Abstract
Members of Picornaviridae and of the Hepacivirus, Pegivirus and Pestivirus genera of Flaviviridae all contain an internal ribosomal entry site (IRES) in the 5′-untranslated region (5′UTR) of their genomes. Each class of IRES has a conserved structure and promotes 5′-end-independent initiation of translation by a different mechanism. Picornavirus 5′UTRs, including the IRES, evolve independently of other parts of the genome and can move between genomes, most commonly by intratypic recombination. We review accumulating evidence that IRESs are genetic entities that can also move between members of different genera and even between families. Type IV IRESs, first identified in the Hepacivirus genus, have subsequently been identified in over 25 genera of Picornaviridae, juxtaposed against diverse coding sequences. In several genera, members have either type IV IRES or an IRES of type I, II or III. Similarly, in the genus Pegivirus, members contain either a type IV IRES or an unrelated type; both classes of IRES also occur in members of the genus Hepacivirus. IRESs utilize different mechanisms, have different factor requirements and contain determinants of viral growth, pathogenesis and cell type specificity. Their dissemination between viruses by horizontal gene transfer has unexpectedly emerged as an important facet of viral evolution.
Keywords:
IRES; Flavivirus; Horizontal gene transfer; Hepacivirus; Pegivirus; Pestivirus; Picornavirus; recombination; translation 1. Recombination as a Motive Force in Viral Evolution
Recombination, defined as the exchange of genetic elements between parental genomes to generate novel chimeras, occurs in divers positive-sense RNA viruses [1]. It can involve replicative or non-replicative mechanisms [2,3] and potentially serves functions that include the purging of deleterious mutations and the creation of advantageous novel genetic combinations to evade host immunity, gain resistance to antiviral agents, alter virulence and expand the host range [1,4,5]. Molecular epidemiological studies, for example, of circulating enteroviruses, have shown that recombination is a frequent occurrence in picornaviruses (e.g., [6,7,8]), and analysis of viral metagenomic data has emphasized the importance of recombination in the acquisition of novel sequences and, consequently, in the evolution of viruses [9,10]. In contrast to nucleotide substitution, which only allows gradual searching through evolutionary fitness space, recombination can lead to large shifts that can create beneficial genetic diversity but may also disrupt favorable combinations of co-adapted alleles [1,11]. This process of movement of genetic information between genomes is known as horizontal gene transfer, and it contrasts with the ‘vertical’ replicative transmission of genetic information from parents to offspring.
Studies of gene acquisition and dissemination by recombination have focused on protein-coding regions (e.g., [9,12,13,14]. For example, recombination between structural genes has influenced the host range expansion of coronaviruses by altering receptor usage [15] and the insertion of a cysteine protease from porcine torovirus (order Nidovirales) into the polyprotein encoded by enterovirus G (order Picornavirales) potentially antagonizes innate immune responses [16].
Less attention has been paid to the acquisition by RNA virus genomes of noncoding elements by horizontal gene transfer, even though such entities are heritable, genetically stable, and have specific functions in key aspects of the viral life cycle that include translation, replication and encapsidation [17,18,19]. Noncoding elements can determine viral host range, cell type specificity and pathogenesis (e.g., [20,21,22,23]. For example, internal ribosomal entry sites (IRESs) are large, structurally complex RNAs that mediate cap- and 5′-end-independent initiation of translation [24,25] and that confer selective advantages on viral mRNAs, particularly during infection. Here, we will discuss IRESs as an example of a class of noncoding RNA element that can move between the genomes of positive-strand RNA viruses by recombination.
2. Viral Infection and Translational Control
With the exception of ‘giant viruses’, viruses do not encode translation factors, and are thus wholly reliant on the host translational apparatus for synthesis of viral proteins [25]. Viral infection leads to competition between viral and cellular mRNAs for access to the protein synthetic machinery, and viruses and their hosts have consequently developed measures and counter measures to promote translation of their own mRNAs.
Translation consists of initiation, elongation, termination and ribosome recycling phases [24]. The canonical initiation process begins with recycling of post-termination ribosomes to generate a pool of free 40S and 60S ribosomal subunits [26]. Aminoacylated initiator tRNA, eukaryotic initiation factor 2 (eIF2) and GTP form a ternary complex (eIF2-TC), which is recruited with eIF1, eIF1A, eIF3 and eIF5 to a 40S subunit, yielding a 43S pre-initiation complex. Eukaryotic mRNAs are post-transcriptionally modified by the addition of a triphosphate-linked 7-methylguanosine (m7GTP) cap at their 5′-termini, and one or two cap-adjacent nucleotides may also be 2′-O-methylated. This ‘cap’ is bound by eIF4F, a heterotrimer comprising the cap-binding protein eIF4E, eIF4G and eIF4A, a RNA helicase. eIF4F recruits the 43S complex to the 5′-end of the mRNA [27], and it then scans in a 5′-3′ direction along the mRNA until it encounters an AUG initiation codon. Establishment of base pairing between the initiation codon and the anticodon of initiator tRNA in the resulting 48S complex triggers eIF5-induced hydrolysis of eIF2-bound GTP, followed by eIF5B-mediated reorientation of tRNA, joining of a 60S subunit and release of initiation factors to form a 80S ribosome that is primed to begin elongation. The elongation process consists of the coordinated decoding of the open reading frame (ORF) and synthesis of the encoded polypeptide.
Cells counter viral infection by activation of innate immune responses, which include synthesis of ‘Interferon-induced protein with tetratricopeptide repeats’ (IFIT) proteins that sequester the 5′-terminal cap of viral mRNAs that lack 2′-O-methylated cap-proximal nucleotides (e.g., [28]), and phosphorylation of the eIF2a subunit by protein kinase R [25]. Viruses may suppress these responses, and in addition, may inhibit cellular translation by abrogating or altering the activity of initiation factors; for example, as a result of their cleavage by virus-encoded proteases (e.g., [29]) or by activation of translational repressors [30]. Viral mRNAs continue to gain access to the translation apparatus in these circumstances by using alternative mechanisms of initiation that require only a subset of initiation factors. Many positive-sense RNA viruses that induce changes in the cellular translation apparatus contain an IRES that promotes cap- and 5′-end-independent initiation of translation. IRESs function by interacting in a specific, non-canonical manner with one or more components of the canonical translation apparatus, leading to recruitment of ribosomes or ribosomal pre-initiation complexes to an internal location on the mRNA [25]. The presence of an IRES in viral mRNAs enables them to escape IFIT-mediated impairment of initiation, and to function despite virus-induced inactivation of one or more initiation factors, and in some instances, despite phosphorylation of eIF2.
3. Classification of Viral IRESs
IRESs occur in a broad range of positive-sense RNA viral genomes, including those of members of Picornaviridae, Flaviviridae, Dicistroviridae and Iflaviridae families, as well as in several retroviruses [19,25,31]. IRESs are classified into a few major groups, each of which has a distinct structural core that is maintained by sequence co-variation [32], and each of which uses a distinct mechanism to initiate translation.
Type I and Type II IRESs are epitomized by poliovirus (PV) (of the genus Enterovirus of Picornaviridae) and encephalomyocarditis virus (EMCV) (of the genus Cardiovirus of Picornaviridae), respectively [25]. These ~450 nt long IRESs have distinct conserved structures (Figure 1A and Figure 2A) but share some functionally important sequence motifs, such as an apical tetraloop in the largest domain and a Yn-Xm-AUG motif at the 3′-border of the IRES in which a pyrimidine-rich tract (Yn; n = 8–10 nt) is separated from an AUG triplet by a spacer (Xm; m = 18–20 nt). Both classes of IRES can promote initiation at this AUG codon as well as at AUG codons that can be as much as ~150 nt downstream and that may or may not be in-frame [33,34,35]. Viruses that contain these types of IRES commonly encode proteases that cleave eIF4G, splitting the N-terminal domain that binds eIF4E from the domains that bind eIF4A and eIF3 (e.g., [29]). Both classes of IRES function in the absence of eF4E and this domain of eIF4G [36,37,38], and instead, depend on a direct, non-canonical interaction with eIF4G’s central domain that recruits eIF4A to the IRES [39,40,41]. Binding of these factors to the IRES and its subsequent restructuring are prerequisites for recruitment of the 43S complex [36,37]. In addition to canonical eIFs, the activity of type I and type II IRESs involves IRES trans-acting factors (ITAFs), which are cellular RNA-binding proteins that are thought to assist the IRES in attaining an active conformation (e.g., [42]). The dependence on ITAFs and the identity of the ITAF(s) required for activity is IRES specific. The pyrimidine tract-binding protein PTB is required by types I and II [43,44,45], whereas the poly(C)-binding protein 2, upstream of N-ras and glycl-tRNA synthetase have only been implicated in type I IRES function and ITAF45/Ebp1 enhances the activity of the type II IRES of foot-and-mouth disease virus (FMDV) (genus Aphthovirus) [38,46,47,48,49]. Differences in expression levels of critical ITAFs are thought to underlie the cell type specificity of both classes of IRES.
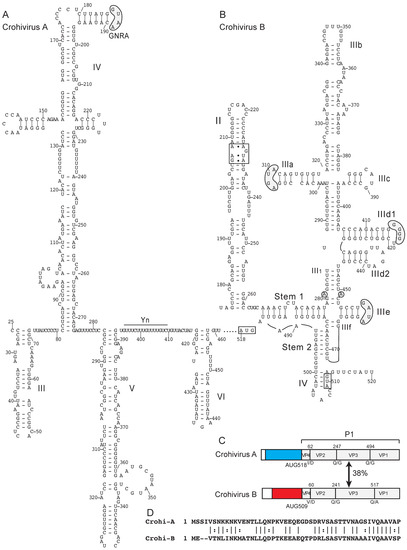
Figure 1.
