Novel HCV Genotype 4d Infectious Systems and Assessment of Direct-Acting Antivirals and Antibody Neutralization
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of HCV Genotype 4d Clone
2.2. Determination of 5′- and 3′UTR of Genotype 4d
2.3. Analysis of Virus Recovered from Cell Culture
2.4. Treatment with Direct-Acting Antivirals (DAAs)
2.5. HCV Neutralization Assay
2.6. Analysis of HCV Viability in Human-Liver Chimeric Mice
3. Results
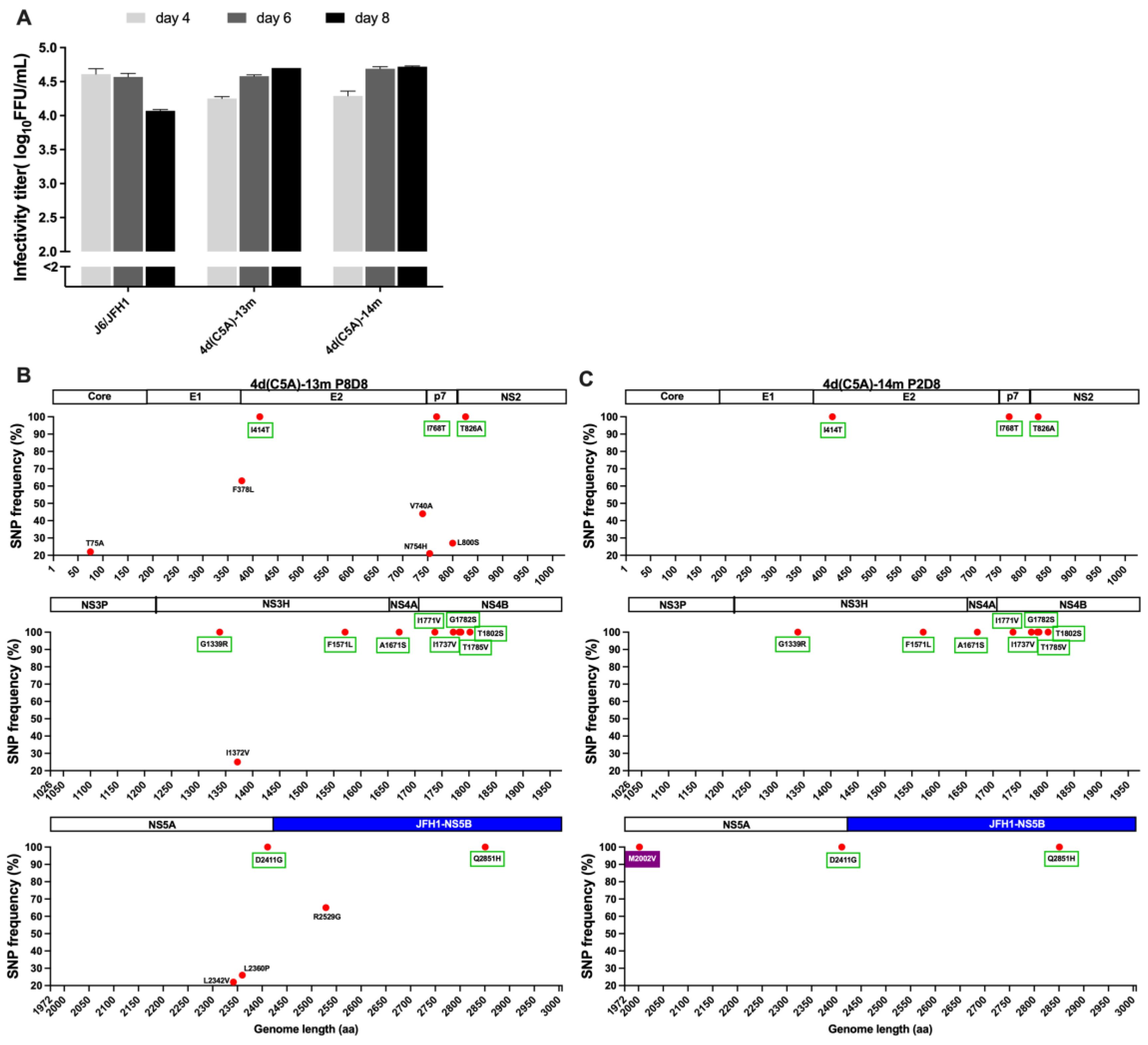
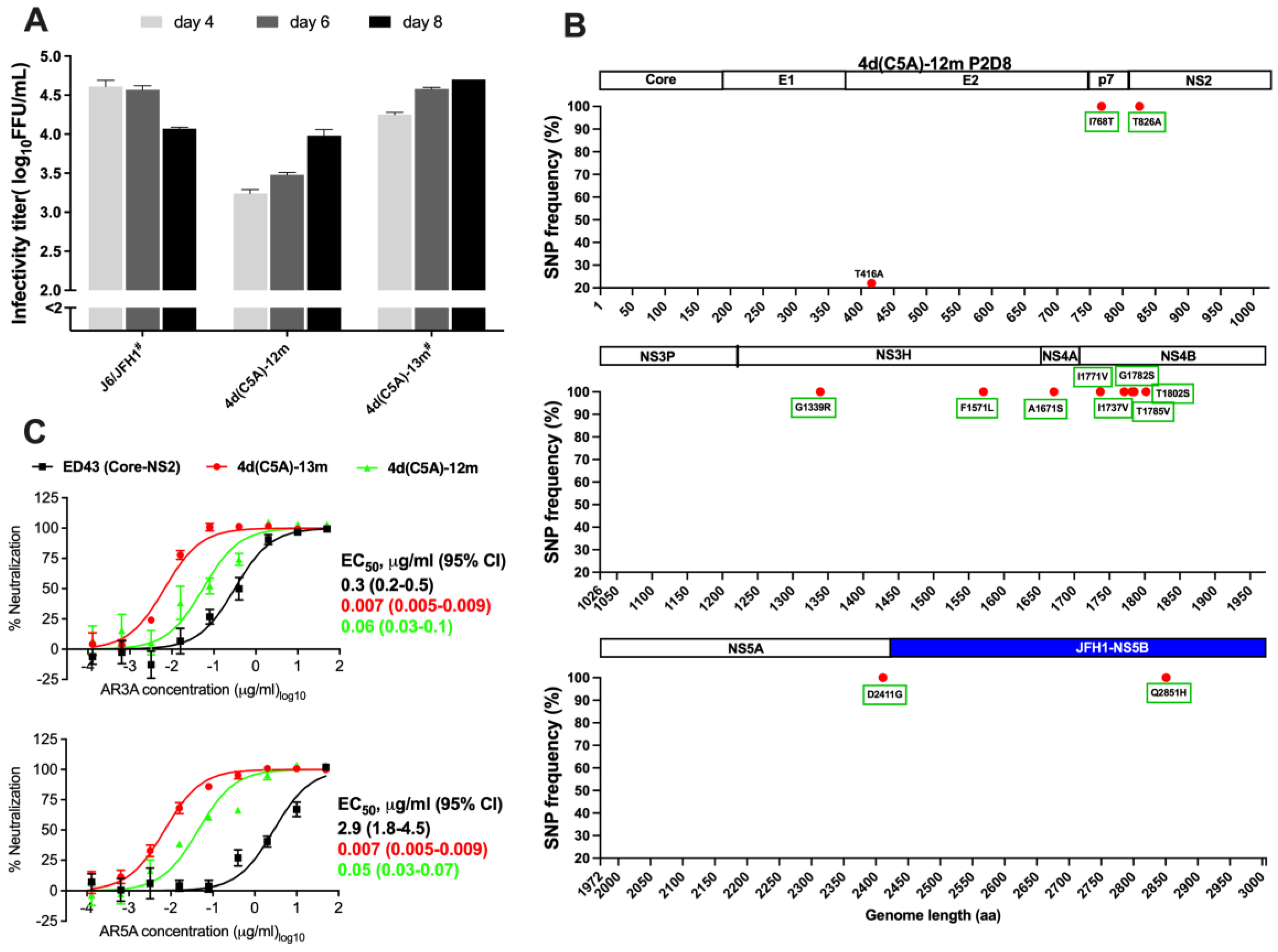
3.1. In Vivo and In Vitro Analysis of HCV Genotype 4d Full-Length Clone
3.2. Development of JFH1-Based Genotype 4d Core-NS5A Infectious System
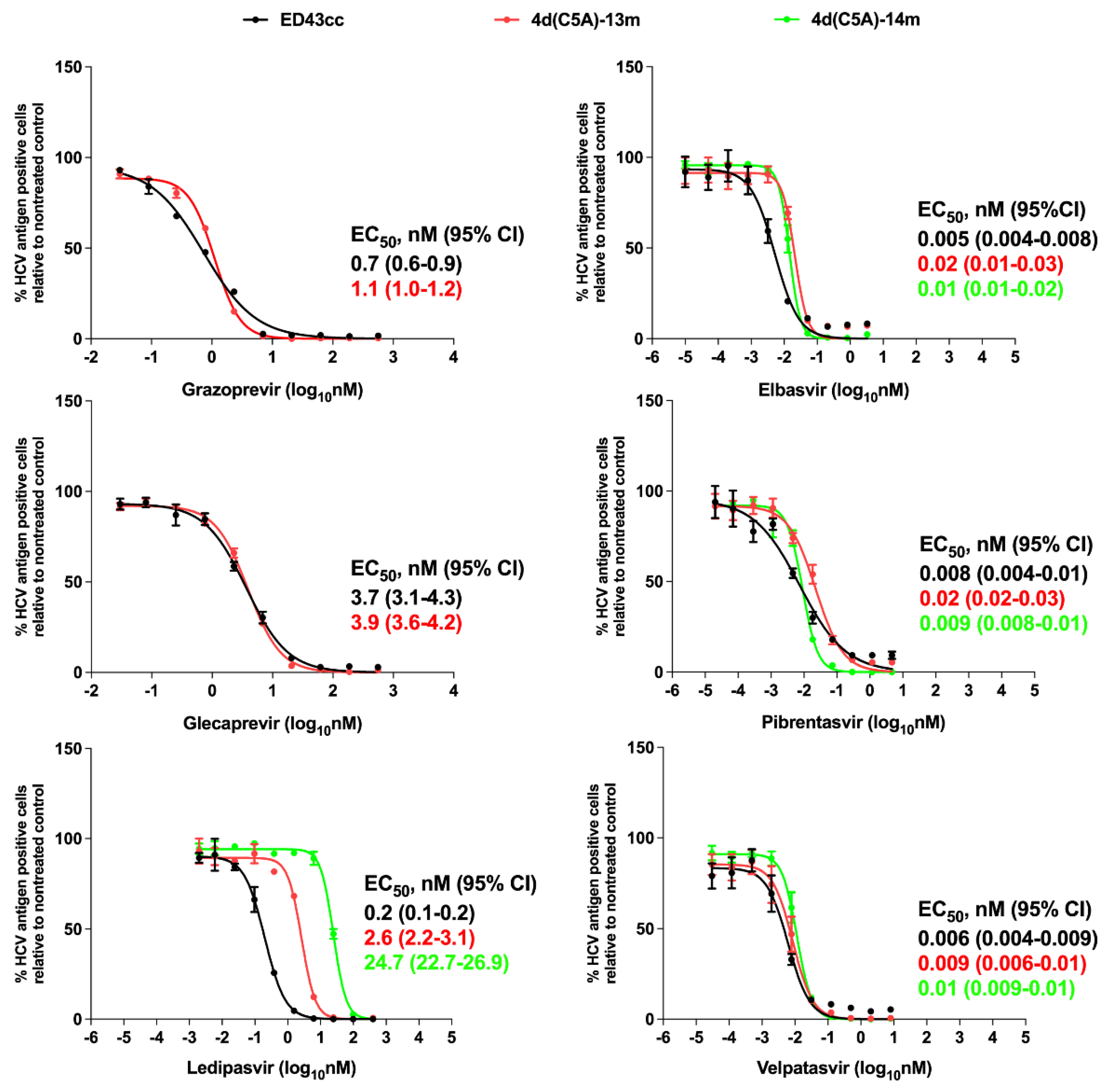
3.3. Sensitivity of 4d Virus to Direct-Acting Antivirals (DAA)
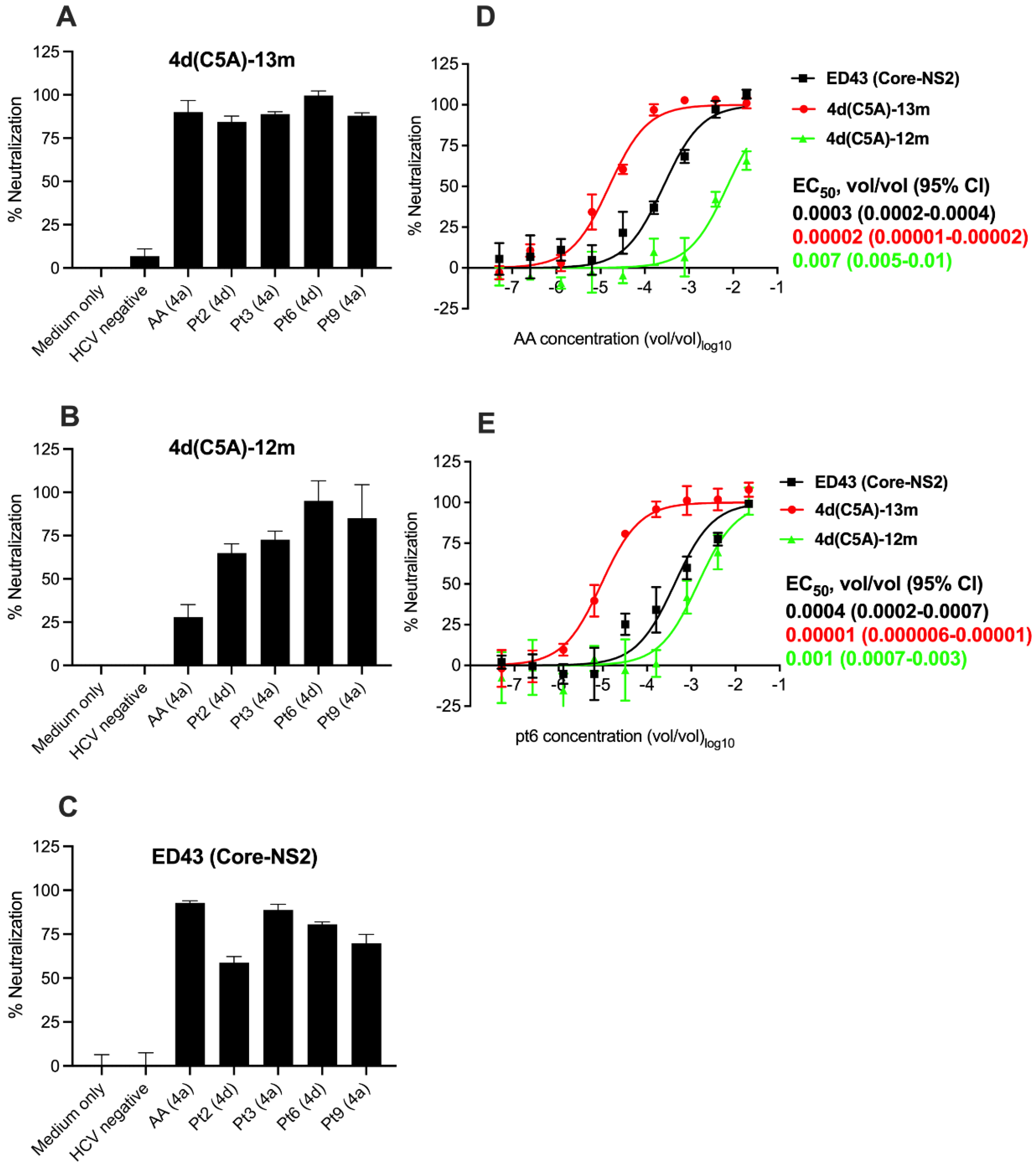
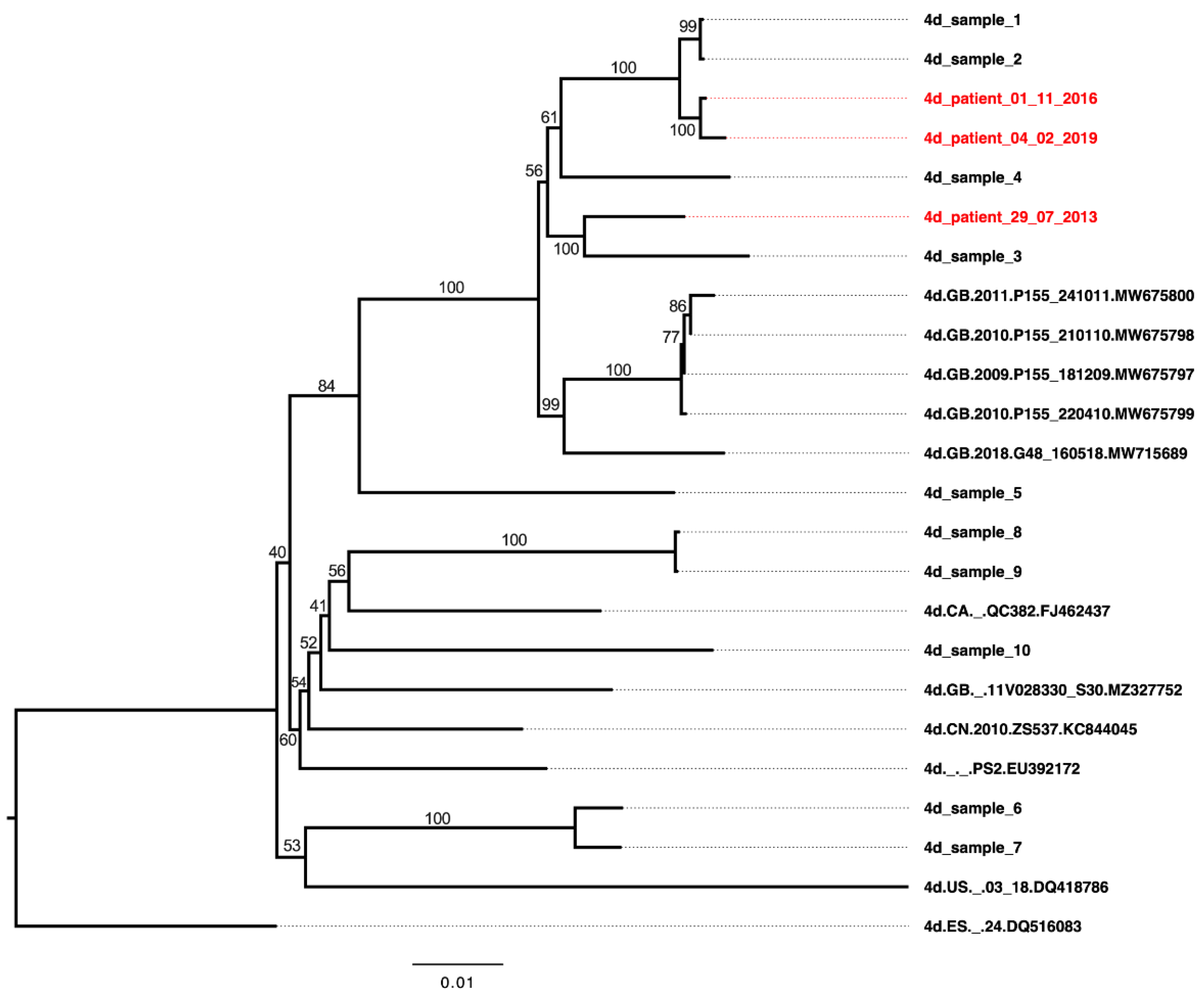
3.4. Sensitivity of HCV Genotype 4d Virus to Monoclonal and Patient Plasma NAbs
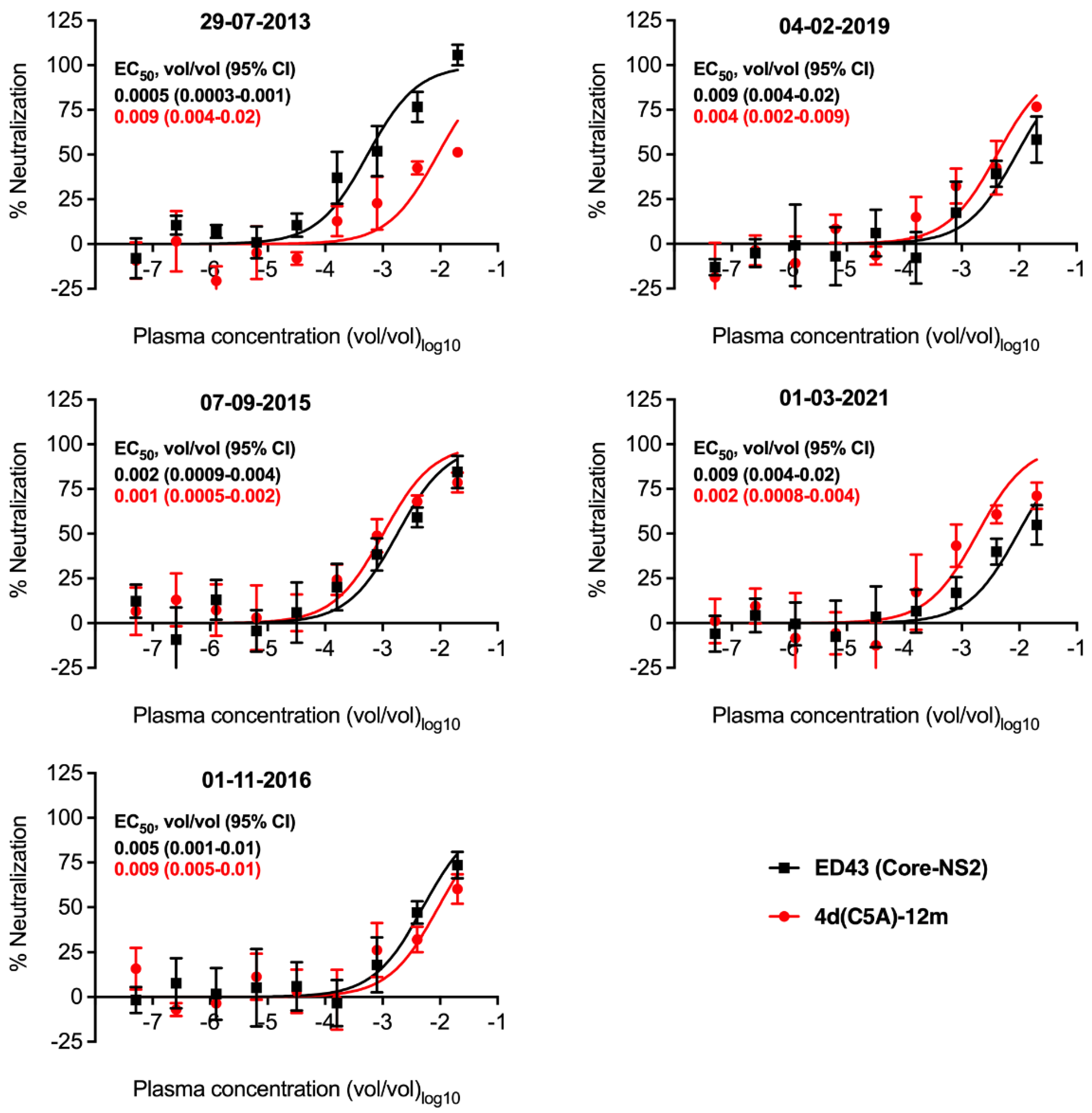
3.5. Neutralization Sensitivity of Genotype 4d Virus to Serial Plasma Samples from DH13 Infected Patient
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubuisson, J.; Cosset, F.L. Virology and cell biology of the hepatitis C virus life cycle: An update. J. Hepatol. 2014, 61, S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Bukh, J. The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J. Hepatol. 2016, 65, S2–S21. [Google Scholar] [CrossRef] [PubMed]
- Borgia, S.M.; Hedskog, C.; Parhy, B.; Hyland, R.H.; Stamm, L.M.; Brainard, D.M.; Subramanian, M.G.; McHutchison, J.G.; Mo, H.; Svarovskaia, E.; et al. Identification of a Novel Hepatitis C Virus Genotype From Punjab, India: Expanding Classification of Hepatitis C Virus Into 8 Genotypes. J. Infect. Dis. 2018, 218, 1722–1729. [Google Scholar] [CrossRef]
- Hedskog, C.; Parhy, B.; Chang, S.; Zeuzem, S.; Moreno, C.; Shafran, S.D.; Borgia, S.M.; Asselah, T.; Alric, L.; Abergel, A.; et al. Identification of 19 Novel Hepatitis C Virus Subtypes-Further Expanding HCV Classification. Open. Forum Infect. Dis. 2019, 6, ofz076. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Ahovegbe, L.; Niebel, M.; Shepherd, J.; Thomson, E.C. Non-epidemic HCV genotypes in low- and middle-income countries and the risk of resistance to current direct-acting antiviral regimens. J. Hepatol. 2021, 75, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Bukh, J.; Purcell, R.H.; Miller, R.H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 1993, 90, 8234–8238. [Google Scholar] [CrossRef]
- Gotte, M.; Feld, J.J. Direct-acting antiviral agents for hepatitis C: Structural and mechanistic insights. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 338–351. [Google Scholar] [CrossRef]
- Baumert, T.F.; Berg, T.; Lim, J.K.; Nelson, D.R. Status of Direct-Acting Antiviral Therapy for Hepatitis C Virus Infection and Remaining Challenges. Gastroenterology 2019, 156, 431–445. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C: Final update of the series(☆). J. Hepatol. 2020, 73, 1170–1218. [Google Scholar] [CrossRef]
