences observed for other polymorphism mutations in naïve and treated patients, respectively. The presence of these mutations in ART-treated though INI-naïve patients is conceivable, since reports have shown that integrase mutations increase their prevalence in ART-treated compared to ART-naïve patients, suggesting that some NNRTIs, NRTIs, or PIs could interact (directly or indirectly) with the integrase inducing or selecting INI secondary mutations [29]. However, the presence of accessory INI-resistance mutations in ART-naïve patients may also result from transmitted drug mutation, mostly acquired by children via mother-to-child transmission, as demonstrated with NNRTI TDR in Africa [7,46]. Although naturally occurring integrase polymorphisms generally have little or no clinical effect on INI susceptibility [29], they are of concern when they evolve into major resistance mutations, as this could facilitate the viral evolution of resistance under INI pressure [35]. Indeed, INI-based therapies, according to WHO guidelines, are currently used, even in children. INIs, such as raltegravir and dolutegravir, are increasingly used in both ART-naïve and -treated patients in Africa. In addition, in a recent study, the authors reported that the major INI-associated mutation represents a public-health concern whether natural or transmitted, since it may affect treatment efficacy and negatively affect prognosis [13]. We also recommend future studies to consider the mutations in the nef 3′-PPT region, which suggest an alternative pathway to resistance to DTG and other INIs [24].
5. Conclusions
Currently, INI-based treatments are associated with excellent clinical results in patients, leading to a healthier lifestyle and reducing the risk of viral transmission. No TDR to INI was detected among the sequences from INI-naïve child patients. These data support using this class of ARTs in West Africa, including in the pediatric population. However, continued surveillance of INI RAMs is recommended to prevent the emergence of resistance to this class of drugs.
Author Contributions
Conceptualization, D.B.F. and L.M.-J.; methodology, D.B.F.; software, D.B.F. and A.B.; validation, D.B.F. and L.M.-J.; formal analysis, D.B.F. and H.D.; investigation, D.B.F., H.D., M.M., I.G. and F.I.D.; resources, J.L.H., A.-G.M., V.C., C.K., R.L.M., A.K., A.I.M., S.L.-N., S.M., C.A.H. and M.S.; data curation, D.B.F., H.D., M.d., M.K.S. and F.I.D.; writing—original draft preparation, D.B.F., J.L.H., L.M.-J., C.S. and H.D.; writing—review and editing: all authors; visualization, D.B.F.; supervision, L.M.-J.; funding acquisition, N/A. All authors have read and agreed to the published version of the manuscript.
Funding
This specific research received no external funding. D.B.F. is independently supported by Fogarty International Center and the Office of the Director of the National Institutes of Health, Career Development Award K43TW011957 and the Agence Nationale de la Recherche sur le SIDA et les Maladies Infectieuses Emergentes (ANRS MIE) ANRS-MIE22295.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethic Committee of National Institute for Research in Public Health, (protocol code 06/2016/CE-INRSP, 27th May 2016).
Informed Consent Statement
Children’s parents and/or legal guardians provided informed consent for the collection of a dried blood spot (DSB) sample and clinical information, using a Scientific Ethics Committee-approved form. This research also involved some retrospective chart review of de-identified patient data and samples from previous studies; it was determined that the research involved no more risk to these subjects.
Data Availability Statement
Data available upon request from the corresponding author.