Two different classes of IRES in the genomes of members of the genus Crohivirus of Picornaviridae. (A) Model of the structure of the type I IRES of crohivirus A strain ZM54 (NCBI ref. seq. NC_025474.1) and (B) Model of the structure of the type IV IRES of crohivirus B clone Bat/CAM/CroV-P25/2013 (NCBI ref. seq. NC_033819.1). Nucleotides are numbered and the initiation codon is boxed. Domains are labeled (A) III – VI and (B) II – IV. The IRES structures have been annotated (A) to show the GNRA tetraloop at the apex of domain IV and the pyrimidine (Yn) tract of the Yn-Xm-AUG motif and (B) to show the loop-E motif in domain II and to highlight sequence motifs in domain III that are conserved in other type IV IRESs, such as the apical loops in subdomains IIIa, IIId and IIIe, and the unpaired purine residues in helix III1 (grey shading). The tertiary base-pairing interaction between one of these unpaired purine residues in helix III1 and a flipped-out pyrimidine residue in the loop of subdomain IIIe is indicated by a blue line. (C) Schematic representations of the 5′UTRs and adjacent coding region for the P1 capsid protein precursor, annotated to show noncoding regions that constitute the type I IRES in crohivirus A (light blue) and the type IV IRES in crohivirus B (red), the N-termini of the structural proteins VP4, VP2, VP3 and VP1 and the protease cleavage sites between them, and the amino acid sequence identity between the two P1 polyproteins. (D) Sequence alignment of the N-terminal regions of the VP4 proteins of crohivirus A and crohivirus B. Identical and related residues are indicated by lines and colons, respectively.
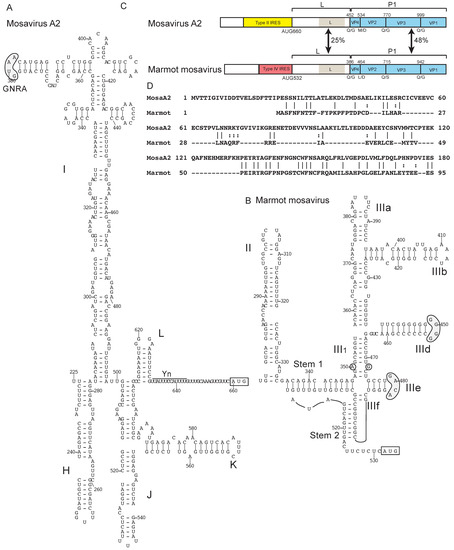
Figure 2.
Two different classes of IRES in the genus Mosavirus of Picornaviridae. (A) Model of the structure of the type II IRES of mosavirus A2 strain SZAL6-MoV/2011/HUN (NCBI ref. seq. NC_023987.1) and (B) model of the structure of the type IV IRES of mosavirus B marmot mosavirus strain HT8 (Genbank: KY855435). Nucleotides are numbered and the initiation codon is boxed. Domains are labeled (A) H – L and (B) II – III. The IRES structures have been annotated (A) to show the GNRA tetraloop at the apex of domain I and the Yn-Xm-AUG motif and (B) to highlight sequence motifs in domain III that are conserved in other type IV IRESs (grey shading). (C) Schematic representations of the 5′UTRs and adjacent coding region for the Leader (L) protein and the P1 capsid protein precursor (light blue), annotated to show noncoding regions that constitute the type II IRES in mosavirus A (yellow) and the type IV IRES in mosavirus B (rose), the L protein and the most conserved elements in it, the N-termini of the structural proteins VP4, VP2, VP3 and VP1 and the protease cleavage sites between them, and the amino acid sequence identity between the two L proteins and between the P1 polyproteins. (D) Sequence alignment of the N-terminal regions of the L proteins of mosavirus A2 and marmot mosavirus. Identical and related residues are indicated by lines and colons, respectively.
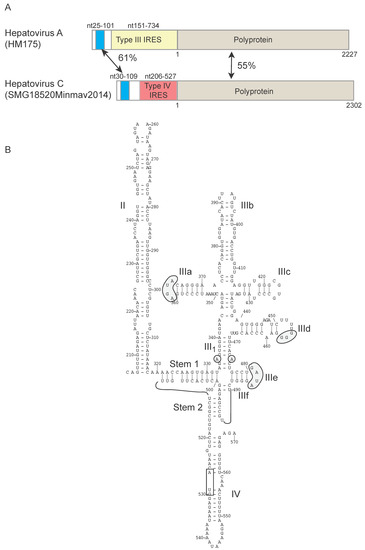
Type III IRESs were first identified in the 734 nt-long 5′UTR of hepatitis A virus (genus Hepatovirus of Picornaviridae) [50]. This 5′UTR comprises six domains: domains IV and V form the essential core of the IRES and are located immediately upstream of a Yn-Xm-AUG motif, in which the AUG triplet is the initiation codon for the viral polyprotein [51]. Initiation activity is weak, is augmented by ITAFs, and depends on binding of eIF4G to the IRES as a constituent of intact eIF4F, but the full set of factors required for initiation on this IRES has not been established [52,53]. The HAV IRES domain that is recognized by eIF4G [54] topologically resembles the J-K domain of type II IRESs to which eIF4G binds [41].
Type IV IRESs were initially identified in hepatitis C virus (HCV) of the Hepacivirus genus, and in bovine viral diarrhea virus (BVDV) and classical swine fever virus (CSFV) of the Pestivirus genus of Flaviviridae [55,56,57] and as described below, variant forms have subsequently been identified in numerous genera of Picornaviridae. In members of Hepacivirus and Pestivirus and in some members of Picornaviridae (e.g., Figure 1B), these ~300 nt-long IRESs contain two major domains (II and III), and in some instances, the minor domain IV, a hairpin structure that includes the initiation codon [58]. The liver-specific microRNA-122 (miR-122), which is a determinant of the hepatotropism of HCV, binds to two sites upstream of the HCV IRES and promotes RNA stability, translation and replication [59]. Domain III consists of a series of stemloops (IIIa – IIIf) that form three- or four-way junctions. It is strongly conserved, particularly at the apex of IIId (which contains a characteristic exposed GGG motif) and in the basal region, which contains a pseudoknot and a stemloop (IIIe) with a universally conserved apical GA[U/C]A tetraloop that base pairs with a bulged nucleotide in the adjacent helix III1 [58,60]. The apical region of domain III contains subdomains IIIa and IIIc that interact with the 40S subunit, for example via the conserved apical ‘AGUA’ loop in IIIa [61,62]; this apical region also binds to eIF3 [63]. Domain II is a flexible 60–75 nt-long hairpin that in many instances contains a loop-E motif [58]. Its role is to facilitate steps in initiation leading up to subunit joining [64,65]. Initiation on these IRESs involves direct binding of eIF3, and multipoint interaction of the 40S subunit with stemloops IIIa, IIIc, IIId, IIIe and the pseudoknot [61,62,63,66,67,68]. Base pairing of the apical GGG motif in IIId with expansion segment 7 (ES7) of 18S ribosomal RNA (rRNA) is critical for the affinity of the IRES-40S interaction [68] and induces a structural rearrangement in the resulting complex that likely primes it for binding of initiator tRNA and start codon recognition [69,70]. These interactions with the 40S subunit enable type IV IRESs to initiate translation without the involvement of eIF4F. Moreover, the IRES-mediated positioning of the initiation codon in the ribosomal P site without prior scanning enables the IRES to promote initiation both by an eIF2-dependent mechanism [66,71] and by an eIF2-independent mechanism in which eIF5B stabilizes binding of initiator tRNA and orients it on the 40S subunit prior to subunit joining [65,72].
Type V IRESs have been identified in Kobuvirus, Oscivirus, Passerivirus and Salivirus genera of Picornaviridae [73,74,75]. Their domain organization resembles that of type I and type II IRESs, with a class-specific domain I at the 3′-border followed by a large cruciform domain J (with an apical GNRA tetraloop) that resembles domain IV of type I IRESs, the eIF4G-binding domain K that contains motifs that are characteristic of type II IRESs, a Yn-Xm-AUG motif at the 3′ border and a hairpin domain that sequesters this AUG triplet, which is the initiation codon for the viral polyprotein. The apparent hybrid nature of type V IRESs suggests that they may be chimeras formed by recombination between type I and type II IRES.
The genomes of members of Dicistroviridae and related viruses are dicistronic and contain IRESs in the 5′UTR and in the intergenic region (IGR) [76]. The 5′UTR IRESs have not been classified but are structurally diverse and utilize distinct mechanisms for initiation [77,78]. IGR IRESs that have been characterized to date fall into two structural groups, exemplified by cricket paralysis virus (CrPV) and Taura syndrome virus (TSV). Both are ~200 nt long and consist of one domain that comprises two nested pseudoknots and a second domain that consists of a single pseudoknot (in CrPV) or of a pseudoknot with a protruding hairpin (in TSV). This domain mimics the anticodon stemloop of tRNA base paired to an mRNA codon. These IRESs bind directly to the 40S subunit and recruit a 60S subunit to form an active initiation complex without the involvement of initiation factors, initiator tRNA or an initiation codon [79,80,81]. The bound IRES occupies the ribosomal A site and must be translocated to the P site by elongation factor 2 (eEF2) before an aminoacyl-tRNA/eIF1A-GTP complex can bind to the A site to allow elongation to occur [82]. IGR IRES function is independent of eIF2 and is upregulated by phosphorylation of eIF2a, likely because of the greater availability of ribosomes when translation of cellular mRNA is impaired [79]. This mechanism therefore exploits the innate immune response to promote viral replication.