- Li, D.K.; Chung, R.T. Overview of Direct-Acting Antiviral Drugs and Drug Resistance of Hepatitis C Virus. Methods Mol. Biol. 2019, 1911, 3–32. [Google Scholar] [CrossRef]
- Newsum, A.M.; Molenkamp, R.; van der Meer, J.T.; Rebers, S.P.; Prins, M.; van der Valk, M.; Schinkel, J. Persistence of NS5B-S282T, a sofosbuvir resistance-associated substitution, in a HIV/HCV-coinfected MSM with risk of onward transmission. J. Hepatol. 2018, 69, 968–970. [Google Scholar] [CrossRef] [PubMed]
- Minosse, C.; Selleri, M.; Giombini, E.; Bartolini, B.; Capobianchi, M.R.; Cerilli, S.; Loiacono, L.; Taibi, C.; D’Offizi, G.; McPhee, F.; et al. Clinical and virological properties of hepatitis C virus genotype 4 infection in patients treated with different direct-acting antiviral agents. Infect. Drug Resist. 2018, 11, 2117–2127. [Google Scholar] [CrossRef]
- Bukh, J. Neutralizing Antibodies Against Hepatitis C Virus and Their Role in Vaccine Immunity. Gastroenterology 2022, 162, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Osburn, W.O.; Snider, A.E.; Wells, B.L.; Latanich, R.; Bailey, J.R.; Thomas, D.L.; Cox, A.L.; Ray, S.C. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology 2014, 59, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, P.; Bukh, J.; Verhoye, L.; Farhoudi, A.; Vanwolleghem, T.; Wang, R.Y.; Desombere, I.; Alter, H.; Purcell, R.H.; Leroux-Roels, G. In vivo evaluation of the cross-genotype neutralizing activity of polyclonal antibodies against hepatitis C virus. Hepatology 2011, 53, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Bukh, J.; Engle, R.E.; Faulk, K.; Wang, R.Y.; Farci, P.; Alter, H.J.; Purcell, R.H. Immunoglobulin with High-Titer In Vitro Cross-Neutralizing Hepatitis C Virus Antibodies Passively Protects Chimpanzees from Homologous, but Not Heterologous, Challenge. J. Virol. 2015, 89, 9128–9132. [Google Scholar] [CrossRef]
- Velázquez-Moctezuma, R.; Augestad, E.H.; Castelli, M.; Holmboe Olesen, C.; Clementi, N.; Clementi, M.; Mancini, N.; Prentoe, J. Mechanisms of Hepatitis C Virus Escape from Vaccine-Relevant Neutralizing Antibodies. Vaccines 2021, 9, 291. [Google Scholar] [CrossRef]
- Velázquez-Moctezuma, R.; Galli, A.; Law, M.; Bukh, J.; Prentoe, J. Hepatitis C Virus Escape Studies of Human Antibody AR3A Reveal a High Barrier to Resistance and Novel Insights on Viral Antibody Evasion Mechanisms. J. Virol. 2019, 93, e01909-18. [Google Scholar] [CrossRef]
- Velázquez-Moctezuma, R.; Galli, A.; Law, M.; Bukh, J.; Prentoe, J. Hepatitis C Virus-Escape Studies for Human Monoclonal Antibody AR4A Reveal Isolate-Specific Resistance and a High Barrier to Resistance. J. Infect. Dis. 2019, 219, 68–79. [Google Scholar] [CrossRef]