Acknowledgments
We would like to acknowledge the Agence Nationale de la Recherche sur le SIDA et les Maladies Infectieuses Emergentes (ANRS MIE) for her support through the Laboratory of virology Saint-Antoine Hospital in Paris and the “Building the Next Generation of Researchers in TB/HIV Diagnostics in Mali (B-NextGen) Mali”, under award number D43TW010350. The authors are also grateful to the Northwestern University’s Institute for Global Health Catalyzer program, the National Institutes of Health, and the HBNU Consortium.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ji, H.; Patterson, A.; Taylor, T.; Rank, C.; Halverson, J.; Capina, R.; Brooks, J.; Sandstrom, P. Prevalence of Primary Drug Resistance Against HIV-1 Integrase Inhibitors in Canada. Am. J. Ther. 2018, 78, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Kolakowska, A.; Maresca, A.F.; Collins, I.J.; Cailhol, J. Update on Adverse Effects of HIV Integrase Inhibitors. Curr. Treat. Options Infect. Dis. 2019, 11, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, S.-X.; He, Q.; Fan, R. Advances in the development of HIV integrase strand transfer inhibitors. Eur. J. Med. Chem. 2021, 225, 113787. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-W.; Shen, M.-C.; Wang, W.-H.; Li, W.-Y.; Wang, J.-H.; Tseng, C.-Y.; Liu, P.-Y.; Wang, L.-S.; Lee, Y.-L.; Chen, Y.-M.A.; et al. High prevalence of HIV-1 transmitted drug resistance and factors associated with time to virological failure and viral suppression in Taiwan. J. Antimicrob. Chemother. 2021, 77, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Park, T.E.; Mohamed, A.; Kalabalik, J.; Sharma, R. Review of integrase strand transfer inhibitors for the treatment of human immunodeficiency virus infection. Expert Rev. Anti-Infect. Ther. 2015, 13, 1195–1212. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, P.; Wensing, A.M.J.; Fun, A.; Nijhuis, M.; Brusselaers, N.; Vandekerckhove, L. Clinical Use of HIV Integrase Inhibitors: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e52562. [Google Scholar] [CrossRef] [PubMed]
- Abram, M.E.; Ram, R.R.; Margot, N.A.; Barnes, T.L.; White, K.L.; Callebaut, C.; Miller, M.D. Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PLoS ONE 2017, 12, e0172206. [Google Scholar] [CrossRef]
- Wainberg, M.A.; Zaharatos, G.J.; Brenner, B.G. Development of Antiretroviral Drug Resistance. N. Engl. J. Med. 2011, 365, 637–646. [Google Scholar] [CrossRef]
- Giovanetti, M.; Ciccozzi, M.; Parolin, C.; Borsetti, A. Molecular Epidemiology of HIV-1 in African Countries: A Comprehensive Overview. Pathogens 2020, 9, 1072. [Google Scholar] [CrossRef]
- Jordan, M.R.; Penazzato, M.; Cournil, A.; Vubil, A.; Jani, I.; Hunt, G.; Carmona, S.; Maphalala, G.; Mthethwa, N.; Watera, C.; et al. Human Immunodeficiency Virus (HIV) Drug Resistance in African Infants and Young Children Newly Diagnosed With HIV: A Multicountry Analysis. Clin. Infect. Dis. 2017, 65, 2018–2025. [Google Scholar] [CrossRef]
- World Health Organization. Updated Recommendations on First-Line and Secondline Antiretroviral Regimens and Post-Exposure Prophylaxis and Recommendations on Early Infant Diagnosis of HIV: Interim Guidelines; WHO: Geneva, Switzerland, 2018; Available online: https://www.who.int/publications/i/item/WHO-CDS-HIV-18.51 (accessed on 2 January 2023).
- Turkova, A.; White, E.; Mujuru, H.A.; Kekitiinwa, A.R.; Kityo, C.M.; Violari, A.; Lugemwa, A.; Cressey, T.R.; Musoke, P.; Variava, E.; et al. Dolutegravir as First- or Second-Line Treatment for HIV-1 Infection in Children. N. Engl. J. Med. 2021, 385, 2531–2543. [Google Scholar] [CrossRef] [PubMed]
- Mazzuti, L.; Melengu, T.; Falasca, F.; Calabretto, M.; Cella, E.; Ciccozzi, M.; Mezzaroma, I.; Iaiani, G.; Spaziante, M.; D’Ettorre, G.; et al. Transmitted drug resistance mutations and trends of HIV-1 subtypes in treatment-naïve patients: A single-centre experience. J. Glob. Antimicrob. Resist. 2019, 20, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Fofana, D.B.; D’Almeida, M.; Lambert-Niclot, S.; Peytavin, G.; Girard, P.M.; Lafia, B.; Zohoun-Guidigbi, L.; Keke, R.K.; Soulie, C.; Marcelin, A.G.; et al. Resistance profile and treatment outcomes in HIV-infected children at virological failure in Benin, West Africa. J. Antimicrob. Chemother. 2018, 73, 3143–3147. [Google Scholar] [CrossRef] [PubMed]
- Margot, N.A.; Hluhanich, R.M.; Jones, G.S.; Andreatta, K.N.; Tsiang, M.; McColl, D.J.; White, K.L.; Miller, M.D. In vitro resistance selections using elvitegravir, raltegravir, and two metabolites of elvitegravir M1 and M4. Antivir. Res. 2011, 93, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Lataillade, M.; Chiarella, J.; Kozal, M.J. Natural Polymorphism of the HIV-1 Integrase Gene and Mutations associated with Integrase Inhibitor Resistance. Antivir. Ther. 2007, 12, 563–570. [Google Scholar] [CrossRef]
- Standford University. HIV drug resistance Database: INI Mutation Pattern and Susceptibility. Updated on 2022-10-15. [Online]. 2022. Available online: https://hivdb.stanford.edu/ (accessed on 2 January 2023).
