Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Cases Description and Herd History
2.2. Sample Processing for Virus Isolation
2.3. Virus Isolation and Propagation in Cell Culture
2.4. Real-Time PCR (qPCR)
2.5. In Situ Hybridization (RNAscope)
2.6. Generation of Polyclonal Antibody
2.7. Indirect Enzyme-Linked Immunosorbent Assays (iELISAs)
2.8. Indirect Immunofluorescence Assay (iIFA)
2.9. Virus Genome Extraction and Sequencing
2.10. PCV3 Genome Assembly, Bioinformatics and Phylogenetic Analysis
2.11. Experimental Inoculation of CD/CD Pigs
2.12. Statistical Analysis
2.13. Data Availability
3. Results
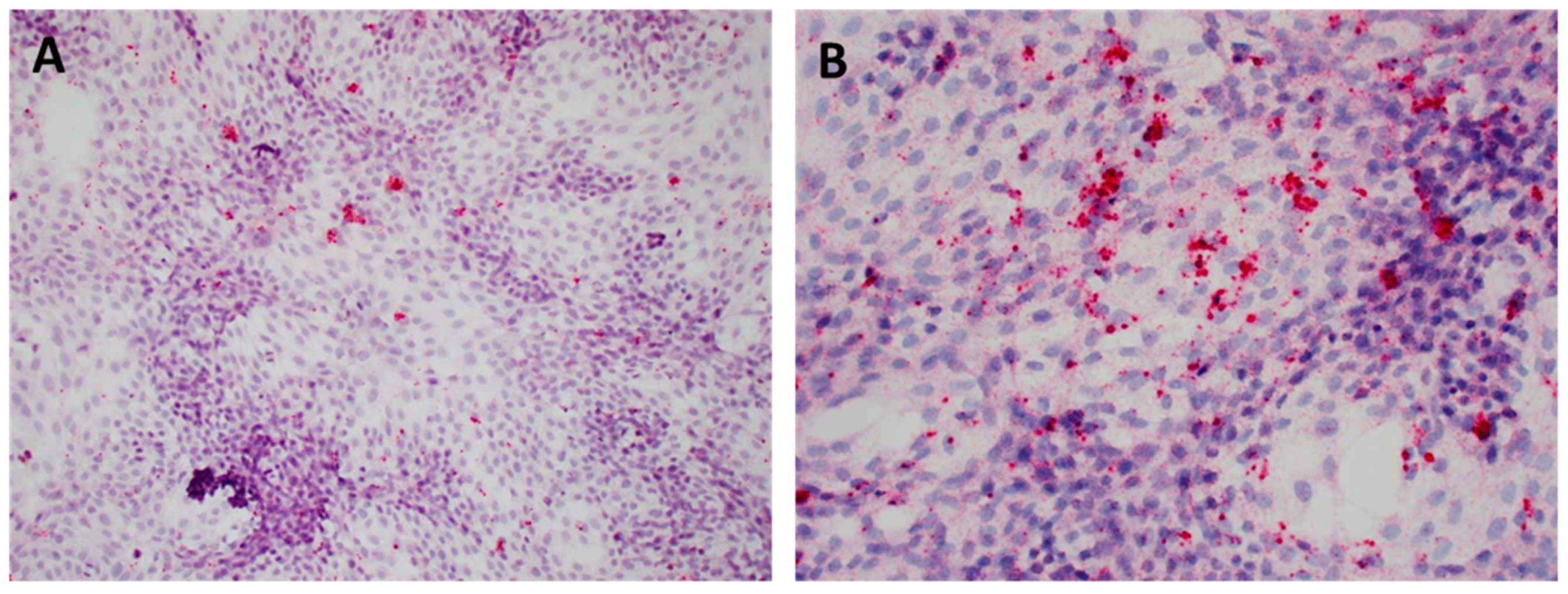
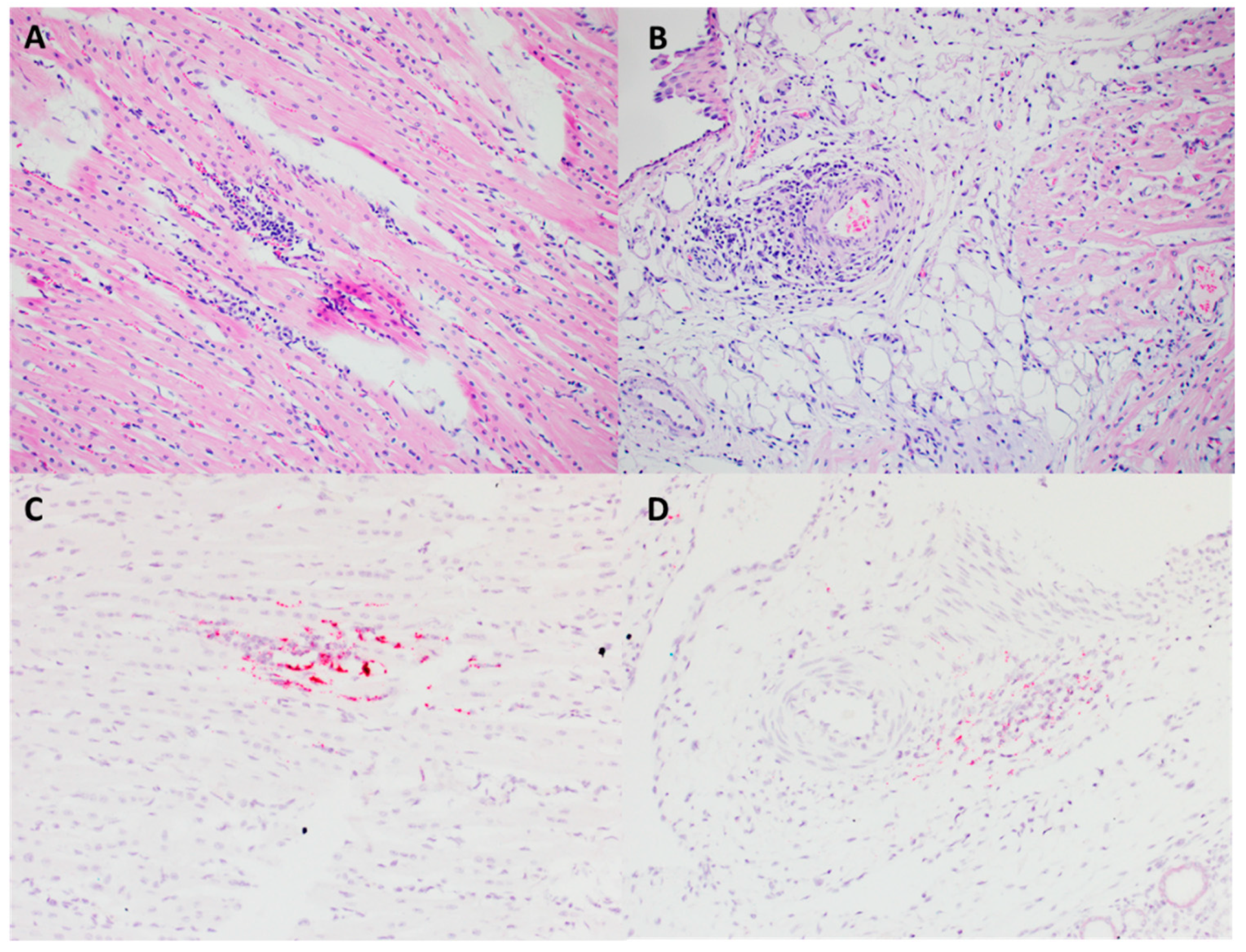
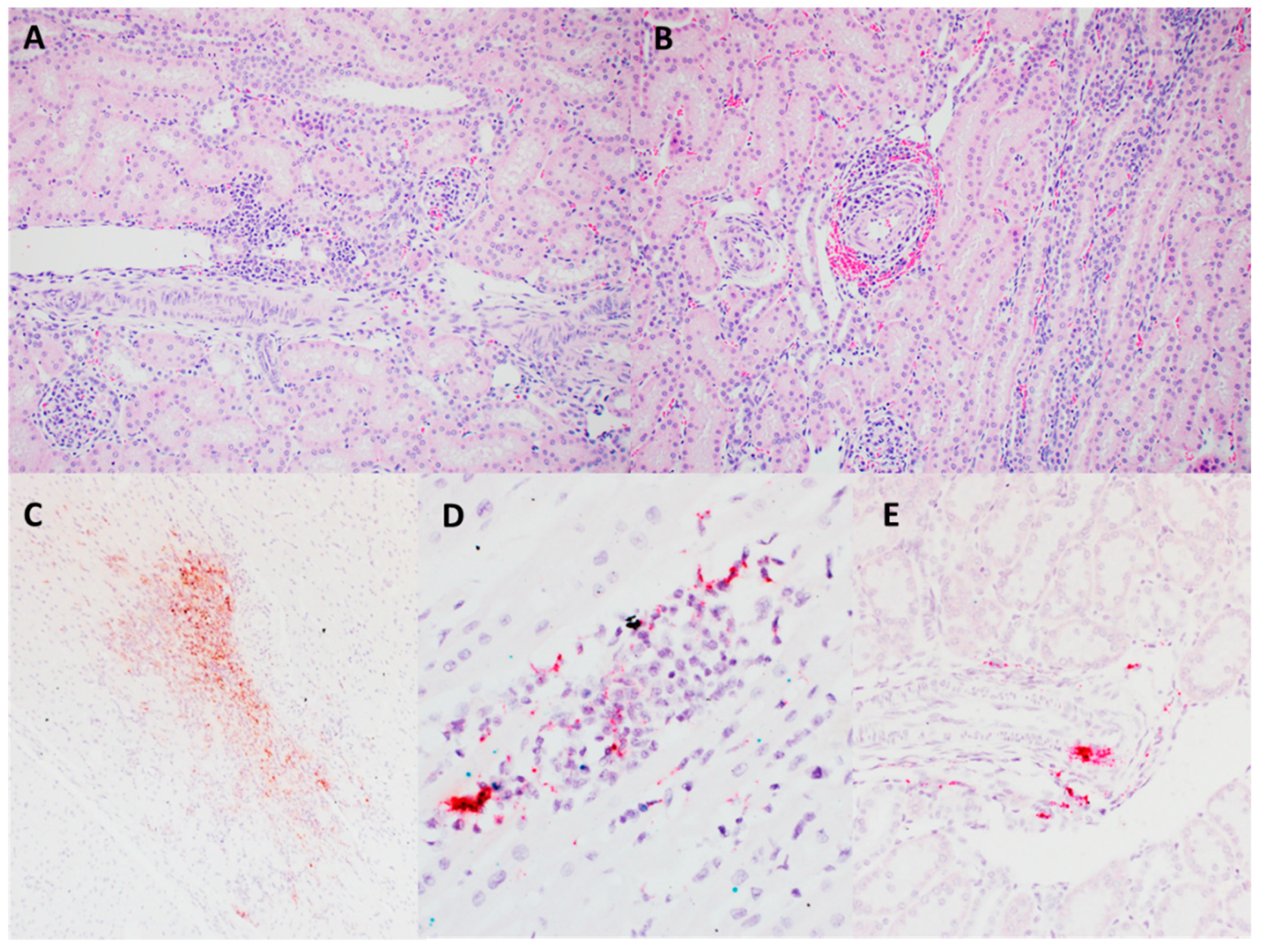
3.1. PCV3 Was Successfully Isolated from Perinatal and Reproductive Cases
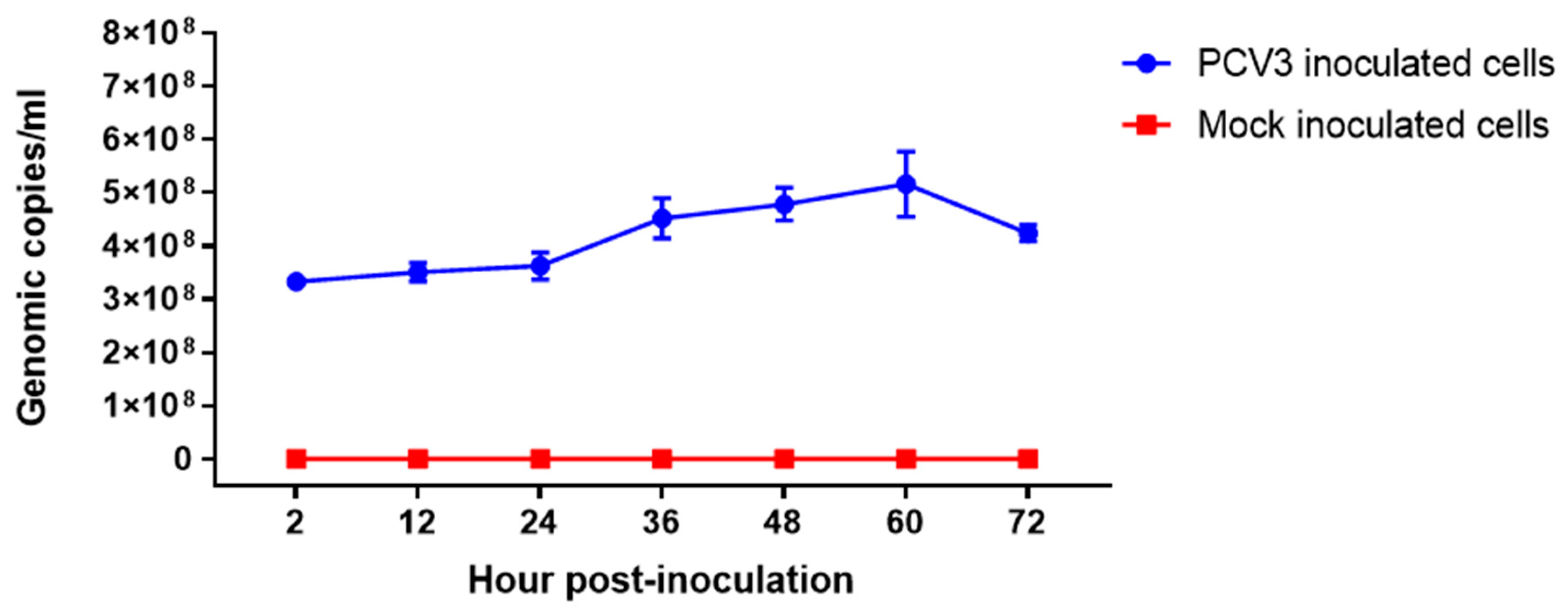
3.2. PCV3 Replicates in PK-15 Cells
3.3. Full Length Sequence and Assembly
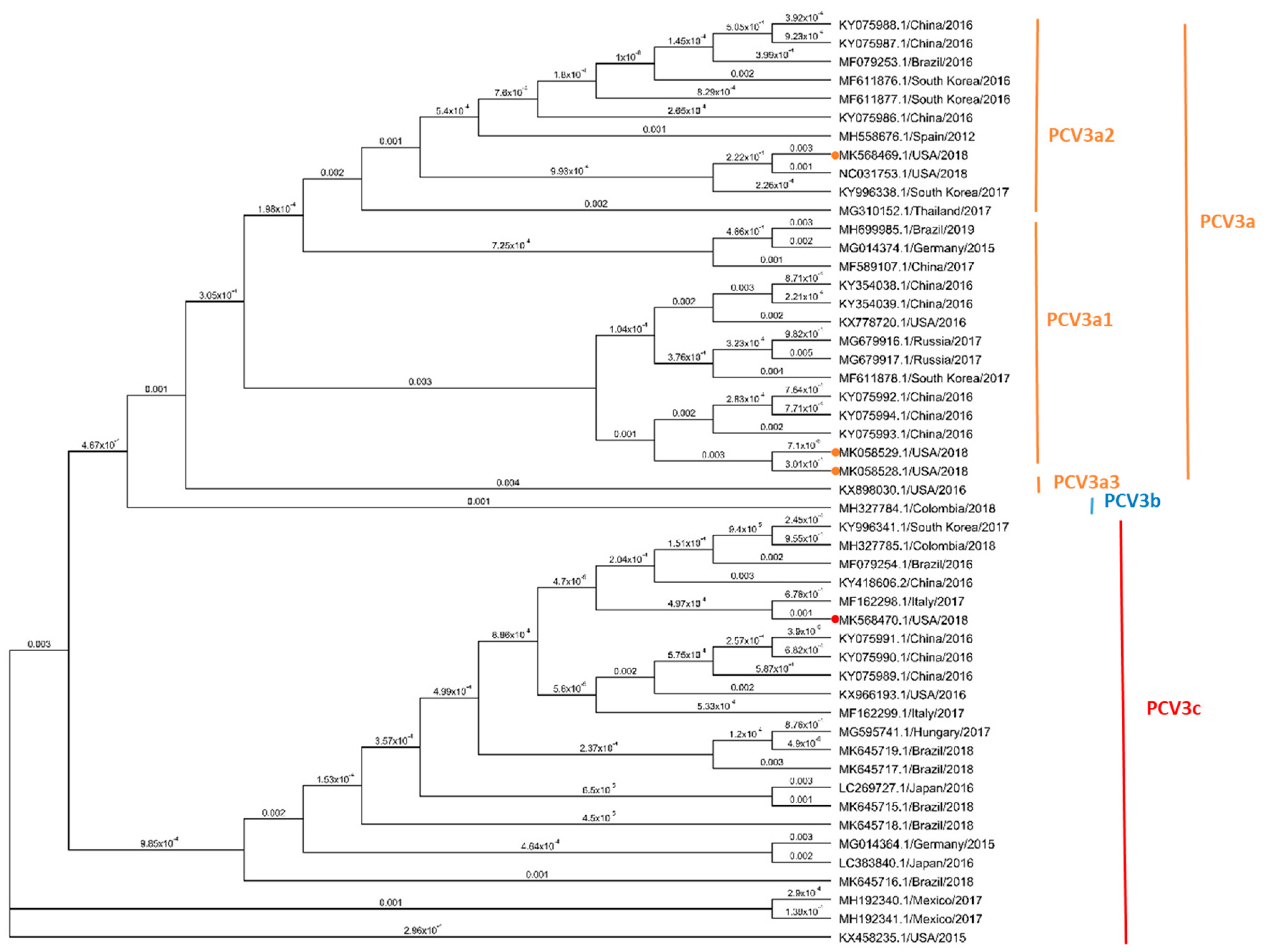
3.4. Phylogenetic Analysis Based on PCV3 Complete Genome Sequences
3.5. Phylogenetic Analysis Based on PCV3 ORF2 Sequences
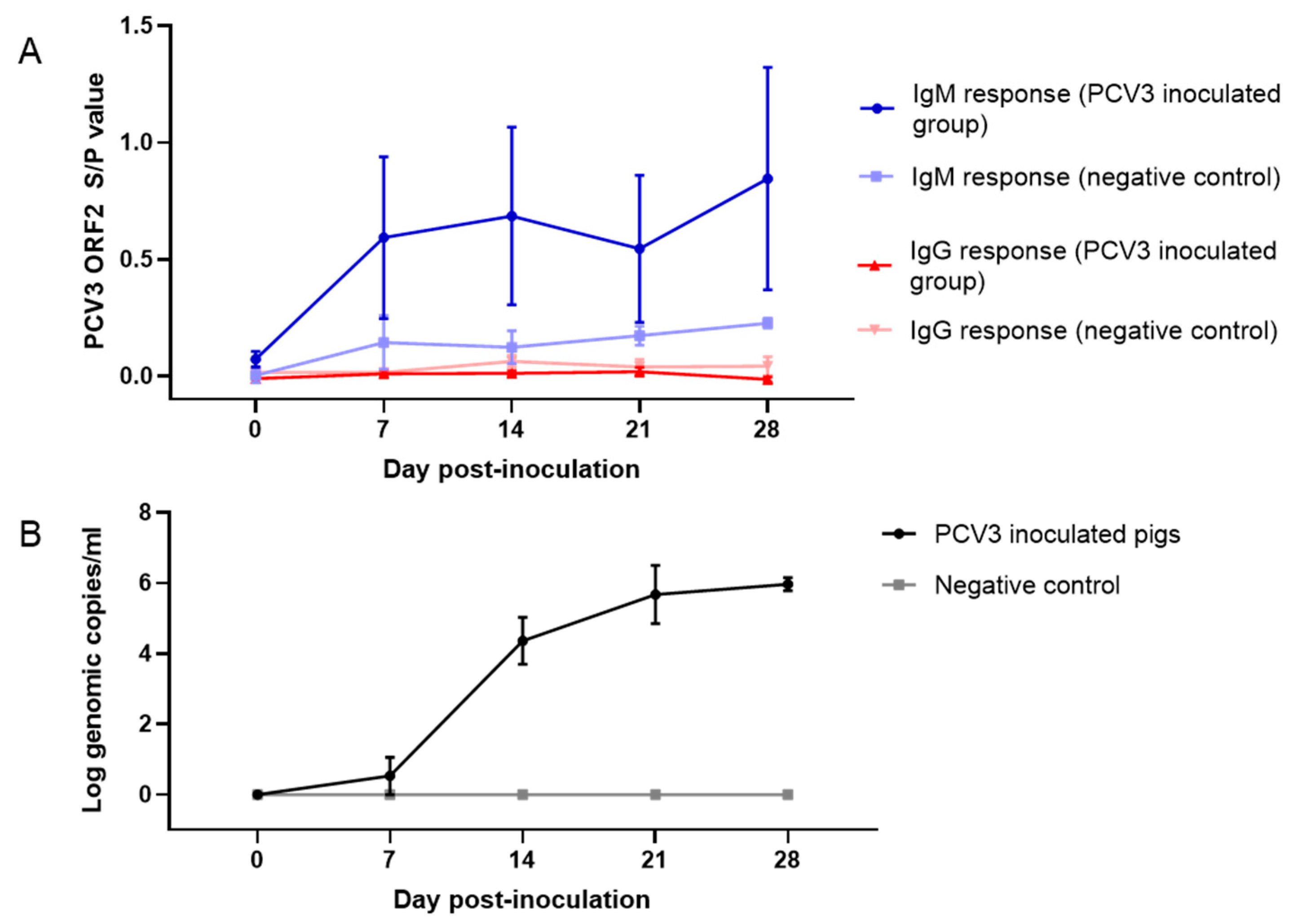
3.6. PCV3 Isolate Replicates in CD/CD Pigs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Breitbart, M.; Delwart, E.; Rosario, K.; Segales, J.; Varsani, A.; Ictv Report, C. ICTV Virus Taxonomy Profile: Circoviridae. J. Gen. Virol. 2017, 98, 1997–1998. [Google Scholar] [CrossRef]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A 1974, 226, 153–167. [Google Scholar] [PubMed]