A few groups of viral IRESs, each with related sequences and conserved structures, have not yet been incorporated into this classification scheme. For example, the 5′UTRs of pegivirus A and pegivirus C, members of the Pegivirus genus of Flaviviridae, contain functional IRESs [83]. The ~600 nt-long 5′UTRs both have a 3′-terminal Yn-Xm-AUG motif, but otherwise appear to be unrelated to other classes of IRES. The mechanism by which they function has not been determined.
5. Experimental Exchange of IRESs between Members of Different Viral Genera and Families
Substitution of the EMCV IRES for the native IRES in PV and HAV [106,107] was tolerated without alteration of viral growth characteristics, whereas replacement of the poliovirus IRES by the HCV IRES and part of the adjacent coding region yielded a genetically stable virus with strongly reduced growth kinetics [108]. Viral IRESs can therefore function in the context of heterologous viral genomes.
However, type I and some type II IRESs have strong requirements for ITAFs that likely underlie cell type-specific differences in IRES activity. Consistently, some IRES exchanges are associated with significant impairment of viral translation in specific cell types and with attenuation of virulence. Replacement of the PV IRES by that of HRV-2 yielded a chimeric virus that grew better than the wild-type virus in human epithelial type 2 (HEp-2) cells, but its growth was almost abrogated in neuronal cells and it lacked neurovirulence in a mouse model and in Cynomolgus monkeys [92,109]. Determinants of tropism for cells of neuronal origin mapped to apical regions of IRES domains V and VI. Replacement of the 5′UTR of a virulent strain of Coxsackie virus B3 that causes severe pancreatic and heart disease in mice by the PV 5′UTR yielded a chimeric virus that had strongly attenuated growth in murine fetal heart fibroblasts and caused transient mild disease in mice even though its growth in HeLa cells was unaffected [93]. The Theiler’s murine encephalomyocarditis virus (TMEV) GDVII strain causes fatal encephalitis in mice following intercranial inoculation, but it was completely attenuated in mice following substitution of its IRES by that of the FMDV O1K strain, even though substitution had little or no effect on translation in vitro and in baby hamster kidney (BHK) 21 cells, and the chimeric viral genome was infectious and the plaque size of progeny was only slightly reduced [48]. Notably, initiation on these two type II IRESs has different factor requirements: they need the same set of canonical initiation factors but whereas the TMEV IRES requires PTB, the FMDV IRES requires PTB and ITAF45/Ebp1.
9. Transfer of Heterologous IRESs between the Pegivirus and other Genera of Flaviviridae
The Pegivirus genus of Flaviviridae contains 11 species, designated Pegivirus A–K, and numerous unclassified members [155]. They infect mammals, and although they commonly cause persistent infections, generally have little or no associated pathogenesis [156]. Pegiviruses have a positive-strand RNA genome with a structure that closely resembles that of hepaciviruses: long untranslated regions flank an open reading frame encoding a single, large polyprotein that is proteolytically processed to yield mature structural and non-structural proteins. The order of genes in hepacivirus and pegivirus genomes is the same, except that pegiviruses do not encode an N-terminal Core protein (although pegivirus F, pegivirus G and pegivirus H encode basic proteins at the equivalent N-terminal location in the polyprotein), and some pegiviruses may encode an additional structural protein (‘X’) between structural and non-structural proteins [157,158,159].
The structurally related 5′UTRs of pegivirus A and C both contain IRESs [83,161]. 5′UTRs with a similar domain organization occur in pegivirus D [162], pegivirus E [163], pegivirus I [164], pegivirus K [163], numerous isolates of Pegivirus B [164], as well as in the 5′UTRs of currently unclassified pegiviruses, for example from pigs and dolphins [165,166]. These 5′UTRs contain two extended hairpin domains (II and IV) flanking a pair of smaller stemloops (IIIa and IIIb), the first of which may form part of a pseudoknot (Figure 6A). The conserved Y-shaped domain Va/Vb is commonly, but not invariably, followed by a pyrimidine tract that forms part of a Yn-Xm-AUG motif. In some pegivirus genomes, the initiation codon for the viral polyprotein is followed by a hairpin domain VI that could potentially enhance translation by preventing further 5′-3′ scanning by ribosomal initiation complexes. Although the combination of a Y-shaped domain and the Yn-Xm-AUG motif resembles the 3′-terminal region of type II IRESs, there is no evidence that pegivirus domain V binds analogously to eIF4G/eIF4A or that the pyrimidine tract acts the landing site for ribosomes. In addition to this structural conservation, several sequence motifs in these pegivirus IRESs are highly conserved, including motifs in domains IIIa/IIIb, at the apex of domain IVa and in domain Vb (Figure 6A). The factor interactions and mechanism of action of this class of pegivirus has not been characterized, and the function of conserved motifs and structures remains unknown.
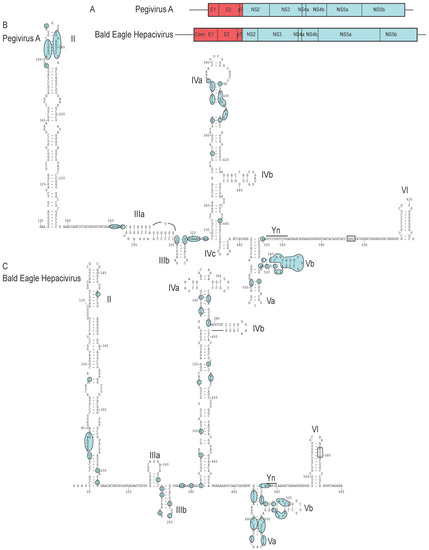
Figure 6.
Structurally related IRESs in Pegivirus A and Bald eagle hepacivirus (A) Schematic representations of pegivirus A and Bald eagle hepacivirus (BeHCV), showing the ORFs encoding the polyprotein in each, the individual proteins that are cleaved from each by cellular and viral proteins, the flanking noncoding regions flanking the ORFs. Note that the IRESs are located adjacent to unrelated coding sequences, namely the pegivirus E1 envelope glycoprotein and the BeHCV Core protein, respectively. (B) Secondary structure model of a segment of the 5′UTR of pegivirus A [83] (NCBI ref. seq. NC_001837.1) and (C) secondary structure model of a segment of the 5′UTR of BeHCV (GenBank: MN062427). A similar model to that shown for BeHCV has been proposed for the 5′UTR of Duck hepacivirus [160]. Domains are labeled II – VI, and nucleotides are numbered at 20 nt. intervals. The initiation codon is boxed and the pyrimidine tract is labeled Yn. The shaded elements indicate nucleotides that are conserved in (B) six or more of pegivirus A (NCBI ref. seq. NC_001837.1), pegivirus B (GenBank: KC796089.1), pegivirus C (GenBank: HM638236.1), pegivirus D (NCBI ref. seq.: NC_038433), pegvirus I (NCBI ref. seq.: NC_038437.1), pegivirus K (NCBI Ref. seq. NC_034442.1) and Dolphin pegivirus isolate DPgV (GenBank: MK059751.1)and (C) in seven or more of Bald eagle hepacivirus isolate NA03-001 (GenBank: MN062427.1), Duck hepacivirus strain HCL-3 (GenBank: MK737641.1), hepacivirus F (NCBI Ref. seq. NC_038427.1), hepacivirus J isolate hepacivirus/RMU10-3382/GER/2010 (NCBI Ref. seq. NC_038429.1), hepacivirus sp. (from Trachemys scripta elegans) (GenBank: MG334001), Jogalong virus isolate P1-1 (GenBank: MN133813.1), Rodent hepacivirus isolate hepacivirus/NLR08-365/NEL/2008 (GenBank: KC411796) and Sigmodontinae hepacivirus strain On/2012 (GenBank: MH370348.1).
Several hepaciviruses also contain pegivirus-like IRESs, as first noted for hepacivirus F, hepacivirus J and other rodent-infecting hepaciviruses [164], for human hepacivirus H [159] and for three Duck hepacivirus strains [160]. We identified additional pegivirus IRES-like sequences in Bald Eagle hepacivirus (BeHV) [167] (Figure 6B), in the putatively avian Jogalong virus [168] and in hepaciviruses from Oliogoryzomys nigripes (black-footed pygmy rice rat) [169] and Trachemys scripta elegans (red-eared terrapin) (GenBank: MG334001). The predicted structure of these elements resembles that of the canonical pegivirus A 5′UTR (Figure 6A), although domains IIIa/IIIb and the apex of domain IVa are truncated in some of the hepacivirus 5′UTRs. However, domain V and the adjacent Yn-Xm-AUG motif are strongly conserved (except in BeHV, which has a short Yn motif and a GUG triplet in place of the AUG triplet), and the most strongly conserved nucleotides in these putative hepacivirus IRESs are located at analogous positions to those in the pegivirus IRESs (Figure 6A,B), suggesting that they have the same functional role. The coding sequences adjacent to these hepacivirus 5‘UTRs encode the Core protein, and their N-terminal regions are non-conserved and variable in length, suggesting that the recombination breakpoint lies in this area of the genome, and that hepaciviruses may have acquired pegivirus-type IRESs by horizontal gene transfer on multiple occasions.