- Velázquez-Moctezuma, R.; Law, M.; Bukh, J.; Prentoe, J. Applying antibody-sensitive hypervariable region 1-deleted hepatitis C virus to the study of escape pathways of neutralizing human monoclonal antibody AR5A. PLoS Pathog. 2017, 13, e1006214. [Google Scholar] [CrossRef]
- Meunier, J.C.; Engle, R.E.; Faulk, K.; Zhao, M.; Bartosch, B.; Alter, H.; Emerson, S.U.; Cosset, F.L.; Purcell, R.H.; Bukh, J. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 2005, 102, 4560–4565. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.H.; Urbanowicz, R.A.; Guest, J.D.; Frumento, N.; Figueroa, A.; Clark, K.E.; Keck, Z.; Cowton, V.M.; Cole, S.J.; Patel, A.H.; et al. An Antigenically Diverse, Representative Panel of Envelope Glycoproteins for Hepatitis C Virus Vaccine Development. Gastroenterology 2022, 162, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, J.M.; Scheel, T.K.; Jensen, T.B.; Lademann, J.B.; Prentoe, J.C.; Knudsen, M.L.; Hoegh, A.M.; Bukh, J. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: Role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 2009, 49, 364–377. [Google Scholar] [CrossRef]
- Ramirez, S.; Bukh, J. Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: Essential tools in testing of antivirals and emerging vaccine strategies. Antivir. Res. 2018, 158, 264–287. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [PubMed]
- Lindenbach, B.D.; Evans, M.J.; Syder, A.J.; Wolk, B.; Tellinghuisen, T.L.; Liu, C.C.; Maruyama, T.; Hynes, R.O.; Burton, D.R.; McKeating, J.A.; et al. Complete replication of hepatitis C virus in cell culture. Science 2005, 309, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Alzua, G.P.; Pihl, A.F.; Offersgaard, A.; Duarte Hernandez, C.R.; Duan, Z.; Feng, S.; Fahnøe, U.; Sølund, C.; Weis, N.; Law, M.; et al. Inactivated genotype 1a, 2a and 3a HCV vaccine candidates induced broadly neutralising antibodies in mice. Gut, 2022; ahead of print. [Google Scholar] [CrossRef]
- Bukh, J. Vaccines against hepatitis C: A travel into neutralisation space. Gut 2021, 70, 1609–1610. [Google Scholar] [CrossRef]
- Bankwitz, D.; Bahai, A.; Labuhn, M.; Doepke, M.; Ginkel, C.; Khera, T.; Todt, D.; Ströh, L.J.; Dold, L.; Klein, F.; et al. Hepatitis C reference viruses highlight potent antibody responses and diverse viral functional interactions with neutralising antibodies. Gut 2021, 70, 1734–1745. [Google Scholar] [CrossRef]
- Pedersen, J.; Carlsen, T.H.; Prentoe, J.; Ramirez, S.; Jensen, T.B.; Forns, X.; Alter, H.; Foung, S.K.; Law, M.; Gottwein, J.; et al. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology 2013, 58, 1587–1597. [Google Scholar] [CrossRef]
- Li, Y.P.; Ramirez, S.; Humes, D.; Jensen, S.B.; Gottwein, J.M.; Bukh, J. Differential sensitivity of 5’UTR-NS5A recombinants of hepatitis C virus genotypes 1-6 to protease and NS5A inhibitors. Gastroenterology 2014, 146, 812–821. [Google Scholar] [CrossRef]