- Segales, J.; Allan, G.M.; Domingo, M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005, 6, 119–142. [Google Scholar] [CrossRef]
- Allan, G.M.; Ellis, J.A. Porcine circoviruses: A review. J. Vet. Diagn. Investig. 2000, 12, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Langohr, I. Current state of knowledge on porcine circovirus type 2-associated lesions. Vet. Pathol. 2013, 50, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.; Pineyro, P.; Shang, P.; Yuan, F.; Guo, R.; Fang, Y.; Byers, E.; Hause, B.M. A Novel Porcine Circovirus Distantly Related to Known Circoviruses Is Associated with Porcine Dermatitis and Nephropathy Syndrome and Reproductive Failure. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Stadejek, T.; Wozniak, A.; Milek, D.; Biernacka, K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound. Emerg. Dis. 2017, 64, 1350–1353. [Google Scholar] [CrossRef]
- Kwon, T.; Yoo, S.J.; Park, C.K.; Lyoo, Y.S. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 2017, 207, 178–180. [Google Scholar] [CrossRef]
- Collins, P.J.; McKillen, J.; Allan, G. Porcine circovirus type 3 in the UK. Vet. Rec. 2017, 181, 599. [Google Scholar] [CrossRef] [PubMed]
- Faccini, S.; Barbieri, I.; Gilioli, A.; Sala, G.; Gibelli, L.R.; Moreno, A.; Sacchi, C.; Rosignoli, C.; Franzini, G.; Nigrelli, A. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound Emerg. Dis. 2017, 64, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Lumyai, M.; Kesdangsakonwut, S.; Teankum, K.; Jittimanee, S.; Thanawongnuwech, R. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Vet. Microbiol. 2018, 215, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Tochetto, C.; Lima, D.A.; Varela, A.P.M.; Loiko, M.R.; Paim, W.P.; Scheffer, C.M.; Herpich, J.I.; Cerva, C.; Schmitd, C.; Cibulski, S.P.; et al. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound. Emerg. Dis. 2018, 65, 5–9. [Google Scholar] [CrossRef]
- Ye, X.; Berg, M.; Fossum, C.; Wallgren, P.; Blomstrom, A.L. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes 2018, 54, 466–469. [Google Scholar] [CrossRef]
- Hayashi, S.; Ohshima, Y.; Furuya, Y.; Nagao, A.; Oroku, K.; Tsutsumi, N.; Sasakawa, C.; Sato, T. First detection of porcine circovirus type 3 in Japan. J. Vet. Med. Sci. 2018, 80, 1468–1472. [Google Scholar] [CrossRef]
- Yuzhakov, A.G.; Raev, S.A.; Alekseev, K.P.; Grebennikova, T.V.; Verkhovsky, O.A.; Zaberezhny, A.D.; Aliper, T.I. First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes 2018, 54, 608–611. [Google Scholar] [CrossRef]
- Franzo, G.; Legnardi, M.; Hjulsager, C.K.; Klaumann, F.; Larsen, L.E.; Segales, J.; Drigo, M. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound. Emerg. Dis. 2018, 65, 602–606. [Google Scholar] [CrossRef]