As a counterpoint to these observations, it is apparent that as well as acting as IRES donors to other members of Flaviviridae, pegiviruses may also have acquired IRESs from members of this family. Thus, a type IV IRES was identified in the pegivirus J genome [170] and our analysis revealed that several other pegivirus genomes also contain type IV IRESs, including pegivirus F, pegivirus H, a member of Pegivirus B (GenBank: KC796087.1) and some currently unclassified rodent pegiviruses [157,158,169,171]. Sequence identity ranges from ~50%–65%, and although some sequences are incomplete, it is evident that they all adopt similar structures (Figure 7), including a pestivirus-like domain IIId2. Domain II is least conserved, and in some instances, the initiation codon is sequestered in domain IV. The loop-E motif and the apical ‘AGUA’ motif in subdomain IIIa is absent in some instances, but these IRESs contain all of the other hallmarks of canonical type IV IRESs. The strongest homology is with the IRES of BVDV, a member of the Pestivirus genus of Flaviviridae (~62% nucleotide identity with Pegivirus B and Pegivirus J).
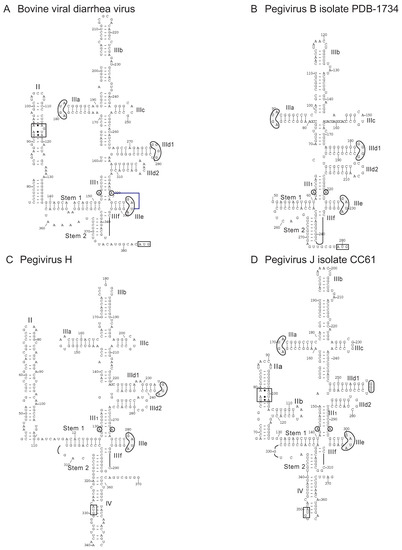
Figure 7.
Models of the type IV IRESs of bovine viral diarrhea virus of the genus Pestivirus and related type IV IRESs in members of the genus Pegivirus of Flaviviridae. The models show the structures of the IRESs of (A) bovine viral diarrhea virus (BVDV isolate Carlito) (GenBank: KP313732.1), (B) pegivirus B isolate PDB-1734 (GenBank: KC796087.1), (C) pegivirus H (human pegivirus 2) (GenBank: KT427409.1) and (D) pegivirus J isolate CC61 (GenBank: KC815311.1), in the last instance as previously proposed [170]. The nomenclature of domains and helices is as in [114]; nucleotides are numbered at intervals and the initiation codon is boxed. The loop-E motif in domain II [58] is labeled and conserved sequence motifs in domain III, such as the apical loops in subdomains IIIa, IIId and IIIe, and the unpaired purine residues in helix III1 are indicated by grey shading. The tertiary base-pairing interaction between the unpaired purine residue in helix III1 and loop IIIe is indicated by blue line. The pegivirus B sequence is incomplete, so that a model of domain I could not be proposed.
The presence of two types of IRES in the genomes of members of the genus Pegivirus that also occur in members of the genus Hepacivirus suggests that pegiviruses can act as donors and as recipients of IRESs to and from other viral genomes.
10. Conclusions
Molecular epidemiological studies established that the junction of the 5′UTR and the open reading frame that encodes the viral polyprotein constitutes a hot spot for recombination in picornaviruses. Intratypic recombination leading to acquisition of novel 5′UTRs by these viruses has been extensively documented, establishing the important concept that the 5′UTR is a distinct genetic unit that evolves independently and at a different rate to other elements of the viral genome (e.g., [84]). Intertypic recombination occurs less efficiently and is consequently less frequently observed but has been associated with the appearance of novel viral species. These recombination events involved transfer of IRESs of the same class within the same genus. However, the observation that a member of the genus Teschovirus of Picornaviridae has a type IV IRES [110], which had until then only been identified in members of the genera Hepacivirus and Pestivirus of Flaviviridae suggested that recombinational transfer of IRESs could even occur between different virus families. Subsequent analyses have revealed type IV IRESs in many other genera of Picornaviridae, and with the identification here of type IV IRESs in a further seven genera (Crohivirus, Diresapivirus, Grusopivirus, Mosavirus, Pemapivirus, Symapivirus and Tropivirus), it is now evident that they are very widely distributed in this family. The presence of structurally distinct IRESs appended to homologous coding sequences and or related IRESs linked to different coding sequences in viral genomes strongly suggests that IRESs have moved between them by horizontal gene transfer. Significantly, several genera of Picornaviridae have one or more members that contain a type IV IRES in addition to other members that contain an IRES of either type I, II or III. The complementary observations [159,161,163,164] summarized and extended here that some members of the genus Pegivirus contain pestivirus-like type IV IRESs (Figure 7) whereas some members of the genus Hepacivirus contain pegivirus-like IRESs (Figure 6) strongly support the conclusion that IRESs are independent genetic entities that can move between viral genomes by horizontal gene transfer. The presence of type IV IRESs in multiple genera of Picornaviridae as well as in three genera of Flaviviridae is of particular interest.
The structures of the type IV IRESs reported previously and identified here in additional genera of Picornaviridae and Flaviviridae confirm previous conclusions made concerning their conserved and variable elements. The most conserved region comprises the pseudoknot and subdomains IIId, IIIe and IIIf, which are critical for binding of the IRES to the 40S subunit and for positioning the initiation codon in the P site and the proximal coding region in the mRNA-binding cleft of the 40S subunit. The pegivirus type IV IRESs have most or all of the key motifs present in canonical type IV IRESs (Figure 7), whereas picornavirus type IV IRESs are more divergent. Thus, the apical GGG motif in subdomain III that base pairs with ES7 of 18S rRNA is reduced to a GG dinucleotide in members of Pemapivirus (Figure 5C) and the GA[U/C]A loop at the apex of subdomain IIIe is not conserved in members of the genera Aalivirus and Megrivirus [130]. Nevertheless, these elements are all still capable of engaging in critical interactions with 18S rRNA and within the IRES, respectively. By contrast, there is significant structural heterogeneity in elements that have accessory rather than essential functions, such as domain II and subdomain IIId2 (e.g., [134,137,172,173]), and several picornavirus type IV IRESs lack one or two of the subdomains that conventionally radiate from the IIIabc four-way junction. Members of the genera Aalivirus, Aquamavirus, Avihepatovirus and Grusapivirus contain a highly conserved ‘8-like’ motif at the apex of a long hairpin immediately upstream of the IRES (e.g., [130,132]) that in avihepatovirus augments IRES function [138]. How it functions, and how initiation complexes assemble on IRESs that lack binding determinants for eIF3 and the 40S subunit in apical regions of domain III, remain to be determined.
A key conclusion arising from the observations reported here and elsewhere (e.g., [130]) is that IRESs appear to have undergone recombinational exchange between viral genomes on multiple occasions. Although experimental studies of the effects of IRES exchanges on viral phenotype have revealed that they can impair viral replication and lead to restricted activity in some cell types, so that a heterologous IRES may not be beneficial immediately after acquisition, additional adaptive mutations in the IRES or other elements of the genome may be necessary to optimize activity in the new genetic and physical environment. Such changes could include the appearance of synonymous substitutions to optimize the proximal coding region for IRES function (e.g., [174]), regeneration of a disrupted Yn-Xm-AUG motif or, in the case of IRESs that depend on specific interactions with initiation factors, substitutions that optimize interactions with the likely somewhat divergent proteins from the new host, or substitutions that mimic ITAF-induced conformational changes.
Although the direction of movement of IRESs between viral genomes has not definitively been established, the widespread distribution of type IV IRESs in two different virus families and in multiple genera of Picornaviridae, and their presence in genera that predominantly utilize a different type of IRES together suggest that type IV IRESs have repeatedly been acquired by members of Picornaviridae and possibly, by members of the genus Pegivirus of Flaviviridae. Several factors could favor acquisition of type IV IRESs. Recombination that involves a replicative mechanism would be limited to viruses that can infect the same cell. In this respect it may be important that type IV IRESs depend on interaction with a widely conserved element in ES7 of 18S rRNA: this may account for the widespread distribution of this class of IRES in viruses that infect amphibians, birds, fish, reptiles and mammals. Exploitation of this type of interaction would lead to fewer host cell restrictions on function than those encountered by IRESs that depend primarily on interactions with initiation factors, and particularly, with ITAFs that may be expressed in a cell type-specific manner. Utilization of a type IV IRESs may therefore be beneficial by minimizing host cell restrictions on translation of viral proteins. Type IV IRESs are also significantly more active than type I and type III IRESs, and moreover, are notably resistant to inhibition by phosphorylation of eIF2 [65,72], a common virus-induced innate immune response.
As noted previously [130], recombination could also lead to the exchange or de novo acquisition of IRES domains or subdomains. The ability of the HCV IRES to function after substitution of domain II by the analogous domain from other type IV IRESs [175,176] indicates that this is feasible. Moreover, we have noted above that domain II is much more variable than domain III, and that homology between IRESs from different viral species is commonly restricted to the PK-IIId-IIIe-IIIf core. Such a scenario, which could involve a combination of non-replicative and replicative recombination process, could account for the capture and dissemination of the extended and highly conserved ‘8-like’ motif that occurs at the apex of domain IIIb in members of the genus Megrivirus but at the apex of a hairpin upstream of domain II in members of the genera Aalivirus, Anativirus, Aquamavirus and Avihepatovirus [129,130,132].
