Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe respiratory illness in humans; the second-largest and most deadly outbreak to date occurred in Saudi Arabia. The dromedary camel is considered a possible host of the virus and also to act as a reservoir, transmitting the virus to humans. Here, we studied evolutionary relationships for 31 complete genomes of betacoronaviruses, including eight newly sequenced MERS-CoV genomes isolated from dromedary camels in Saudi Arabia. Through bioinformatics tools, we also used available sequences and 3D structure of MERS-CoV spike glycoprotein to predict MERS-CoV epitopes and assess antibody binding affinity. Phylogenetic analysis showed the eight new sequences have close relationships with existing strains detected in camels and humans in Arabian Gulf countries. The 2019-nCov strain appears to have higher homology to both bat coronavirus and SARS-CoV than to MERS-CoV strains. The spike protein tree exhibited clustering of MERS-CoV sequences similar to the complete genome tree, except for one sequence from Qatar (KF961222). B cell epitope analysis determined that the MERS-CoV spike protein has 24 total discontinuous regions from which just six epitopes were selected with score values of >80%. Our results suggest that the virus circulates by way of camels crossing the borders of Arabian Gulf countries. This study contributes to finding more effective vaccines in order to provide long-term protection against MERS-CoV and identifying neutralizing antibodies.
1. Introduction
Coronaviruses are a family of single-stranded, positive-sense RNA genomes that range from 26,000 to 32,000 basepairs in length. This family can infect both humans and animals, and has been found in many mammal hosts, including bats, camels, and pigs [1]. Coronaviruses are also known to have high genetic diversity influenced by mutation and recombination, which can lead to the development of new viruses. Before 2003, human coronaviruses were primarily known for causing common colds. In 2003, severe acute respiratory syndrome coronavirus (SARS-CoV) was first diagnosed in southern China as an epidemic, and went on to infect almost 8000 people in 29 countries, with a fatality rate of approximately 10% [2,3]. Almost ten years later, in 2012, another novel and deadly human coronavirus emerged, named the Middle East respiratory syndrome coronavirus (MERS-CoV); it infected more than 1700 people and exhibited a high mortality rate of ∼36% [4,5]. On 30 December 2019, a third coronavirus outbreak emerged in Wuhan, China, designated as the 2019 novel coronavirus (2019-nCoV), which is still ongoing and has now spread over 213 countries, becoming a pandemic. According to the daily report of the World Health Organization (WHO) on 17 April 2020, 1,995,983 confirmed cases and 131,037 deaths worldwide are attributed to the pandemic so far. These novel betacoronaviruses are global threats with the potential for further outbreaks that cause substantial health and economic consequences. Better understanding of their evolutionary history is necessary in order to determine their origin and spread and to effectively design interventions and neutralizing antibodies.
Of the betacoronaviruses that have caused major outbreaks to date, MERS-CoV is notable for having the highest mortality rate and for being centered in the Middle East. It was first detected in Saudi Arabia in 2012, in a patient with a fatal respiratory infection [6]. The viral respiratory illness caused by MERS-CoV is known as Middle East respiratory syndrome (MERS), and its symptoms range from fever, cough, and shortness of breath to severe illness and respiratory failure. The symptoms typically develop within 2–14 days, but some patients do not demonstrate any clinical symptoms despite testing positive for MERS-CoV in a laboratory assay. Severe symptoms are more likely to appear in older people, those with weakened immune systems, and those with chronic diseases such as cancer or diabetes. Since the initial identification of MERS, Saudi Arabia has continued to report the largest number of cases. As of February 2019, 1983 cases and 783 associated deaths have been detected in Saudi Arabia, producing a case fatality rate of 35.7% [7].