- Ramirez, S.; Mikkelsen, L.S.; Gottwein, J.M.; Bukh, J. Robust HCV Genotype 3a Infectious Cell Culture System Permits Identification of Escape Variants With Resistance to Sofosbuvir. Gastroenterology 2016, 151, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.V.; Ramirez, S.; Gottwein, J.M.; Fahnoe, U.; Li, Y.P.; Pedersen, J.; Bukh, J. HCV Genotype 6a Escape From and Resistance to Velpatasvir, Pibrentasvir, and Sofosbuvir in Robust Infectious Cell Culture Models. Gastroenterology 2018, 154, 2194–2208. [Google Scholar] [CrossRef]
- Pham, L.V.; Pedersen, M.S.; Fahnøe, U.; Fernandez-Antunez, C.; Humes, D.; Schønning, K.; Ramirez, S.; Bukh, J. HCV genome-wide analysis for development of efficient culture systems and unravelling of antiviral resistance in genotype 4. Gut 2022, 71, 627–642. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Q.; Rong, L.; Rong, D.; Yang, Y.; Hao, J.; Zhang, Z.; Ma, L.; Rao, G.; Zhou, Y.; et al. Development of cell culture infectious clones for hepatitis C virus genotype 1b and transcription analysis of 1b-infected hepatoma cells. Antivir. Res. 2021, 193, 105136. [Google Scholar] [CrossRef]
- Ramirez, S.; Fernandez-Antunez, C.; Mikkelsen, L.S.; Pedersen, J.; Li, Y.P.; Bukh, J. Cell Culture Studies of the Efficacy and Barrier to Resistance of Sofosbuvir-Velpatasvir and Glecaprevir-Pibrentasvir against Hepatitis C Virus Genotypes 2a, 2b, and 2c. Antimicrob. Agents Chemother. 2020, 64, e01888-19. [Google Scholar] [CrossRef]
- Chen, M.; Zheng, F.; Yuan, G.; Duan, X.; Rong, L.; Liu, J.; Feng, S.; Wang, Z.; Wang, M.; Feng, Y.; et al. Development of an Infectious Cell Culture System for Hepatitis C Virus Genotype 6a Clinical Isolate Using a Novel Strategy and Its Sensitivity to Direct-Acting Antivirals. Front. Microbiol. 2018, 9, 2950. [Google Scholar] [CrossRef]
- Chen, M.; Xu, Y.; Li, N.; Yin, P.; Zhou, Q.; Feng, S.; Wu, T.; Wei, L.; Wang, H.; Fu, Y.; et al. Development of full-length cell-culture infectious clone and subgenomic replicon for a genotype 3a isolate of hepatitis C virus. J. Gen. Virol. 2021, 102, 001704. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Ramirez, S.; Jensen, S.B.; Purcell, R.H.; Gottwein, J.M.; Bukh, J. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc. Natl. Acad. Sci. USA 2012, 109, 19757–19762. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Ramirez, S.; Gottwein, J.M.; Scheel, T.K.; Mikkelsen, L.; Purcell, R.H.; Bukh, J. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc. Natl. Acad. Sci. USA 2012, 109, E1101–E1110. [Google Scholar] [CrossRef]
- Li, Y.P.; Ramirez, S.; Mikkelsen, L.; Bukh, J. Efficient infectious cell culture systems of the hepatitis C virus (HCV) prototype strains HCV-1 and H77. J. Virol. 2015, 89, 811–823. [Google Scholar] [CrossRef]
- Ramirez, S.; Li, Y.P.; Jensen, S.B.; Pedersen, J.; Gottwein, J.M.; Bukh, J. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology 2014, 59, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.S.; Meunier, J.C.; Takikawa, S.; Faulk, K.; Engle, R.E.; Bukh, J.; Purcell, R.H.; Emerson, S.U. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. USA 2008, 105, 4370–4375. [Google Scholar] [CrossRef] [PubMed]