- Fux, R.; Sockler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G.; Ritzmann, M.; Eddicks, M. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018, 15, 25. [Google Scholar] [CrossRef]
- Vargas-Bermudez, D.S.; Campos, F.S.; Bonil, L.; Mogollon, D.; Jaime, J. First detection of porcine circovirus type 3 in Colombia and the complete genome sequence demonstrates the circulation of PCV3a1 and PCV3a2. Vet. Med. Sci. 2019, 5, 182–188. [Google Scholar] [CrossRef]
- Deim, Z.; Dencso, L.; Erdelyi, I.; Valappil, S.K.; Varga, C.; Posa, A.; Makrai, L.; Rakhely, G. Porcine circovirus type 3 detection in a Hungarian pig farm experiencing reproductive failures. Vet. Rec. 2019, 185, 84. [Google Scholar] [CrossRef] [PubMed]
- Savic, B.; Milicevic, V.; Radanovic, O.; Zdravkovic, N.; Stevancevic, O.; Kureljusic, B.; Nesic, K. Identification and genetic characterization of porcine circovirus 3 on pig farms in Serbia. Arch. Virol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Mi, S.; Luo, Q.; Guo, H.; Tu, C.; Zhu, G.; Gong, W. Retrospective study of porcine circovirus type 2 infection reveals a novel genotype PCV2f. Transbound Emerg. Dis. 2018, 65, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Wang, X.; Gao, Q.; Huan, C.; Wang, W.; Gao, S.; Liu, X. Retrospective survey and phylogenetic analysis of porcine circovirus type 3 in Jiangsu province, China, 2008 to 2017. Arch. Virol. 2018, 163, 2531–2538. [Google Scholar] [CrossRef] [PubMed]