The exact origin of MERS-CoV is still uncertain, and further studies remain necessary to confirm the source of its transmission to humans [8]. Notably, Arabian camels (Camelus dromedarius) from Saudi Arabia and other Gulf countries are found to be seropositive for MERS-CoV at high rates [9,10]. Furthermore, a retrospective study carried out on archived sera of dromedary camels from 1983 to 1997 revealed neutralizing antibodies whose presence implies a long-term circulation of MERS-CoV among Arabian camels [11]. These camels are thought to be an intermediate host for the virus, with its ancestral origin being associated with African bats [12,13]. Furthermore, data from many investigations about the MERS-CoV outbreak suggests that camels could be a source of human infections [14,15,16]. C. dromedarius is a member of the camelid family, plays a significant role in transportation, is a source of essential products such as meat and milk [17], and is overall a central part of the heritage and way of life of most residents of Arabian Peninsula countries [18]. Although many MERS-CoV patients had no direct contact with camels [19,20], an indirect path of infection has been proposed through camel workers, who have direct and prolonged contact. Additionally, travelers who visit the Middle East are vulnerable to infection and may seed the virus in other regions, including Europe, North America, and Southeast Asia [21]; for example, phylogenetic analysis of a South Korean outbreak in 2015 traced its evolutionary history to Saudi Arabian strains [22]. Accordingly, in-depth genetic analysis of strains within Saudi Arabia would provide an essential reference and resource in the face of further outbreaks. Of particular research interest is the MERS-CoV spike protein, which enables virus entry into host cells via binding to cellular receptor dipeptidyl peptidase-4 (DPP4), and therefore is considered a key target for vaccine design [23].
This study aimed to analyze the evolutionary history of eight newly sequenced MERS-CoV isolated from dromedary camels in Saudi Arabia, along with representative members of the genus Betacoronavirus, based on both complete genomes and spike protein sequences. We applied the time-framed Bayesian evolution analysis approach implemented in BEAST to determine the evolutionary relationships between these sequences. We also utilized the Immune Epitope Database and Analysis Resource (IEDB) to collect known epitope sites from other betacoronaviruses, map equivalent protein sites in MERS-CoV, and predict likely epitopes. This study provides insights into the origin of MERS-CoV and likely means of transmission through Gulf countries.
2. Results
2.1. Genome Organization
The consensus MERS-CoV genome obtained through analyzing isolates from eight infected camels from different locations across Saudi Arabia was 30,118 nt in length with a GC content of 41%. The eight constituent genomes shared >99% identity. The camel MERS-CoV genome is similar to that of human MERS-CoV, containing ten ORFs (ORF1ab, ORF3, spike [S], ORF5, ORF4a, ORF4b, envelope [E], membrane [M], nucleocapsid [N], and ORF8b). BLAST comparison to other camel and human MERS-CoV strains available from NCBI revealed that all share ∼99% identity with one another. The comparative analysis of homologous sequences of spike proteins and genes has been performed for the 31 Betacoronavirus strains (Table 1). The MERS-CoV strains were nearly identical across their spike proteins and genes (>99.50% and >81%, respectively). Indicative of a very recent emergence into the human population.

Table 1.
List of viruses from genus Betacoronavirus used in this study showing their S protein and S gene homologous identities.
2.2. Phylogenetic Analysis
Phylogenetic analysis was performed on the complete genomes and spike proteins of the eight new MERS-CoV sequences and 23 representative viruses from the genus Betacoronavirus. The eight camel-derived MERS-CoV sequences clustered in a distinct clade and were all relatively close to other MERS-CoV sequences from human and camel hosts (Figure 1). The 2019-nCoV falls in a distinct clade and shows a closer relationship to Bat-SARS-Like (GU190215.1) than the MERS-CoV strains. In agreement with the complete genome results, analysis based on the spike proteins showed that 2019-nCoV has a closer relationship to Bat-SARS-Like and SARS-CoV strains than MERS-CoV strains (Figure 2).
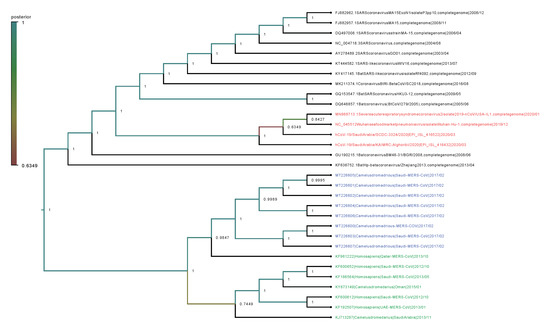
Figure 1.
Phylogenetic analysis of the complete genomes of our eight MERS-CoV isolates and representative viruses of the genus Betacoronavirus. The new eight isolates are in blue, other MERS-CoV strains in green, and 2019-nCoV in red. The numbers at nodes represent the posterior probabilities of their clustering.