Author Contributions
Conceptualization, C.U.T.H.; Investigation, Y.A., A.G.B. and C.U.T.H.; Data Curation, C.U.T.H.; Writing—Original Draft Preparation, C.U.T.H.; Writing—Review & Editing, A.G.B., Y.A., T.V.P. and C.U.T.H.; Visualization, C.U.T.H.; Supervision, T.V.P. and C.U.T.H.; Project Administration, C.U.T.H.; Funding Acquisition, T.V.P. and C.U.T.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants the National Institutes of Health (NIH) (R01 AI123406 to C.H., R35 GM122602 to T.P.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Agol, V.I.; Gmyl, A.P. Emergency services of viral RNAs: Repair and remodeling. Microbiol. Mol. Biol. Rev. 2018, 82, e00067-17. [Google Scholar] [CrossRef] [PubMed]
- Bentley, K.; Evans, D.J. Mechanism and consequences of positive-strand RNA virus recombination. J. Gen. Virol. 2018, 99, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Kempf, B.J.; Watkins, C.L.; Peersen, O.B.; Barton, D.J. Picornavirus RNA recombination counteracts error catastrophe. J. Virol. 2019, 93, e00652-19. [Google Scholar] [CrossRef]
- Xiao, Y.; Rouzine, I.M.; Bianco, S.; Acevedo, A.; Goldstein, E.F.; Farkov, M.; Brodsky, L.; Andino, R. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 2016, 19, 493–503. [Google Scholar] [CrossRef]
- Lukashev, A.N. Role of recombination in evolution of enteroviruses. Rev. Med. Virol. 2005, 15, 157–167. [Google Scholar] [CrossRef]
- Muslin, C.; Mac Kain, A.; Bessaud, M.; Blondel, B.; Delpeyroux, F. Recombination in enteroviruses, a multi-step modular evolutionary process. Viruses 2019, 11, 859. [Google Scholar] [CrossRef]
- Nikolaidis, M.; Mimouli, K.; Kyriakopoulou, Z.; Tsimpidis, M.; Tsakogiannis, D.; Markoulatos, P.; Amoutzias, G.D. Large-scale genomic analysis reveals recurrent patterns of intertypic recombination in human enteroviruses. Virology 2019, 526, 72–80. [Google Scholar] [CrossRef]
- Dolja, V.V.; Koonin, E.V. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res. 2018, 244, 36–52. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Shi, M.; Holmes, E.C. Using metagenomics to characterize an expanding virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef]
- Krupovic, M.; Koonin, E.V. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci. Rep. 2014, 4, 5347. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.G.; Neftzler, N.E.; White, P.A. Ancient recombination events and the origins of hepatitis E virus. BMC Evol. Biol. 2016, 16, 210. [Google Scholar] [CrossRef]
- Pankovics, P.; Boros, Á.; Kiss, T.; Engelmann, P.; Reuter, G. Genetically highly divergent RNA virus with astrovirus-like (5′-end) and hepevirus-like (3′-end) genome organization in carnivorous birds, European roller (Coracias garrulus). Infect. Genet. Evol. 2019, 71, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Misra, S.; Hause, B.; Fang, Y. A naturally occurring recombinant Enterovirus expresses a Torovirus deubiquitinase. J. Virol. 2017, 91, e00450-17. [Google Scholar] [CrossRef]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ untranslated regions of the Flaviviral genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef]
- Truniger, V.; Miras, M.; Aranda, M.A. Structural and functional diversity of plant virus 3′-cap-independent translation enhancers (3′-CITEs). Front. Plant Sci. 2017, 8, 2047. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, Z.A.; Kieft, J.S. Viral RNA structure-based strategies to manipulate translation. Nat Rev. Microbiol. 2019, 17, 110–123. [Google Scholar] [CrossRef]
- Wimmer, E.; Hellen, C.U.; Cao, X. Genetics of poliovirus. Annu. Rev. Genet. 1993, 27, 353–436. [Google Scholar] [CrossRef]
- Shiroki, K.; Ishii, T.; Aoki, T.; Ota, Y.; Yang, W.X.; Komatsu, T.; Ami, Y.; Arita, M.; Abe, S.; Hashizume, S.; et al. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J. Virol. 1997, 71, 1–8. [Google Scholar] [CrossRef]
- Miras, M.; Sempere, R.N.; Kraft, J.J.; Miller, W.A.; Aranda, M.A.; Truniger, V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014, 202, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Villordo, S.M.; Carballeda, J.M.; Filomatori, C.V.; Gamarnik, A.V. RNA structure duplications and Flavivirus host adaptation. Trends Microbiol. 2016, 24, 270–283. [Google Scholar] [CrossRef]
- Jan, E.; Mohr, I.; Walsh, D. A Cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 2016, 3, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.T. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032656. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Hellen, C.U.; Pestova, T.V. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev. 2016, 30, 1573–1588. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Sweeney, T.R.; Skabkin, M.A.; Skabkina, O.V.; Hellen, C.U.; Pestova, T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014, 42, 3228–3245. [Google Scholar] [CrossRef]
- Gradi, A.; Svitkin, Y.V.; Imataka, H.; Sonenberg, N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 1998, 95, 11089–11094. [Google Scholar] [CrossRef]
- Gingras, A.C.; Svitkin, Y.; Belsham, G.J.; Pause, A.; Sonenberg, N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA 1996, 93, 5578–5583. [Google Scholar] [CrossRef]
- De Breyne, S.; Ohlmann, T. Focus on translation initiation of the HIV-1 mRNAs. Int. J. Mol. Sci. 2018, 20, 101. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Howell, M.T.; Kaminski, A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 1990, 15, 477–483. [Google Scholar] [CrossRef]
- Clarke, B.E.; Sangar, D.V.; Burroughs, J.N.; Newton, S.E.; Carroll, A.R.; Rowlands, D.J. Two initiation sites for foot-and-mouth disease virus polyprotein in vivo. J. Gen. Virol. 1985, 66, 2615–2626. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U.; Wimmer, E. A conserved AUG triplet in the 5′ nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology 1994, 204, 729–737. [Google Scholar] [CrossRef]
- Lulla, V.; Dinan, A.M.; Hosmillo, M.; Chaudhry, Y.; Sherry, L.; Irigoyen, N.; Nayak, K.M.; Stonehouse, N.J.; Zilbauer, M.; Goodfellow, I.; et al. An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat. Microbiol. 2019, 4, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996, 16, 6859–6869. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Shatsky, I.N.; Hellen, C.U. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996, 16, 6870–6878. [Google Scholar] [CrossRef]
- Sweeney, T.R.; Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014, 33, 76–92. [Google Scholar] [CrossRef]
- Lomakin, I.B.; Hellen, C.U.; Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000, 20, 6019–6029. [Google Scholar] [CrossRef]
- De Breyne, S.; Yu, Y.; Unbehaun, A.; Pestova, T.V.; Hellen, C.U. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. USA 2009, 106, 9197–9202. [Google Scholar] [CrossRef]
- Imai, S.; Kumar, P.; Hellen, C.U.; D’Souza, V.M.; Wagner, G. An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 2016, 23, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Abaeva, I.S.; Marintchev, A.; Pestova, T.V.; Hellen, C.U. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 2011, 39, 4851–4865. [Google Scholar] [CrossRef]
- Hellen, C.U.; Witherell, G.W.; Schmid, M.; Shin, S.H.; Pestova, T.V.; Gil, A.; Wimmer, E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7642–7646. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.; Howell, M.T.; Patton, J.G.; Jackson, R.J. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 1993, 74, 1775–1788. [Google Scholar] [CrossRef] [PubMed]
- Borovjagin, A.; Pestova, T.; Shatsky, I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994, 351, 299–302. [Google Scholar] [CrossRef]
- Blyn, L.B.; Towner, J.S.; Semler, B.L.; Ehrenfeld, E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997, 71, 6243–6246. [Google Scholar] [CrossRef]
- Hunt, S.L.; Hsuan, J.J.; Totty, N.; Jackson, R.J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999, 13, 437–448. [Google Scholar] [CrossRef]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000, 14, 2028–2045. [Google Scholar]
- Andreev, D.E.; Hirnet, J.; Terenin, I.M.; Dmitriev, S.E.; Niepmann, M.; Shatsky, I.N. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012, 40, 5602–5614. [Google Scholar] [CrossRef]
- Brown, E.A.; Day, S.P.; Jansen, R.W.; Lemon, S.M. The 5′ nontranslated region of hepatitis A virus RNA: Secondary structure and elements required for translation in vitro. J. Virol. 1991, 65, 5828–5838. [Google Scholar] [CrossRef]
- Brown, E.A.; Zajac, A.J.; Lemon, S.M. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994, 68, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Baugh, J.M.; Pilipenko, E.V. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell 2004, 16, 575–586. [Google Scholar] [CrossRef]
- Avanzino, B.C.; Fuchs, G.; Fraser, C.S. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc. Natl. Acad. Sci. USA 2017, 114, 9611–9616. [Google Scholar] [CrossRef] [PubMed]