- Sukmak, M.; Thanantong, N.; Poolperm, P.; Boonsoongnern, A.; Ratanavanichrojn, N.; Jirawattanapong, P.; Woonwong, Y.; Soda, N.; Kaminsonsakul, T.; Phuttapatimok, S.; et al. The retrospective identification and molecular epidemiology of porcine circovirus type 3 (PCV3) in swine in Thailand from 2006 to 2017. Transbound Emerg. Dis. 2018. [Google Scholar] [CrossRef]
- Fu, X.; Fang, B.; Ma, J.; Liu, Y.; Bu, D.; Zhou, P.; Wang, H.; Jia, K.; Zhang, G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg. Dis. 2018, 65, e296–e303. [Google Scholar] [CrossRef]
- Arruda, B.; Pineyro, P.; Derscheid, R.; Hause, B.; Byers, E.; Dion, K.; Long, D.; Sievers, C.; Tangen, J.; Williams, T.; et al. PCV3-associated disease in the United States swine herd. Emerg. Microbes Infect. 2019, 8, 684–698. [Google Scholar] [CrossRef]
- Ku, X.; Chen, F.; Li, P.; Wang, Y.; Yu, X.; Fan, S.; Qian, P.; Wu, M.; He, Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg. Dis. 2017, 64, 703–708. [Google Scholar] [CrossRef]
- Zhai, S.L.; Zhou, X.; Zhang, H.; Hause, B.M.; Lin, T.; Liu, R.; Chen, Q.L.; Wei, W.K.; Lv, D.H.; Wen, X.H.; et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol. J. 2017, 14, 222. [Google Scholar] [CrossRef]