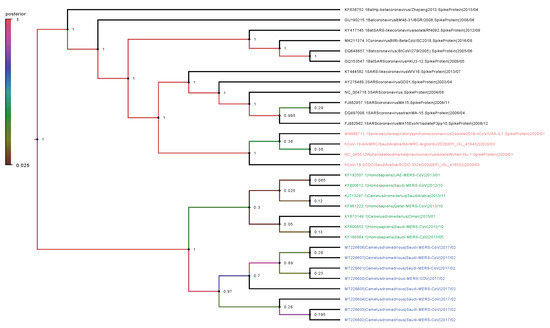
Figure 2.
Phylogenetic analysis of spike proteins of our eight MERS-CoV isolates and representative viruses of the genus Betacoronavirus. The new eight isolates are in blue, other MERS-CoV strains in green, and 2019-nCoV in red. The numbers at nodes represent the posterior probabilities of their clustering.
We observed very high conservation between the new eight sequenced MERS-CoV genomes, with sequence identity >99%, despite the different geographical locations of sampling. In addition, a specific sample collected in 2015 from a human in Qatar was revealed to be more closely related to the eight new MERS-CoV isolates than to other UAE and Oman MERS-CoV strains. Genetically, the MERS-CoV genome can overall be considered more diverse than those of other betacoronavirus family members. Phylogenetic analysis of the MERS-CoV spike protein and its counterparts showed that the four clades formed four well-supported branches (Figure 2).
2.3. MERS-CoV B Cell Epitope Analysis
B cell epitopes are significant in the defense of the immune system against viral diseases, allowing B cells to identify different viral infections and activate responses against them. There has been many studies aiming to design MERS-Cov vaccine [24,25,26]. In this study, epitope analysis of MERS-CoV showed that the MERS-CoV spike protein has 24 discontinuous regions in total, from which just six epitopes with score values of >0.8 could be selected. The higher the score, the greater the potential of a true discontinuous epitope; the highest probability obtained was 0.898. Table 2 details the peptides of these high-scoring discontinuous epitopes and their sequence locations, epitope lengths, and associated scores. Their positions on the 3D structure of the MERS-CoV spike protein (PDB ID: 5X5F) are illustrated in Figure 3.

Table 2.
Summary of predicted B cell epitopes present on the outer surface of the MERS-CoV, 2019-nCov, and SARS-CoV spike proteins.
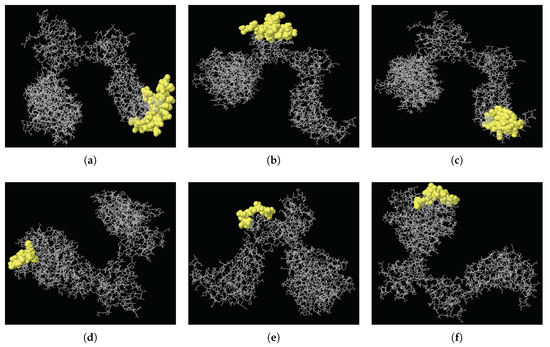
Figure 3.
Three-dimensional locations of discontinuous epitopes (a–f) predicted in the MERS-CoV spike protein. The epitopes are shown in yellow, with the bulk of the MERS-CoV spike protein as grey sticks.
3. Discussion
MERS-CoV is a zoonotic virus for which ongoing studies indicate major roles in transmission for direct contact with live camels and with humans having symptomatic MERS [27,28,29], with dromedary camels being considered intermediate hosts that are the main source of human infections. The second largest outbreak to date occurred in Saudi Arabia, and MERS-CoV is widespread among humans and camels in the Gulf countries [30], where it remains a potential hazard to regional and global health and welfare. The present study seeks to determine the evolutionary relationships between eight new MERS-CoV isolates obtained from Arabian camels in different regions of Saudi Arabia and 23 other previously sequenced betacoronavirus strains, based on phylogenetic trees constructed with BEAST for the complete viral genome and the spike protein.
In the phylogenetic analysis, the eight new strains clustered in a separate clade, exhibiting close relationships with strains obtained from MERS-CoV-infected camels and humans in Saudi Arabia, the United Arab Emirates (UAE), Qatar, and Oman. This finding supports the expectation that the virus circulates via camels crossing borders with Oman and other Gulf countries, and is also consistent with the report of another study that movement of camels between Saudi Arabia, the UAE, and Oman contributes to the spread of the virus throughout the Arabian Peninsula [31]. Based on these results, it is recommended that restrictions be implemented to prevent these animals from migrating between Gulf countries.