- Ambrosioni, J.; Nicolás, D.; Manzardo, C.; Agüero, F.; Blanco, J.L.; Mosquera, M.D.M.; Peñafiel, J.; Gatell, J.M.; Marcos, M.A.; Miró, J.M. Integrase strand-transfer inhibitor polymorphic and accessory resistance substitutions in patients with acute/recent HIV infection. J. Antimicrob. Chemother. 2016, 72, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Stekler, J.D.; McKernan, J.; Milne, R.; Tapia, K.A.; Mykhalchenko, K.; Holte, S.; Maenza, J.; Stevens, C.E.; Buskin, S.E.; Mullins, J.I.; et al. Lack of Resistance to Integrase Inhibitors among Antiretroviral-Naive Subjects with Primary HIV-1 Infection, 2007–2013. Antivir. Ther. 2015, 20, 77–80. [Google Scholar] [CrossRef]
- Gutiérrez, C.; Hernández-Novoa, B.; Pérez-Elías, M.J.; Moreno, A.M.; Holguín, Á.; Dronda, F.; Casado, J.L.; Moreno, S. Prevalence of Primary Resistance Mutations to Integrase Inhibitors in Treatment-Naïve and -Experienced Patients Infected with B and Non-B HIV-1 Variants. HIV Clin. Trials 2013, 14, 10–16. [Google Scholar] [CrossRef]
- Madyadi, A.; Dhoro, M.; Shamu, T.; Washaya, T.; Kouamou, M.V.; Chimukangara, B.; Katzenstein, D.; Manasa, J. HIV-1 Genetic Diversity and Natural Polymorphisms of the Integrase Gene in Integrase Inhibitor-Naive Patients in Harare, Zimbabwe. AIDS Res. Hum. Retroviruses 2021, 37, 954–961. [Google Scholar] [CrossRef]
- McCluskey, S.M.; Kamelian, K.; Musinguzi, N.; Kigozi, S.; Yap, B.O.U.M.; Bwana, M.B.; Muzoora, C.; Brumme, Z.L.; Carrington, M.; Carlson, J.; et al. Pre-treatment integrase inhibitor resistance is uncommon in ART-naïve individuals with HIV-1 subtype A1 and D infections in Uganda Suzanne. AIDS 2022, 35, 1083–1089. [Google Scholar] [CrossRef]
- Alaoui, N.; El Alaoui, M.A.; Touil, N.; El Annaz, H.; Melloul, M.; Tagajdid, R.; Hjira, N.; Boui, M.; El Fahime, E.M.; Mrani, S. Prevalence of resistance to integrase strand-transfer inhibitors (INSTIs) among untreated HIV-1 infected patients in Morocco. BMC Res. Notes 2018, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.; Tagny, C.T.; Mbanya, D.; Fonsah, J.Y.; Nchindap, E.; Kenmogne, L.; Jihyun, M.; Njamnshi, A.K.; Kanmogne, G.D. Variability in HIV-1 Integrase Gene and 3′-Polypurine Tract Sequences in Cameroon Clinical Isolates, and Implications for Integrase Inhibitors Efficacy. Int. J. Mol. Sci. 2020, 21, 1553. [Google Scholar] [CrossRef] [PubMed]
- Inzaule, S.C.; Hamers, R.L.; Noguera-Julian, M.; Casadellà, M.; Parera, M.; de Wit, T.F.R.; Paredes, R. Primary resistance to integrase strand transfer inhibitors in patients infected with diverse HIV-1 subtypes in sub-Saharan Africa. J. Antimicrob. Chemother. 2018, 73, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.J.; Rhee, S.-Y.; Shafer, R.W. Integrase Strand Transfer Inhibitor Resistance in Integrase Strand Transfer Inhibitor-Naive Persons. AIDS Res. Hum. Retroviruses 2021, 37, 736–743. [Google Scholar] [CrossRef]
- Arimide, D.A.; Szojka, Z.I.; Zealiyas, K.; Gebreegziabxier, A.; Adugna, F.; Sasinovich, S.; Björkman, P.; Medstrand, P. Pre-Treatment Integrase Inhibitor Resistance and Natural Polymorphisms among HIV-1 Subtype C Infected Patients in Ethiopia. Viruses 2022, 14, 729. [Google Scholar] [CrossRef]