- Shen, H.; Liu, X.; Zhang, P.; Wang, L.; Liu, Y.; Zhang, L.; Liang, P.; Song, C. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg. Dis. 2018, 65, 264–266. [Google Scholar] [CrossRef]
- Chen, G.H.; Tang, X.Y.; Sun, Y.; Zhou, L.; Li, D.; Bai, Y.; Mai, K.J.; Li, Y.Y.; Wu, Q.W.; Ma, J.Y. Development of a SYBR green-based real-time quantitative PCR assay to detect PCV3 in pigs. J. Virol. Methods 2018, 251, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Kedkovid, R.; Woonwong, Y.; Arunorat, J.; Sirisereewan, C.; Sangpratum, N.; Kesdangsakonwut, S.; Tummaruk, P.; Teankum, K.; Assavacheep, P.; Jittimanee, S.; et al. Porcine circovirus type 3 (PCV3) shedding in sow colostrum. Vet. Microbiol. 2018, 220, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg. Dis. 2017, 64, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Yu, S.; Gallup, J.M.; Evans, R.B.; Fenaux, M.; Pallares, F.; Thacker, E.L.; Brockus, C.W.; Ackermann, M.R.; Thomas, P.; et al. Effect of vaccination with selective bacterins on conventional pigs infected with type 2 porcine circovirus. Vet. Pathol. 2003, 40, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, Y.; Xia, X.Q.; Chen, Q.; Bade, S.A.; Yoon, K.J.; Harmon, K.M.; Gauger, P.C.; Main, R.G.; Li, G. High-throughput whole genome sequencing of Porcine reproductive and respiratory syndrome virus from cell culture materials and clinical specimens using next-generation sequencing technology. J. Vet. Diagn. Investig. 