Within the Betacoronavirus genus, SARS-CoV and MERS-CoV are distinct from 2019-nCoV [32]; the 2019-nCoV genome and spike glycoprotein, respectively, showed 48.83% and 41.95% sequence identity with human-derived MERS-CoV [33]. The 2019-nCoV virus was found to be most closely related to Bat-SARS-Like [1]. This confirms the identification of bats as reservoirs for coronaviruses, including MERS-CoV and 2019-nCoV.
In conclusion, this study determined the evolutionary relationships of newly sequenced MERS-CoV, along with 23 previously sequenced isolates of the genus Betacoronavirus. We also performed genomic characterization of MERS-CoV, highlighting its origin and likely means of transmission through Gulf countries. In addition, we analyzed the MERS-CoV spike protein for suitable epitopic candidates and identified six viral peptides with good antigen presentation scores (>80%).
4. Materials and Methods
4.1. Sample Collection
The samples were collected from 30 dromedary camels at different locations in Saudi Arabia, following the guidelines of the Food and Agriculture Organization of the United Nations. The camel was appropriately restrained, and the camel nasal wing was then held and raised by the thumb and index finger, followed by gentle insertion of the nasal swab by the upper half of the nasal vestibule as deep as possible until the swab is handled. The swab was then laterally rotated, transferred into a tube containing virus transport media, and kept at 4 °C in preparation for PCR screening.
4.2. RNA Extraction
Total RNA was successfully extracted from eight samples of camel nasal fluids using a MagMax-96 viral RNA extraction kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s protocol. The final DNAase-treated RNA product was stored at −80 °C.
4.3. Sequence Preparation
Eight complete genomes of MERS-CoV from C. dromedarius have been sequenced in our lab. The samples came from multiple geographical locations in Saudi Arabia: four from Jeddah city, one from Northern Saudi Arabia, two from Mecca city, and one from Riyadh city (Figure 4). For the sake of comparison, we also retrieved 21 complete genomes of betacoronavirus strains from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/Database/nph-select2.cgi), along with those of two 2019-nCoV strains isolated in Saudi Arabia (https://www.gisaid.org). In total, 31 betacoronavirus sequences were analyzed (Table 3).
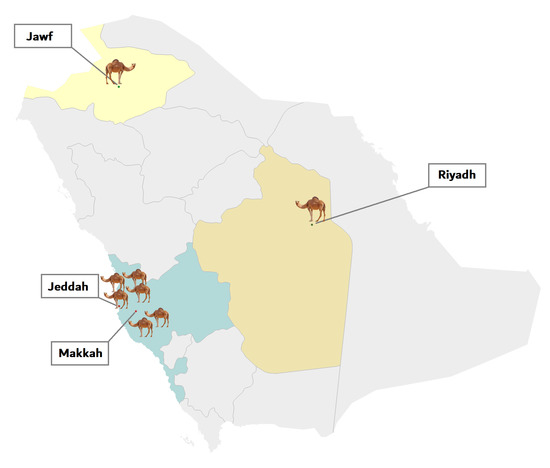
Figure 4.
Geographical distribution of dromedaries in Saudi Arabia infected with MERS-CoV viruses that were isolated in the present study. The number of dromedaries corresponds to the number of isolates from the sampling location.

Table 3.
List of viruses from genus Betacoronavirus with sequences used in this study.
4.4. RNA Quantification and cDNA Conversion
The concentration of each RNA sample was measured on a Qubit 3.0 using the Qubit RNA BR assay kit (Thermo Fisher Scientific, Waltham, MA, USA) and reverse transcribed with a SuperScript II cDNA kit (Thermo Fisher Scientific, Waltham, MA, USA). A minimum RNA concentration of 100 ng/uL was used for cDNA conversion.