- Rhee, S.-Y.; Tzou, P.; Shafer, R. Temporal Trends in HIV-1 Mutations Used for the Surveillance of Transmitted Drug Resistance. Viruses 2021, 13, 879. [Google Scholar] [CrossRef]
- Ceccherini-Silberstein, F.; Malet, I.; Fabeni, L.; Dimonte, S.; Svicher, V.; D’Arrigo, R.; Artese, A.; Costa, G.; Bono, S.; Alcaro, S.; et al. Specific HIV-1 integrase polymorphisms change their prevalence in untreated versus antiretroviral-treated HIV-1-infected patients, all naive to integrase inhibitors. J. Antimicrob. Chemother. 2010, 65, 2305–2318. [Google Scholar] [CrossRef]
- Low, A.; Prada, N.; Topper, M.; Vaida, F.; Castor, D.; Mohri, H.; Hazuda, D.; Muesing, M.; Markowitz, M. Natural Polymorphisms of Human Immunodeficiency Virus Type 1 Integrase and Inherent Susceptibilities to a Panel of Integrase Inhibitors. Antimicrob. Agents Chemother. 2009, 53, 4275–4282. [Google Scholar] [CrossRef]
- Hachiya, A.; Ode, H.; Matsuda, M.; Kito, Y.; Shigemi, U.; Matsuoka, K.; Imamura, J.; Yokomaku, Y.; Iwatani, Y.; Sugiura, W. Natural polymorphism S119R of HIV-1 integrase enhances primary INSTI resistance. Antivir. Res. 2015, 119, 84–88. [Google Scholar] [CrossRef]
- Tang, M.W.; Liu, T.F.; Shafer, R.W. The HIVdb System for HIV-1 Genotypic Resistance Interpretation. Intervirology 2012, 55, 98–101. [Google Scholar] [CrossRef]
- Doyle, T.; Dunn, D.T.; Ceccherini-Silberstein, F.; De Mendoza, C.; Garcia, F.; Smit, E.; Fearnhill, E.; Marcelin, A.-G.; Martinez-Picado, J.; Kaiser, R.; et al. Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J. Antimicrob. Chemother. 2015, 70, 3080–3086. [Google Scholar] [CrossRef] [PubMed]
- Anstett, K.; Cutillas, V.; Fusco, R.; Mesplède, T.; Wainberg, M.A. Polymorphic substitution E157Q in HIV-1 integrase increases R263K-mediated dolutegravir resistance and decreases DNA binding activity. J. Antimicrob. Chemother. 2016, 71, 2083–2088. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.-Y.; Sankaran, K.; Varghese, V.; Winters, M.A.; Hurt, C.B.; Eron, J.J.; Parkin, N.; Holmes, S.P.; Holodniy, M.; Shafer, R.W. HIV-1 Protease, Reverse Transcriptase, and Integrase Variation. J. Virol. 2016, 90, 6058–6070. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Geretti, A.M.; Zahonero, N.; Booth, C.; Strang, A.; Soriano, V.; De Mendoza, C. Integrase variability and susceptibility to HIV integrase inhibitors: Impact of subtypes, antiretroviral experience and duration of HIV infection. J. Antimicrob. Chemother. 2009, 65, 320–326. [Google Scholar] [CrossRef]
- Sichtig, N.; Sierra, S.; Kaiser, R.; Däumer, M.; Reuter, S.; Schülter, E.; Altmann, A.; Fätkenheuer, G.; Dittmer, U.; Pfister, H.; et al. Evolution of raltegravir resistance during therapy. J. Antimicrob. Chemother. 2009, 64, 25–32. [Google Scholar] [CrossRef]