- Koirala, D.; Shao, Y.; Koldobskaya, Y.; Fuller, J.R.; Watkins, A.M.; Shelke, S.A.; Pilipenko, E.V.; Das, R.; Rice, P.A.; Piccirilli, J.A. A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat. Commun. 2019, 10, 3629. [Google Scholar] [CrossRef] [PubMed]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [CrossRef]
- Poole, T.L.; Wang, C.; Popp, R.A.; Potgieter, L.N.; Siddiqui, A.; Collett, M.S. Pestivirus translation initiation occurs by internal ribosome entry. Virology 1995, 206, 750–754. [Google Scholar] [CrossRef]
- Rijnbrand, R.; van der Straaten, T.; van Rijn, P.A.; Spaan, W.J.; Bredenbeek, P.J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J. Virol. 1997, 71, 451–457. [Google Scholar] [CrossRef]
- Lukavsky, P.J. Structure and function of HCV IRES domains. Virus Res. 2009, 139, 166–171. [Google Scholar] [CrossRef]
- Sarnow, P.; Sagan, S.M. Unraveling the mysterious interactions between Hepatitis C virus RNA and liver-specific MicroRNA-122. Annu. Rev. Virol. 2016, 3, 309–332. [Google Scholar] [CrossRef]
- Easton, L.E.; Locker, N.; Lukavsky, P.J. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009, 37, 5537–5549. [Google Scholar] [CrossRef]
- Quade, N.; Boehringer, D.; Leibundgut, M.; van den Heuvel, J.; Ban, N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nat. Commun. 2015, 6, 7646. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Collier, M.; Loerke, J.; Ismer, J.; Schmidt, A.; Hilal, T.; Sprink, T.; Yamamoto, K.; Mielke, T.; Bürger, J.; et al. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015, 34, 3042–3058. [Google Scholar] [CrossRef] [PubMed]
- Hashem, Y.; des Georges, A.; Dhote, V.; Langlois, R.; Liao, H.Y.; Grassucci, R.A.; Pestova, T.V.; Hellen, C.U.; Frank, J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 2013, 503, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Locker, N.; Easton, L.E.; Lukavsky, P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007, 26, 795–805. [Google Scholar] [CrossRef]
- Pestova, T.V.; de Breyne, S.; Pisarev, A.V.; Abaeva, I.S.; Hellen, C.U. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J. 2008, 27, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998, 12, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Kolupaeva, V.G.; Pestova, T.V.; Hellen, C.U. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 2000, 6, 1791–1807. [Google Scholar] [CrossRef]
- Kieft, J.S.; Zhou, K.; Jubin, R.; Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 2001, 7, 194–206. [Google Scholar] [CrossRef]
- Malygin, A.A.; Kossinova, O.A.; Shatsky, I.N.; Karpova, G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013, 41, 8706–8714. [Google Scholar] [CrossRef]
- Angulo, J.; Ulryck, N.; Deforges, J.; Chamond, N.; Lopez-Lastra, M.; Masquida, B.; Sargueil, B. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016, 44, 1309–1325. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology 1999, 258, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Shatsky, I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008, 15, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Sweeney, T.R.; Kafasla, P.; Jackson, R.J.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011, 30, 4423–4436. [Google Scholar] [CrossRef]
- Sweeney, T.R.; Dhote, V.; Yu, Y.; Hellen, C.U. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 2012, 86, 1468–1486. [Google Scholar] [CrossRef] [PubMed]
- Pankovics, P.; Boros, Á.; Phan, T.G.; Delwart, E.; Reuter, G. A novel passerivirus (family Picornaviridae) in an outbreak of enteritis with high mortality in estrildid finches (Uraeginthus sp.). Arch. Virol. 2018, 163, 1063–1071. [Google Scholar] [CrossRef]
- Wilson, J.E.; Powell, M.J.; Hoover, S.E.; Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000, 20, 4990–4999. [Google Scholar] [CrossRef]
- Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES. Nucleic Acids Res. 2016, 44, 2362–2377. [Google Scholar] [CrossRef]
- Gross, L.; Vicens, Q.; Einhorn, E.; Noireterre, A.; Schaeffer, L.; Kuhn, L.; Imler, J.L.; Eriani, G.; Meignin, C.; Martin, F. The IRES 5′UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Res. 2017, 45, 8993–9004. [Google Scholar] [CrossRef]
- Wilson, J.E.; Pestova, T.V.; Hellen, C.U.; Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell 2000, 102, 511–520. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003, 17, 181–186. [Google Scholar] [CrossRef]
- Jan, E.; Kinzy, T.G.; Sarnow, P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 15410–15415. [Google Scholar] [CrossRef]
- Fernández, I.S.; Bai, X.C.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 2014, 157, 823–831. [Google Scholar] [CrossRef]
- Simons, J.N.; Desai, S.M.; Schultz, D.E.; Lemon, S.M.; Mushahwar, I.K. Translation initiation in GB viruses A and C: Evidence for internal ribosome entry and implications for genome organization. J. Virol. 1996, 70, 6126–6135. [Google Scholar] [CrossRef]
- Santti, J.; Hyypiä, T.; Kinnunen, L.; Salminen, M. Evidence of recombination among enteroviruses. J. Virol. 1999, 73, 8741–8749. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Lashkevich, V.A.; Ivanova, O.E.; Koroleva, G.A.; Hinkkanen, A.E.; Ilonen, J. Recombination in circulating enteroviruses. J. Virol. 2003, 77, 10423–10431. [Google Scholar] [CrossRef]
- Oberste, M.S.; Maher, K.; Pallansch, M.A. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 2004, 78, 855–867. [Google Scholar] [CrossRef]
- McIntyre, C.L.; McWilliam Leitch, E.C.; Savolainen-Kopra, C.; Hovi, T.; Simmonds, P. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J. Virol. 2010, 84, 10297–10310. [Google Scholar] [CrossRef] [PubMed]
- Muslin, C.; Joffret, M.L.; Pelletier, I.; Blondel, B.; Delpeyroux, F. Evolution and emergence of enteroviruses through intra- and inter-species recombination: Plasticity and phenotypic impact of modular genetic exchanges in the 5′ untranslated region. PLoS Pathog. 2015, 11, e1005266. [Google Scholar] [CrossRef] [PubMed]
- Pöyry, T.; Kinnunen, L.; Hyypiä, T.; Brown, B.; Horsnell, C.; Hovi, T.; Stanway, G. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 1996, 77, 1699–1717. [Google Scholar] [CrossRef]
- Rohll, J.B.; Percy, N.; Ley, R.; Evans, D.J.; Almond, J.W.; Barclay, W.S. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 1994, 68, 4384–4391. [Google Scholar] [CrossRef] [PubMed]
- Semler, B.L.; Johnson, V.H.; Tracy, S. A chimeric plasmid from cDNA clones of poliovirus and coxsackievirus produces a recombinant virus that is temperature-sensitive. Proc. Natl. Acad. Sci. USA 1986, 83, 1777–17781. [Google Scholar] [CrossRef]
- Gromeier, M.; Alexander, L.; Wimmer, E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 1996, 93, 2370–2375. [Google Scholar] [CrossRef]
- Chapman, N.M.; Ragland, A.; Leser, J.S.; Höfling, K.; Willian, S.; Semler, B.L.; Tracy, S. A group B coxsackievirus/poliovirus 5′ nontranslated region chimera can act as an attenuated vaccine strain in mice. J. Virol. 2000, 74, 4047–4056. [Google Scholar] [CrossRef] [PubMed]
- Schibler, M.; Gerlach, D.; Martinez, Y.; Van Belle, S.; Turin, L.; Kaiser, L.; Tapparel, C. Experimental human rhinovirus and enterovirus interspecies recombination. J. Gen. Virol. 2012, 93, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Kyriakopoulou, Z.; Pliaka, V.; Amoutzias, G.D.; Markoulatos, P. Recombination among human non-polio enteroviruses: Implications for epidemiology and evolution. Virus Genes 2015, 50, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-F.; Naguib, T.; Yang, S.-J.; Nasr, E.; Jorba, J.; Ahmed, N.; Campagnoli, R.; van der Avoort, H.; Shimizu, H.; Yoneyama, T.; et al. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 2003, 77, 8366–8377. [Google Scholar] [CrossRef] [PubMed]
- Burns, C.C.; Shaw, J.; Jorba, J.; Bukbuk, D.; Adu, F.; Gumede, N.; Pate, M.A.; Abanida, E.A.; Gasasira, A.; Iber, J.; et al. Multiple independent emergences of type 2 vaccine-derived polioviruses during a large outbreak in northern Nigeria. J. Virol. 2013, 87, 4907–4922. [Google Scholar] [CrossRef] [PubMed]
- Kirkegaard, K.; Baltimore, D. The mechanism of RNA recombination in poliovirus. Cell 1986, 47, 433–443. [Google Scholar] [CrossRef]
- Smura, T.; Blomqvist, S.; Paananen, A.; Vuorinen, T.; Sobotová, Z.; Bubovica, V.; Ivanova, O.; Hovi, T.; Roivainen, M. Enterovirus surveillance reveals proposed new serotypes and provides new insight into enterovirus 5′-untranslated region evolution. J. Gen. Virol. 2007, 88, 2520–2526. [Google Scholar] [CrossRef]
- Tapparel, C.; Junier, T.; Gerlach, D.; Belle, S.V.; Turin, L.; Cordey, S.; Mühlemann, K.; Regamey, N.; Aubert, J.; Soccal, P.M.; et al. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg. Infect. Dis. 2009, 15, 719–726. [Google Scholar] [CrossRef]