- Scheel, T.K.; Gottwein, J.M.; Jensen, T.B.; Prentoe, J.C.; Hoegh, A.M.; Alter, H.J.; Eugen-Olsen, J.; Bukh, J. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. USA 2008, 105, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Fahnøe, U.; Pham, L.V.; Fernandez-Antunez, C.; Costa, R.; Rivera-Rangel, L.R.; Galli, A.; Feng, S.; Mikkelsen, L.S.; Gottwein, J.M.; Scheel, T.K.H.; et al. Versatile SARS-CoV-2 Reverse-Genetics Systems for the Study of Antiviral Resistance and Replication. Viruses 2022, 14, 172. [Google Scholar] [CrossRef]
- Li, Y.P.; Gottwein, J.M.; Scheel, T.K.; Jensen, T.B.; Bukh, J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proc. Natl. Acad. Sci. USA 2011, 108, 4991–4996. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.E.; Wolfisberg, R.; Fahnøe, U.; Sharma, H.; Renshaw, R.W.; Nielsen, L.; Nishiuchi, E.; Holm, C.; Dubovi, E.; Rosenberg, B.R.; et al. Equine pegiviruses cause persistent infection of bone marrow and are not associated with hepatitis. PLoS Pathog. 2020, 16, e1008677. [Google Scholar] [CrossRef]
- Fahnoe, U.; Bukh, J. Full-Length Open Reading Frame Amplification of Hepatitis C Virus. Methods Mol. Biol. 2019, 1911, 85–91. [Google Scholar] [CrossRef]
- Gottwein, J.M.; Scheel, T.K.; Jensen, T.B.; Ghanem, L.; Bukh, J. Differential efficacy of protease inhibitors against HCV genotypes 2a, 3a, 5a, and 6a NS3/4A protease recombinant viruses. Gastroenterology 2011, 141, 1067–1079. [Google Scholar] [CrossRef]
- Prentoe, J.; Bukh, J. In Vitro Neutralization Assay Using Cultured Hepatitis C Virus. Methods Mol. Biol. 2019, 1911, 433–439. [Google Scholar] [CrossRef]
- Meuleman, P.; Vanlandschoot, P.; Leroux-Roels, G. A simple and rapid method to determine the zygosity of uPA-transgenic SCID mice. Biochem. Biophys. Res. Commun. 2003, 308, 375–378. [Google Scholar] [CrossRef]
- Meuleman, P.; Libbrecht, L.; De Vos, R.; de Hemptinne, B.; Gevaert, K.; Vandekerckhove, J.; Roskams, T.; Leroux-Roels, G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 2005, 41, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Maillard, P.; Walic, M.; Meuleman, P.; Roohvand, F.; Huby, T.; Le Goff, W.; Leroux-Roels, G.; Pécheur, E.I.; Budkowska, A. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS ONE 2011, 6, e26637. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, C.; Yusim, K.; Boykin, L.; Richardson, R. The Los Alamos hepatitis C sequence database. Bioinformatics 2005, 21, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Combet, C.; Garnier, N.; Charavay, C.; Grando, D.; Crisan, D.; Lopez, J.; hne-Garcia, A.; Geourjon, C.; Bettler, E.; Hulo, C.; et al. euHCVdb: The European hepatitis C virus database. Nucleic Acids Res. 2007, 35, D363–D366. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, J.M.; Scheel, T.K.; Callendret, B.; Li, Y.P.; Eccleston, H.B.; Engle, R.E.; Govindarajan, S.; Satterfield, W.; Purcell, R.H.; Walker, C.M.; et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): Genetic analyses and in vivo pathogenesis studies. J. Virol. 2010, 84, 5277–5293. [Google Scholar] [CrossRef]