4.5. Library Preparation and Sequencing
High-quality cDNA was subjected to Ampliseq library preparation as per the manufacturer’s protocol using MERS Ampliseq Panels (Thermo Fisher Scientific, Waltham, MA, USA) with a mean insert size of 200 bp. Each library was assigned to a distinct barcode using the Ion Xpress Barcode Adapters 1–16 kit and purified using Agencourt AMpure XP beads (Beckman Coulter, Brea, CA, USA). Purified libraries were efficiently quantified on a StepOnePlus Real-Time PCR system using an Ion Library TaqMan quantification kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s protocol. Libraries were normalized to 25 pM, and template-positive Ion Sphere particles (ISPs) were obtained by pooling all ten normalized libraries and performing clonal amplification on a OneTouch 2 system using the Ion PGM HiQ OT2 200 kit (Thermo Fisher Scientific, Waltham, MA, USA). Template-positive ISPs were then loaded on Ion 318 chips and sequenced on an Ion PGM instrument using the Ion PGM HiQ Sequencing kit (Thermo Fisher Scientific, Waltham, MA, USA). The resulting data were preprocessed (base calling, base quality recalibration, alignment and consensus sequence assembly) using Torrent Suite Server v 5.6. Ninety percent of the consensus sequences were obtained from this data; the remaining gaps were filled by Sanger sequencing on a 3730xl Genetic Analyzer using custom-designed primers (Macrogen LLC, Seoul, Korea) and BigDye Terminator v 3.1 Cycle Sequencing chemistry (Thermo Fisher Scientific, Waltham, MA, USA).
4.6. Gap Filling and Sanger Sequencing
To fill the gaps that remained after mapping Ion reads, we designed 11 primer sets (Table 4) against the NCBI MERS-CoV reference genome assembly (NC_019843.3). Primers were designed and checked using Primer Express software (v3.0) (Applied Biosystems, Foster City, CA, USA) and NetPrimer (Premier Biosoft, Palo Alto, CA, USA), with gap flanking regions incorporated into the primers. Sequences for all the major gaps were amplified by PCR using PlatinumTM Hot Start PCR Master Mix 2X (Thermo Fisher Scientific, Waltham, MA, USA). PCR products were sequenced using the BigDye® Terminator Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 3730xl DNA Analyzer. Most minor gaps were sample-specific; to ensure complete genome coverage for phylogenetic analysis, these gaps were closed manually after performing multiple sequence alignment (MSA) on the fully-sequenced genomes. MSA was carried out using Geneious software [34].

Table 4.
Primer sets used for filling sequence gaps.
4.7. Phylogenetic Trees and Evolutionary Dynamics
We first performed multiple sequence alignment of the 31 strains using MAFFT (v7) [35]. Next, we estimated the phylogenetic relationships using a time-framed Bayesian evolution analysis approach via a Markov Chain Monte Carlo (MCMC) inference method; this approach was implemented in the BEAST package (v1.8.2) [36]. For analysis of whole genomes, we used the GTR+I substitution model, a strict clock model, and a constant-size tree prior. For the spike protein dataset, we used the Blosum62+Gamma substitution model, a strict clock model, and a constant-size tree prior. MCMC analyses were run for 20 million iterations, sampling every 100 thousand iterations after a 10% burn-in. We assessed each run’s convergence using Tracer (v1.6) [37]. The BEAST package TreeAnnotator was utilized to summarize a maximum clade credibility (MCC) tree for every dataset using MCMC tree samples. Phylogenetic clustering patterns were visualized and analyzed with FigTree (v1.4.3) (http://tree.bio.ed.ac.uk/software/figtree/).
4.8. Prediction of MERS-CoV B Cell Epitopes
The IEDB [38] and BCPRED [39] servers were utilized to predict B cell epitopes of the MERS-CoV spike protein. IEDB software can predict both linear and discontinuous epitopes using a protein 3D structure (PDB format) as input. The software associates each epitope with its score, which is defined as a Protrusion Index (PI) method. For example, score value 0.8 means the predicted epitope would include within 80% of the protein amino acids and 20% of the protein amino acids will be outside of the epitope. Only epitopes that were located on the outer surface of the protein were chosen. Six residues in length was regarded as adequate to prompt a defensive immune reaction. We focused on discontinuous antigenic epitopes as they are increasingly explicit and have higher dominant attributes over linear epitopes [40,41].
Author Contributions
Conceptualization, B.A.A. and M.M.M.; Data curation, B.A.A., M.M.M. and M.A.A. (Manal A. Alshehri); Formal analysis, B.A.A., M.M.M. and S.N.A.; Funding acquisition, M.B.A.-F.; Investigation, B.A.A. and M.M.M.; Methodology, B.A.A., M.M.M., S.N.A. and M.A.A. (Mussad A. Altammami); Project administration, B.A.A., M.M.M. and M.B.A.-F.; Resources, B.A.A., M.M.M., M.S.N. and M.A.B.; Software, B.A.A. and S.N.A.; Supervision, B.A.A. and M.M.M.; Validation, B.A.A., M.M.M. and S.N.A.; Visualization, B.A.A., M.M.M. and S.N.A.; Writing—original draft, B.A.A., M.M.M. and S.N.A.; Writing—review & editing, B.A.A. and M.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Life Science and Environment Research Institute (grant 20-0098), King Abdulaziz City for Science and Technology, Saudi Arabia.