- Yozwiak, N.L.; Skewes-Cox, P.; Gordon, A.; Saborio, S.; Kuan, G.; Balmaseda, A.; Ganem, D.; Harris, E.; DeRisi, J.L. Human enterovirus 109: A novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 2010, 84, 9047–9058. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Pankovics, P.; Knowles, N.J.; Reuter, G. Natural interspecies recombinant bovine/porcine enterovirus in sheep. J. Gen. Virol. 2012, 93, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Heath, L.; van der Walt, E.; Varsani, A.; Martin, D.P. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. J. Virol. 2006, 80, 11827–11832. [Google Scholar] [CrossRef]
- Benschop, K.S.; de Vries, M.; Minnaar, R.P.; Stanway, G.; van der Hoek, L.; Wolthers, K.C.; Simmonds, P. Comprehensive full-length sequence analyses of human parechoviruses: Diversity and recombination. J. Gen. Virol. 2010, 91, 145–154. [Google Scholar] [CrossRef]
- Zoll, J.; Galama, J.M.; van Kuppeveld, F.J. Identification of potential recombination breakpoints in human parechoviruses. J. Virol. 2009, 83, 3379–3383. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alexander, L.; Lu, H.H.; Wimmer, E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: Genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. USA 1994, 91, 1406–1410. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.Y.; Tesar, M.; Summers, D.F.; Ehrenfeld, E. Replication of hepatitis A viruses with chimeric 5′ nontranslated regions. J. Virol. 1996, 70, 2861–2868. [Google Scholar] [CrossRef]
- Lu, H.H.; Wimmer, E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 1996, 93, 1412–1417. [Google Scholar] [CrossRef]
- Gromeier, M.; Bossert, B.; Arita, M.; Nomoto, A.; Wimmer, E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 1999, 73, 958–964. [Google Scholar] [CrossRef]
- Pisarev, A.V.; Chard, L.S.; Kaku, Y.; Johns, H.L.; Shatsky, I.N.; Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004, 78, 4487–4497. [Google Scholar] [CrossRef]
- Bakhshesh, M.; Groppelli, E.; Willcocks, M.M.; Royall, E.; Belsham, G.J.; Roberts, L.O. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 2008, 82, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Chard, L.S.; Bordeleau, M.E.; Pelletier, J.; Tanaka, J.; Belsham, G.J. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 2006, 87, 927–936. [Google Scholar] [CrossRef]
- Chard, L.S.; Kaku, Y.; Jones, B.; Nayak, A.; Belsham, G.J. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006, 80, 1271–1279. [Google Scholar] [CrossRef]
- Hellen, C.U.; de Breyne, S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007, 81, 5850–5863. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Tsai, H.J. Sequence analysis of a duck picornavirus isolate indicates that it together with porcine enterovirus type 8 and simian picornavirus type 2 should be assigned to a new picornavirus genus. Virus Res. 2007, 129, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Victoria, J.; Simmonds, P.; Wang, C.; Shafer, R.W.; Nims, R.; Nielsen, O.; Delwart, E. A highly divergent picornavirus in a marine mammal. J. Virol. 2008, 82, 311–320. [Google Scholar] [CrossRef]
- Reuter, G.; Boldizsár, A.; Pankovics, P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 2009, 154, 101–108. [Google Scholar] [CrossRef]
- Honkavuori, K.S.; Shivaprasad, H.L.; Briese, T.; Street, C.; Hirschberg, D.L.; Hutchison, S.K.; Lipkin, W.I. Novel picornavirus in Turkey poults with hepatitis, California, USA. Emerg. Inf. Dis. 2011, 17, 480–487. [Google Scholar] [CrossRef]
- Kofstad, T.; Jonassen, C.M. Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: Characterization of novel Astroviruses and Picornaviruses. PLoS ONE 2011, 6, e25964. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Lai, K.K.; Huang, Y.; Yip, C.C.; Shek, C.T.; Lee, P.; Lam, C.S.; Chan, K.H.; Yuen, K.Y. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 2011, 85, 8819–8828. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Yip, C.C.; Choi, G.K.; Wu, Y.; Bai, R.; Fan, R.Y.; Lai, K.K.; Chan, K.H.; Yuen, K.Y. Identification of a novel feline picornavirus from the domestic cat. J. Virol. 2012, 86, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Pankovics, P.; Boros, A.; Reuter, G. Novel picornavirus in domesticated common quail (Coturnix coturnix) in Hungary. Arch. Virol. 2012, 157, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Pankovics, P.; Boros, Á.; Tóth, Z.; Phan, T.G.; Delwart, E.; Reuter, G. Genetic characterization of a second novel picornavirus from an amphibian host, smooth newt (Lissotriton vulgaris). Arch. Virol. 2017, 162, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Kiss, T.; Kiss, O.; Pankovics, P.; Kapusinszky, B.; Delwart, E.; Reuter, G. Genetic characterization of a novel picornavirus distantly related to the marine mammal-infecting aquamaviruses in a long-distance migrant bird species, European roller (Coracias garrulus). J. Gen. Virol. 2012, 94, 2029–2035. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Nemes, C.; Pankovics, P.; Kapusinszky, B.; Delwart, E.; Reuter, G. Genetic characterization of a novel picornavirus in turkeys (Meleagris gallopavo) distinct from turkey galliviruses and megriviruses and distantly related to the members of the genus Avihepatovirus. J. Gen. Virol. 2013, 94, 1496–1509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boros, Á.; Fenyvesi, H.; Pankovics, P.; Biró, H.; Phan, T.G.; Delwart, E.; Reuter, G. Secondary structure analysis of swine pasivirus (family Picornaviridae) RNA reveals a type-IV IRES and a parechovirus-like 3′ UTR organization. Arch. Virol. 2015, 160, 1363–1366. [Google Scholar] [CrossRef]
- Boros, Á.; Pankovics, P.; Simmonds, P.; Kiss, T.; Phan, T.G.; Delwart, E.; Reuter, G. Genomic analysis of a novel picornavirus from a migratory waterfowl, greater white-fronted goose (Anser albifrons). Arch. Virol. 2018, 163, 1087–1090. [Google Scholar] [CrossRef]
- Ng, T.F.; Mesquita, J.R.; Nascimento, M.S.; Kondov, N.O.; Wong, W.; Reuter, G.; Knowles, N.J.; Vega, E.; Esona, M.D.; Deng, X.; et al. Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 2014, 171, 102–111. [Google Scholar] [CrossRef]
- Wang, X.; Liu, N.; Wang, F.; Ning, K.; Li, Y.; Zhang, D. Genetic characterization of a novel duck-origin picornavirus with six 2A proteins. J. Gen. Virol. 2014, 95, 1289–1296. [Google Scholar] [CrossRef]
- Asnani, M.; Kumar, P.; Hellen, C.U. Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology 2015, 478, 61–74. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Lukashev, A.N.; van den Brand, J.M.; Gmyl, A.P.; Brünink, S.; Rasche, A.; Seggewiβ, N.; Feng, H.; Leijten, L.M.; et al. Evolutionary origins of hepatitis A virus in small mammals. Proc. Natl. Acad. Sci. USA 2015, 112, 15190–15195. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, S.; Shan, T.; Wang, X.; Deng, X.; Delwart, E.; Zhang, W. A novel picornavirus in feces of a rainbow lorikeet (Trichoglossus moluccanus) shows a close relationship to members of the genus Avihepatovirus. Arch. Virol. 2019, 164, 1911–1914. [Google Scholar] [CrossRef] [PubMed]
- De Breyne, S.; Yu, Y.; Pestova, T.V.; Hellen, C.U. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA 2008, 14, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Willcocks, M.M.; Locker, N.; Gomwalk, Z.; Royall, E.; Bakhshesh, M.; Belsham, G.J.; Idamakanti, N.; Burroughs, K.D.; Reddy, P.S.; Hallenbeck, P.L.; et al. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: A picornavirus with a pestivirus-like IRES. J. Virol. 2011, 85, 4452–4461. [Google Scholar] [CrossRef]
- Jaafar, Z.A.; Oguro, A.; Nakamura, Y.; Kieft, J.S. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife 2016, 5, e21198. [Google Scholar] [CrossRef]
- Rijnbrand, R.; Abell, G.; Lemon, S.M. Mutational analysis of the GB virus B internal ribosome entry site. J. Virol. 2000, 74, 773–783. [Google Scholar] [CrossRef]
- Willcocks, M.M.; Zaini, S.; Chamond, N.; Ulryck, N.; Allouche, D.; Rajagopalan, N.; Davids, N.A.; Fahnøe, U.; Hadsbjerg, J.; Rasmussen, T.B.; et al. Distinct roles for the IIId2 sub-domain in pestivirus and picornavirus internal ribosome entry sites. Nucleic Acids Res. 2017, 45, 13016–13028. [Google Scholar] [CrossRef]
- Pan, M.; Yang, X.; Zhou, L.; Ge, X.; Guo, X.; Liu, J.; Zhang, D.; Yang, H. Duck Hepatitis A virus possesses a distinct type IV internal ribosome entry site element of picornavirus. J.Virol. 2012, 86, 1129–1144. [Google Scholar] [CrossRef]