- Armenia, D.; Fabeni, L.; Alteri, C.; Di Pinto, D.; Di Carlo, D.; Bertoli, A.; Gori, C.; Carta, S.; Fedele, V.; Forbici, F.; et al. HIV-1 integrase genotyping is reliable and reproducible for routine clinical detection of integrase resistance mutations even in patients with low-level viraemia. J. Antimicrob. Chemother. 2015, 70, 1865–1873. [Google Scholar] [CrossRef]
- Abram, M.E.; Hluhanich, R.M.; Goodman, D.D.; Andreatta, K.N.; Margot, N.A.; Ye, L.; Niedziela-Majka, A.; Barnes, T.L.; Novikov, N.; Chen, X.; et al. Impact of Primary Elvitegravir Resistance-Associated Mutations in HIV-1 Integrase on Drug Susceptibility and Viral Replication Fitness. Antimicrob. Agents Chemother. 2013, 57, 2654–2663. [Google Scholar] [CrossRef]
- Molina, J.-M.; LaMarca, A.; Andrade-Villanueva, J.; Clotet, B.; Clumeck, N.; Liu, Y.-P.; Zhong, L.; Margot, N.; Cheng, A.K.; Chuck, S.L. Efficacy and safety of once daily elvitegravir versus twice daily raltegravir in treatment-experienced patients with HIV-1 receiving a ritonavir-boosted protease inhibitor: Randomised, double-blind, phase 3, non-inferiority study. Lancet Infect. Dis. 2012, 12, 27–35. [Google Scholar] [CrossRef]
- Cooper, D.A.; Steigbigel, R.T.; Gatell, J.M.; Rockstroh, J.K.; Katlama, C.; Yeni, P.; Lazzarin, A.; Clotet, B.; Kumar, P.N.; Eron, J.E.; et al. Subgroup and Resistance Analyses of Raltegravir for Resistant HIV-1 Infection. N. Engl. J. Med. 2008, 359, 355–365. [Google Scholar] [CrossRef]
- Eron, J.J.; Clotet, B.; Durant, J.; Katlama, C.; Kumar, P.; Lazzarin, A.; Poizot-Martin, I.; Richmond, G.; Soriano, V.; Ait-Khaled, M.; et al. Safety and Efficacy of Dolutegravir in Treatment-Experienced Subjects with Raltegravir-Resistant HIV Type 1 Infection: 24-Week Results of the VIKING Study. J. Infect. Dis. 2012, 207, 740–748. [Google Scholar] [CrossRef]
- Vavro, C.; Ruel, T.; Wiznia, A.; Montañez, N.; Nangle, K.; Horton, J.; Buchanan, A.M.; Stewart, E.L.; Palumbo, P. Emergence of Resistance in HIV-1 Integrase with Dolutegravir Treatment in a Pediatric Population from the IMPAACT P1093 Study. Antimicrob. Agents Chemother. 2022, 66, e01645-21. [Google Scholar] [CrossRef] [PubMed]
- Varghese, V.; Liu, T.F.; Rhee, S.-Y.; Libiran, P.; Trevino, C.; Fessel, W.J.; Shafer, R.W. HIV-1 Integrase Sequence Variability in Antiretroviral Naïve Patients and in Triple-Class Experienced Patients Subsequently Treated with Raltegravir. AIDS Res. Hum. Retroviruses 2010, 26, 1323–1326. [Google Scholar] [CrossRef] [PubMed]
- Diarra, H.; Makhulu, E.E.; Odhiambo, P.O.; Irekwa, R.M.; Kinyua, J.; Herren, J.K.; Mobegi, V.A. Molecular Investigation of Genetic Signatures of Selection in Plasmodium falciparum Actin-Binding Protein Coronin, Cysteine Desulfurase, and Plasmepsin 2 Gene in Mbita Field Isolates, Western Kenya. Open J. Genet. 2021, 11, 120–144. [Google Scholar] [CrossRef]
- Boerma, R.S.; Sigaloff, K.C.E.; Akanmu, A.S.; Inzaule, S.; van Hensbroek, M.B.; de Wit, T.F.R.; Calis, J.C. Alarming increase in pretreatment HIV drug resistance in children living in sub-Saharan Africa: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2016, 72, 365–371. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).