Acknowledgments
The authors would like to thank Philippe Lemey at the Rega Institute for Medical Research, Catholic University of Leuven, for his valuable recommendations on this project; and Amer S. Alharthi at the General Directorate for Research and Innovation, King Abdulaziz City for Science and Technology, for his technical support. The authors would also like to thank the technicians at the Integrated Gulf Biosystems for their logistic and technical support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Miłek, J.; Blicharz-Domańska, K. Coronaviruses in avian species–review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, X.; Irwin, D.M.; Shen, Y. Emergence of SARS-like coronavirus poses new challenge in China. J. Infect. 2020, 80, 350–371. [Google Scholar] [CrossRef]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Séron, K.; Labitt, R.N.; Danneels, A.; Palmer, K.E.; Whittaker, G.R.; Dubuisson, J.; Belouzard, S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016, 133, 1–8. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- World Health Organization. Middle East Respiratory Syndrome Coronavirus (MERS-CoV). 2018. Available online: http://www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov) (accessed on 28 August 2018).
- Hemida, M.G.; Al-Naeem, A.; Perera, R.A.; Chin, A.W.; Poon, L.L.; Peiris, M. Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg. Infect. Dis. 2015, 21, 699. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G.; Chu, D.K.; Poon, L.L.; Perera, R.A.; Alhammadi, M.A.; Ng, H.Y.; Siu, L.Y.; Guan, Y.; Alnaeem, A.; Peiris, M. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014, 20, 1231. [Google Scholar] [CrossRef]
- Perera, R.; Wang, P.; Gomaa, M.; El-Shesheny, R.; Kandeil, A.; Bagato, O.; Siu, L.; Shehata, M.; Kayed, A.; Moatasim, Y.; et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Eurosurveillance 2013, 18, 20574. [Google Scholar] [CrossRef]
- Müller, M.A.; Corman, V.M.; Jores, J.; Meyer, B.; Younan, M.; Liljander, A.; Bosch, B.J.; Lattwein, E.; Hilali, M.; Musa, B.E.; et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg. Infect. Dis. 2014, 20, 2093. [Google Scholar] [CrossRef]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Munster, V.J. Animal models of Middle East respiratory syndrome coronavirus infection. Antivir. Res. 2015, 122, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Cotten, M.; Meyer, B.; Watson, S.J.; Alsahafi, A.J.; Al Rabeeah, A.A.; Corman, V.M.; Sieberg, A.; Makhdoom, H.Q.; Assiri, A.; et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg. Infect. Dis. 2014, 20, 1012. [Google Scholar] [CrossRef] [PubMed]
- Alagaili, A.N.; Briese, T.; Mishra, N.; Kapoor, V.; Sameroff, S.C.; de Wit, E.; Munster, V.J.; Hensley, L.E.; Zalmout, I.S.; Kapoor, A.; et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio 2014, 5, e00884-14. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Wu, H.; Guang, X.; Al-Fageeh, M.B.; Cao, J.; Pan, S.; Zhou, H.; Zhang, L.; Abutarboush, M.H.; Xing, Y.; Xie, Z.; et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat. Commun. 2014, 5, 5188. [Google Scholar] [CrossRef]
- Fowler, M. Medicine and Surgery of Camelids; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Conzade, R.; Grant, R.; Malik, M.; Elkholy, A.; Elhakim, M.; Samhouri, D.; Ben Embarek, P.; Van Kerkhove, M. Reported direct and indirect contact with dromedary camels among laboratory-confirmed MERS-CoV cases. Viruses 2018, 10, 425. [Google Scholar] [CrossRef]
- Cauchemez, S.; Nouvellet, P.; Cori, A.; Jombart, T.; Garske, T.; Clapham, H.; Moore, S.; Mills, H.L.; Salje, H.; Collins, C.; et al. Unraveling the drivers of MERS-CoV transmission. Proc. Natl. Acad. Sci. USA 2016, 113, 9081–9086. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Memish, Z.A. Middle East respiratory syndrome coronavirus: Transmission and phylogenetic evolution. Trends Microbiol. 2014, 22, 573–579. [Google Scholar] [CrossRef]