- Kolykhalov, A.A.; Feinstone, S.M.; Rice, C.M. Identification of a highly conserved sequence element at the 3’ terminus of hepatitis C virus genome RNA. J. Virol. 1996, 70, 3363–3371. [Google Scholar] [CrossRef]
- Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin. Infect. Dis. 2018, 67, 1477–1492. [CrossRef]
- Camus, G.; Xu, S.; Han, B.; Lu, J.; Dvory-Sobol, H.; Yu, M.; Cheng, G.; Miller, M.D.; Doehle, B.P.; Mo, H. Establishment of robust HCV genotype 4d, 5a, and 6a replicon systems. Virology 2018, 514, 134–141. [Google Scholar] [CrossRef]
- Schnell, G.; Krishnan, P.; Tripathi, R.; Beyer, J.; Reisch, T.; Irvin, M.; Dekhtyar, T.; Lu, L.; Ng, T.I.; Xie, W.; et al. Hepatitis C virus genetic diversity by geographic region within genotype 1-6 subtypes among patients treated with glecaprevir and pibrentasvir. PLoS ONE 2018, 13, e0205186. [Google Scholar] [CrossRef]
- Prentoe, J.; Velázquez-Moctezuma, R.; Foung, S.K.; Law, M.; Bukh, J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology 2016, 64, 1881–1892. [Google Scholar] [CrossRef]
- Carlsen, T.H.; Pedersen, J.; Prentoe, J.C.; Giang, E.; Keck, Z.Y.; Mikkelsen, L.S.; Law, M.; Foung, S.K.; Bukh, J. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology 2014, 60, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Giang, E.; Dorner, M.; Prentoe, J.C.; Dreux, M.; Evans, M.J.; Bukh, J.; Rice, C.M.; PLoSs, A.; Burton, D.R.; Law, M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. USA 2012, 109, 6205–6210. [Google Scholar] [CrossRef] [PubMed]
- Keck, M.L.; Wrensch, F.; Pierce, B.G.; Baumert, T.F.; Foung, S.K.H. Mapping Determinants of Virus Neutralization and Viral Escape for Rational Design of a Hepatitis C Virus Vaccine. Front. Immunol. 2018, 9, 1194. [Google Scholar] [CrossRef] [PubMed]
- Torrents de la Peña, A.; Sliepen, K.; Eshun-Wilson, L.; Newby, M.L.; Allen, J.D.; Zon, I.; Koekkoek, S.; Chumbe, A.; Crispin, M.; Schinkel, J.; et al. Structure of the hepatitis C virus E1E2 glycoprotein complex. Science 2022, 378, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Xu, C.; Ding, Q.; Li, R.; Xiang, Y.; Chung, J.; Zhong, J. A single point mutation in E2 enhances hepatitis C virus infectivity and alters lipoprotein association of viral particles. Virology 2009, 395, 67–76. [Google Scholar] [CrossRef]
- Augestad, E.H.; Castelli, M.; Clementi, N.; Ströh, L.J.; Krey, T.; Burioni, R.; Mancini, N.; Bukh, J.; Prentoe, J. Global and local envelope protein dynamics of hepatitis C virus determine broad antibody sensitivity. Sci. Adv. 2020, 6, eabb5938. [Google Scholar] [CrossRef]
- Pedersen, M.S.; Fahnøe, U.; Hansen, T.A.; Pedersen, A.G.; Jenssen, H.; Bukh, J.; Schønning, K. A near full-length open reading frame next generation sequencing assay for genotyping and identification of resistance-associated variants in hepatitis C virus. J. Clin. Virol. 2018, 105, 49–56. [Google Scholar] [CrossRef]