2017, 29, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, L.; Zheng, Y.; Zhang, J.; Guo, B.; Yoon, K.J.; Gauger, P.C.; Harmon, K.M.; Main, R.G.; Li, G. Metagenomic analysis of the RNA fraction of the fecal virome indicates high diversity in pigs infected by porcine endemic diarrhea virus in the United States. Virol. J. 2018, 15, 95. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Li, G.; He, W.; Zhu, H.; Bi, Y.; Wang, R.; Xing, G.; Zhang, C.; Zhou, J.; Yuen, K.Y.; Gao, G.F.; et al. Origin, Genetic Diversity, and Evolutionary Dynamics of Novel Porcine Circovirus 3. Adv. Sci. (Weinh) 2018, 5, 1800275. [Google Scholar] [CrossRef] [PubMed]
- Hamel, A.L.; Lin, L.L.; Nayar, G.P. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 1998, 72, 5262–5267. [Google Scholar] [CrossRef] [PubMed]
- Larochelle, R.; Magar, R.; D’Allaire, S. Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) strains from cases presenting various clinical conditions. Virus Res. 2002, 90, 101–112. [Google Scholar] [CrossRef]
- Meehan, B.M.; McNeilly, F.; Todd, D.; Kennedy, S.; Jewhurst, V.A.; Ellis, J.A.; Hassard, L.E.; Clark, E.G.; Haines, D.M.; Allan, G.M. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 1998, 79 Pt 9, 2171–2179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Y.; Chen, Q.X.; Ye, J.X.; Shen, H.G.; Chen, T.F.; Shang, S.B. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet. Res. Commun. 