- Kim, J.I.; Kim, Y.J.; Lemey, P.; Lee, I.; Park, S.; Bae, J.Y.; Kim, D.; Kim, H.; Jang, S.I.; Yang, J.S.; et al. The recent ancestry of Middle East respiratory syndrome coronavirus in Korea has been shaped by recombination. Sci. Rep. 2016, 6, 18825. [Google Scholar] [CrossRef]
- Du, L.; Yang, Y.; Zhou, Y.; Lu, L.; Li, F.; Jiang, S. MERS-CoV spike protein: A key target for antivirals. Expert Opin. Ther. Targets 2017, 21, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ul Qamar, M.T.; Saleem, S.; Ashfaq, U.A.; Bari, A.; Anwar, F.; Alqahtani, S. Epitope-based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: An immune-informatics study. J. Transl. Med. 2019, 17, 362. [Google Scholar] [CrossRef] [PubMed]
- Goo, J.; Jeong, Y.; Park, Y.S.; Yang, E.; Jung, D.I.; Rho, S.; Park, U.; Sung, H.; Park, P.G.; Choi, J.A.; et al. Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Res. 2020, 197863. [Google Scholar] [CrossRef] [PubMed]
- Park, B.K.; Lee, S.I.; Bae, J.Y.; Park, M.S.; Lee, Y.; Kwon, H.J. Production of a Monoclonal Antibody targeting the M protein of MERS-CoV for detection of MERS-CoV using a synthetic peptide epitope formulated with a CpG–DNA–liposome complex. Int. J. Pept. Res. Ther. 2019, 25, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Paden, C.; Yusof, M.; Al Hammadi, Z.; Queen, K.; Tao, Y.; Eltahir, Y.; Elsayed, E.; Marzoug, B.; Bensalah, O.; Khalafalla, A.; et al. Zoonotic origin and transmission of Middle East respiratory syndrome coronavirus in the UAE. Zoonoses Public Health 2018, 65, 322–333. [Google Scholar] [CrossRef]
- Wernery, U.; Lau, S.K.; Woo, P.C. Middle East respiratory syndrome (MERS) coronavirus and dromedaries. Vet. J. 2017, 220, 75–79. [Google Scholar] [CrossRef]
- Killerby, M.E.; Biggs, H.M.; Midgley, C.M.; Gerber, S.I.; Watson, J.T. Middle East respiratory syndrome coronavirus transmission. Emerg. Infect. Dis. 2020, 26, 191. [Google Scholar] [CrossRef]
- Nowotny, N.; Kolodziejek, J. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels, Oman, 2013. Eurosurveillance 2014, 19, 20781. [Google Scholar] [CrossRef]
- Yusof, M.F.; Eltahir, Y.M.; Serhan, W.S.; Hashem, F.M.; Elsayed, E.A.; Marzoug, B.A.; Abdelazim, A.S.; Bensalah, O.K.A.; Al Muhairi, S.S. Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi emirate, United Arab Emirates. Virus Genes 2015, 50, 509–513. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020. [Google Scholar] [CrossRef]
- Hemida, M.G.; Abduallah, M.M.B. The SARS-CoV-2 outbreak from a one health perspective. One Health 2020, 100127. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.; Xie, D.; Drummond, A. Tracer v1. 6. Available online: http://tree.bio.ed.ac.uk/software/tracer (accessed on 26 May 2020).
- Peters, B.; Sidney, J.; Bourne, P.; Bui, H.H.; Buus, S.; Doh, G.; Fleri, W.; Kronenberg, M.; Kubo, R.; Lund, O.; et al. The immune epitope database and analysis resource: From vision to blueprint. PLoS Biol. 2005, 3, e91. [Google Scholar] [CrossRef] [PubMed]
- EL-Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. Interdiscip. J. 2008, 21, 243–255. [Google Scholar] [CrossRef]
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. Conformational B-cell epitope prediction on antigen protein structures: A review of current algorithms and comparison with common binding site prediction methods. PLoS ONE 2013, 8, e62249. [Google Scholar] [CrossRef]
- Haste Andersen, P.; Nielsen, M.; Lund, O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006, 15, 2558–2567. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).