- Yamane, D.; McGivern, D.R.; Wauthier, E.; Yi, M.; Madden, V.J.; Welsch, C.; Antes, I.; Wen, Y.; Chugh, P.E.; McGee, C.E.; et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat. Med. 2014, 20, 927–935. [Google Scholar] [CrossRef]
- Asselah, T.; Reesink, H.; Gerstoft, J.; De Ledinghen, V.; Pockros, P.J.; Robertson, M.; Hwang, P.; Asante-Appiah, E.; Wahl, J.; Nguyen, B.; et al. Efficacy of elbasvir and grazoprevir in participants with hepatitis C virus genotype 4 infection: A pooled analysis. Liver Int. 2018, 38, 1583–1591. [Google Scholar] [CrossRef]
- Pham, L.V.; Jensen, S.B.; Fahnoe, U.; Pedersen, M.S.; Tang, Q.; Ghanem, L.; Ramirez, S.; Humes, D.; Serre, S.B.N.; Schonning, K.; et al. HCV genotype 1-6 NS3 residue 80 substitutions impact protease inhibitor activity and promote viral escape. J. Hepatol. 2019, 70, 388–397. [Google Scholar] [CrossRef]
- Shiha, G.; Esmat, G.; Hassany, M.; Soliman, R.; Elbasiony, M.; Fouad, R.; Elsharkawy, A.; Hammad, R.; Abdel-Razek, W.; Zakareya, T.; et al. Ledipasvir/sofosbuvir with or without ribavirin for 8 or 12 weeks for the treatment of HCV genotype 4 infection: Results from a randomised phase III study in Egypt. Gut 2019, 68, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Nishio, A.; Hasan, S.; Park, H.; Park, N.; Salas, J.H.; Salinas, E.; Kardava, L.; Juneau, P.; Frumento, N.; Massaccesi, G.; et al. Serum neutralization activity declines but memory B cells persist after cure of chronic hepatitis C. Nat. Commun. 2022, 13, 5446. [Google Scholar] [CrossRef] [PubMed]


| Primer ID | Sequence 5′-3′ |
|---|---|
| 4d-1320-R | GCAGTTCTGTTGATGTGCCAGCTC |
| 4d-540-R | CTAGTCGCGCGCACACCCAATCTAG |
| 4drX-9584-RT | CATGATCTGCAGAGAGACC |
| 4dr-8530-F | CTCGACACACTCCAGTCAAC |
| 4drX-9562-R | GTTACGGCACTCTCTGCAGTC |
| TS-O-00178 | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTTTTTVN |
| 3UTR-9476-F1 | GGTGGCTCCATCTTAGCCCTAG |
| AUAP | GGCCACGCGTCGACTAGTAC |
| qPCR-forward | AGYGTTGGGTYGCGAAAG |
| qPCR-reverse | CACTCGCAAGCRCCCT |
| qPCR-probe | FAM-CCTTGTGGTACTGCCTGA-MGB |
| Predominant aa in GT4 (100) | Predominant aa in GT4a (24) | Predominant aa in GT4d (11) | Predominant aa in GT4r (6) | DH13 Wildtype | 4d(C5A)-13m [DH13(C5A)-13m] | 4d_Patient 01_11_2016 |
|---|---|---|---|---|---|---|
| L13 | L | M | L | M | M | M |
| S15 | S | G | G | G | G | G |
| V18 | I | I | V | I | I | I |
| V33 | V | I | V | I | I | I |
| V48 | V | I | I | I | I | I |
| A101 | A | S | A | S | S | S |
| M147 | M | L | M | L | L | L |
| A150 | A | V | A | V | V | V |
| V151 | A | A | V | A | A | A |
| T185 | T | S | T | S | S | S |
| Predominant aa in GT4 (100) | Predominant aa in GT4a (24) | Predominant aa in GT4d (11) | Predominant aa in GT4r (6) | DH13 Wildtype | 4d(C5A)-13m [DH13(C5A)-13m] | 4d(C5A)-14m [DH13(C5A)-14m] | 4d_Patient 01_11_2016 |