- Berry, K.E.; Waghray, S.; Mortimer, S.A.; Bai, Y.; Doudna, J.A. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 2011, 19, 1456–1466. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2018, 556, 197–202. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Chen, X.; Tian, J.H.; Chen, L.J.; Li, K.; Wang, W.; Eden, J.S.; Shen, J.J.; Liu, L.; et al. The evolutionary history of vertebrate RNA viruses. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Smits, S.L.; Raj, V.S.; Oduber, M.D.; Schapendonk, C.M.; Bodewes, R.; Provacia, L.; Stittelaar, K.J.; Osterhaus, A.D.; Haagmans, B.L. Metagenomic analysis of the ferret fecal viral flora. PLoS ONE 2013, 8, e71595. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G.; Pankovics, P.; Knowles, N.J.; Boros, Á. Two closely related novel picornaviruses in cattle and sheep in Hungary from 2008 to 2009, proposed as members of a new genus in the family Picornaviridae. J. Virol. 2012, 86, 13295–13302. [Google Scholar] [CrossRef]
- Yinda, C.K.; Zell, R.; Deboutte, W.; Zeller, M.; Conceição-Neto, N.; Heylen, E.; Maes, P.; Knowles, N.J.; Ghogomu, S.M.; Van Ranst, M.; et al. Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit bats. BMC Genom. 2017, 18, 249. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G.; Boros, A.; Kiss, T.; Delwart, E.; Pankovics, P. Complete genome characterization of mosavirus (family Picornaviridae) identified in droppings of a European roller (Coracias garrulus) in Hungary. Arch. Virol. 2014, 159, 2723–2729. [Google Scholar] [CrossRef]
- Luo, X.L.; Lu, S.; Jin, D.; Yang, J.; Wu, S.S.; Xu, J. Marmota himalayana in the Qinghai-Tibetan plateau as a special host for bi-segmented and unsegmented picobirnaviruses. Emerg. Microbes Infect. 2018, 7, 20. [Google Scholar] [CrossRef]
- Cohen, J.I.; Ticehurst, J.R.; Purcell, R.H.; Buckler-White, A.; Baroudy, B.M. Complete nucleotide sequence of wild-type hepatitis A virus: Comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 1987, 61, 50–59. [Google Scholar] [CrossRef]
- Wille, M.; Shi, M.; Klaassen, M.; Hurt, A.C.; Holmes, E.C. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 2019, 13, 2603–2616. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, L.; Ren, X.; He, G.; Zhang, J.; Yang, J.; Qian, Z.; Dong, J.; Sun, L.; Zhu, Y.; et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016, 10, 609–620. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, S.; Liu, D.; Zhou, C.; Li, W.; Lin, Y.; Wang, X.; Shen, Q.; Wang, H.; Li, C.; et al. The fecal virome of red-crowned cranes. Arch. Virol. 2019, 164, 3–16. [Google Scholar] [CrossRef]
- Zhang, W.; Kataoka, M.; Doan, H.Y.; Ami, Y.; Suzuki, Y.; Muramatsu, M.; Li, T.C. Characterization of a novel simian sapelovirus islated from Cynomolgus monkeys using PLC/PRF/5 cells. Sci. Rep. 2019, 9, 20221. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Ren, X.; Yang, L.; Hu, Y.; Yang, J.; He, G.; Zhang, J.; Dong, J.; Sun, L.; Du, J.; et al. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012, 86, 10999–11112. [Google Scholar] [CrossRef]
- Wille, M.; Eden, J.S.; Shi, M.; Klaassen, M.; Hurt, A.C.; Holmes, E.C. Virus-virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol. 2018, 27, 5263–5278. [Google Scholar] [CrossRef] [PubMed]
- Boros, A.; Pankovics, P.; Adonyi, A.; Fenyvesi, H.; Day, J.M.; Phan, T.G.; Delwart, E.; Reuter, G. A diarrheic chicken simultaneously co-infected with multiple picornaviruses: Complete genome analysis of avian picornaviruses representing up to six genera. Virology 2015, 489, 63–74. [Google Scholar] [CrossRef]
- Smith, D.B.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, A.S.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; et al. Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family. J. Gen. Virol. 2015, 97, 2894–2907. [Google Scholar] [CrossRef]
- Thézé, J.; Lowes, S.; Parker, J.; Pybus, O.G. Evolutionary and phylogenetic analysis of the Hepaciviruses and Pegiviruses. Genome Biol. Evol. 2015, 7, 2996–3008. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quan, P.L.; Firth, C.; Conte, J.M.; Williams, S.H.; Zambrana-Torrelio, C.M.; Anthony, S.J.; Ellison, J.A.; Gilbert, A.T.; Kuzmin, I.V.; Niezgoda, M.; et al. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. USA 2013, 110, 8194–8199. [Google Scholar] [CrossRef]
- Berg, M.G.; Lee, D.; Coller, K.; Frankel, M.; Aronsohn, A.; Cheng, K.; Forberg, K.; Marcinkus, M.; Naccache, S.N.; Dawson, G.; et al. Discovery of a novel human pegivirus in blood associated with Hepatitis C virus co-infection. PLoS Pathog. 2015, 11, e1005325. [Google Scholar] [CrossRef]
- Kapoor, A.; Kumar, A.; Simmonds, P.; Bhuva, N.; Singh Chauhan, L.; Lee, B.; Sall, A.A.; Jin, Z.; Morse, S.S.; Shaz, B.; et al. Virome analysis of transfusion recipients reveals a novel human virus that shares genomic features with Hepaciviruses and Pegiviruses. mBio 2015, 6, e01466-15. [Google Scholar] [CrossRef]
- Chu, L.; Jin, M.; Feng, C.; Wang, X.; Zhang, D. A highly divergent hepacivirus-like flavivirus in domestic ducks. J. Gen. Virol. 2019, 100, 1234–1240. [Google Scholar] [CrossRef]
- Simons, J.N.; Leary, T.P.; Dawson, G.J.; Pilot-Matias, T.J.; Muerhoff, A.S.; Schlauder, G.G.; Desai, S.M.; Mushahwar, I.K. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1995, 1, 564–569. [Google Scholar] [CrossRef]
- Chandriani, S.; Skewes-Cox, P.; Zhong, W.; Ganem, D.E.; Divers, T.J.; Van Blaricum, A.J.; Tennant, B.C.; Kistler, A.L. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc. Natl. Acad. Sci. USA 2013, 110, E1407–E1415. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Simmonds, P.; Cullen, J.M.; Scheel, T.K.; Medina, J.L.; Giannitti, F.; Nishiuchi, E.; Brock, K.V.; Burbelo, P.D.; Rice, C.M.; et al. Identification of a pegivirus (GB virus-like virus) that infects horses. J. Virol. 2013, 87, 7185–71890. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Lukashev, A.N.; Gmyl, A.; Coutard, B.; Adam, A.; Ritz, D.; Leijten, L.M.; van Riel, D.; et al. Evidence for novel hepaciviruses in rodents. PLoS Pathog. 2013, 9, e1003438. [Google Scholar] [CrossRef]
- Baechlein, C.; Grundhoff, A.; Fischer, N.; Alawi, M.; Hoeltig, D.; Waldmann, K.H.; Becher, P. Pegivirus infection in domestic pigs, Germany. Emerging Infect. Dis. 2016, 22, 1312–1314. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.C.S.; Subramaniam, K.; McCulloch, S.D.; Goldstein, J.D.; Schaefer, A.M.; Fair, P.A.; Reif, J.S.; Bossart, G.D.; Waltzek, T.B. Genomic characterization of a novel pegivirus species from free-ranging bottlenose dolphins (Tursiops truncatus) in the Indian River Lagoon, Florida. Virus Res. 2019, 263, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, T.L.; Sibley, S.D.; Pinkerton, M.E.; Dunn, C.D.; Long, L.J.; White, L.C.; Strom, S.M. Multidecade mortality and a homolog of Hepatitis C virus in Bald Eagles (Haliaeetus leucocephalus), the national bird of the USA. Sci. Rep. 2019, 9, 14953. [Google Scholar] [CrossRef]
- Williams, S.H.; Levy, A.; Yates, R.A.; Somaweera, N.; Neville, P.J.; Nicholson, J.; Lindsay, M.D.A.; Mackenzie, J.S.; Jain, K.; Imrie, A.; et al. Discovery of Jogalong virus, a novel hepacivirus identified in a Culex annulirostris (Skuse) mosquito from the Kimberley region of Western Australia. PLoS ONE 2020, 15, e0227114. [Google Scholar] [CrossRef]
- De Souza, W.M.; Fumagalli, M.J.; Sabino-Santos, G., Jr.; Motta Maia, F.G.; Modha, S.; Teixeira Nunes, M.R.; Murcia, P.R.; Moraes Figueiredo, L.T. A novel hepacivirus in wild rodents from South America. Viruses 2019, 11, 297. [Google Scholar] [CrossRef]
- Kapoor, A.; Simmonds, P.; Scheel, T.K.; Hjelle, B.; Cullen, J.M.; Burbelo, P.D.; Chauhan, L.V.; Duraisamy, R.; Sanchez Leon, M.; Jain, K.; et al. Identification of rodent homologs of hepatitis C virus and pegiviruses. MBio 2013, 4, e00216-13. [Google Scholar] [CrossRef]
- Van Nguyen, D.; Van Nguyen, C.; Bonsall, D.; Ngo, T.T.; Carrique-Mas, J.; Pham, A.H.; Bryant, J.E.; Thwaites, G.; Baker, S.; Woolhouse, M.; et al. Detection and characterization of homologues of human hepatitis viruses and pegiviruses in rodents and bats in Vietnam. Viruses 2018, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.P.; Jackson, R.J. Pestivirus internal ribosome entry site (IRES) structure and function: Elements in the 5′ untranslated region important for IRES function. J. Virol. 2002, 76, 5024–5033. [Google Scholar] [CrossRef]
- Friis, M.B.; Rasmussen, T.B.; Belsham, G.J. Modulation of translation initiation efficiency in classical swine fever virus. J. Virol. 2012, 86, 8681–8692. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.E.; Kaminski, A.; Kettinen, H.J.; Grace, K.; Clarke, B.E.; Carroll, A.R.; Rowlands, D.J.; Jackson, R.J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995, 14, 6010–6020. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.E.M.; Dalebout, T.J.; Eerligh, P.; Bredenbeek, P.J.; Spaan, W.J.M. Analysis of hepatitis C virus/classical swine fever virus chimeric 5′NTRs: Sequences within the hepatitis C virus IRES are required for viral RNA replication. J. Gen. Virol. 2003, 84, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Boerneke, M.A.; Dibrov, S.M.; Gu, J.; Wyles, D.L.; Hermann, T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl. Acad. Sci. USA 2014, 111, 15952–15957. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).