|---|---|---|---|---|---|---|---|
| S2 | E | R | S | R | R | R | C |
| V8 | V | I | V/I # | I | I | I | I |
| M31 | M | M | L | M | M | V | M |
| E46 | E | V | E | V | V | V | V |
| I67 | I | V | V | V | V | V | V |
| G98 | G | S | G | S | S | S | S |
| D126 | D | E | D | E | E | E | E |
| L168 | L | M | L | M | M | M | M |
| S174 | S | S | S | F | F | F | T |
| S176 | S | T | S | T | T | T | A |
| S181 | S | T | T | T | T | T | T |
| Predominant aa in GT4 (100) | Predominant aa in GT4a (24) | Predominant aa in GT4d (11) | Predominant aa in GT4r (6) | DH13 Wildtype | 4d(C5A)-12m [DH13(C5A)-12m] | 4d(C5A)-13m [DH13(C5A)-13m] | 4d_Patient 01_11_2016 |
|---|---|---|---|---|---|---|---|
| I414 | I | I | I | I | I | T | I |
| N434 | N | N | Q | N | N | N | N |
| L438 | L | I | I | L | L | L | L |
| S440 | S | S | G | S | S | S | S |
| Date of Sampling | History of Infection | HCV RNA Titer IU/mL | NGS Analysis | NAb EC50 Value (Genotype) |
|---|---|---|---|---|
| 29-07-2013 | First infection with genotype 4d detected | 5.48 × 106 | DH13 | 0.0005 (4a) vs. 0.009 (4d) |
| 07-09-2015 | Before DAA grazoprevir/elbasvir treatment | 8.7 × 103 | No PCR products 1 | 0.002 (4a) vs. 0.001 (4d) |
| 01-11-2016 | New infection with genotype 4d detected | 1.9 × 106 | New 4d isolate (Genbank accession number OP555739) 2 | 0.005 (4a) vs. 0.009 (4d) |
| 04-02-2019 | Before re-treatment with DAA grazoprevir/elbasvir | 3.7 × 107 | New 4d isolate (Genbank accession number OP555740) 2 | 0.009 (4a) vs. 0.004 (4d) |
| 01-03-2021 | Post-treatment | 0 | 0.009 (4a) vs. 0.002 (4d) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, L.V.; Velázquez-Moctezuma, R.; Fahnøe, U.; Collignon, L.; Bajpai, P.; Sølund, C.; Weis, N.; Holmbeck, K.; Prentoe, J.; Bukh, J. Novel HCV Genotype 4d Infectious Systems and Assessment of Direct-Acting Antivirals and Antibody Neutralization. Viruses 2022, 14, 2527. https://doi.org/10.3390/v14112527
Pham LV, Velázquez-Moctezuma R, Fahnøe U, Collignon L, Bajpai P, Sølund C, Weis N, Holmbeck K, Prentoe J, Bukh J. Novel HCV Genotype 4d Infectious Systems and Assessment of Direct-Acting Antivirals and Antibody Neutralization. Viruses. 2022; 14(11):2527. https://doi.org/10.3390/v14112527
Chicago/Turabian StylePham, Long V., Rodrigo Velázquez-Moctezuma, Ulrik Fahnøe, Laura Collignon, Priyanka Bajpai, Christina Sølund, Nina Weis, Kenn Holmbeck, Jannick Prentoe, and Jens Bukh. 2022. "Novel HCV Genotype 4d Infectious Systems and Assessment of Direct-Acting Antivirals and Antibody Neutralization" Viruses 14, no. 11: 2527. https://doi.org/10.3390/v14112527
APA StylePham, L. V., Velázquez-Moctezuma, R., Fahnøe, U., Collignon, L., Bajpai, P., Sølund, C., Weis, N., Holmbeck, K., Prentoe, J., & Bukh, J. (2022). Novel HCV Genotype 4d Infectious Systems and Assessment of Direct-Acting Antivirals and Antibody Neutralization. Viruses, 14(11), 2527. https://doi.org/10.3390/v14112527