2006, 30, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.; Milek, D.; Baska, P.; Stadejek, T. Does porcine circovirus type 3 (PCV3) interfere with porcine circovirus type 2 (PCV2) vaccine efficacy? Transbound Emerg. Dis. 2019, 66, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Tischer, I.; Peters, D.; Rasch, R.; Pociuli, S. Replication of porcine circovirus: Induction by glucosamine and cell cycle dependence. Arch. Virol. 1987, 96, 39–57. [Google Scholar] [CrossRef]
- Zhu, Y.; Lau, A.; Lau, J.; Jia, Q.; Karuppannan, A.K.; Kwang, J. Enhanced replication of porcine circovirus type 2 (PCV2) in a homogeneous subpopulation of PK15 cell line. Virology 2007, 369, 423–430. [Google Scholar] [CrossRef]
- Meerts, P.; Misinzo, G.; McNeilly, F.; Nauwynck, H.J. Replication kinetics of different porcine circovirus 2 strains in PK-15 cells, fetal cardiomyocytes and macrophages. Arch. Virol. 2005, 150, 427–441. [Google Scholar] [CrossRef]
- Fenaux, M.; Opriessnig, T.; Halbur, P.G.; Meng, X.J. Immunogenicity and pathogenicity of chimeric infectious DNA clones of pathogenic porcine circovirus type 2 (PCV2) and nonpathogenic PCV1 in weanling pigs. J. Virol. 2003, 77, 11232–11243. [Google Scholar] [CrossRef]
- Beach, N.M.; Juhan, N.M.; Cordoba, L.; Meng, X.J. Replacement of the replication factors of porcine circovirus (PCV) type 2 with those of PCV type 1 greatly enhances viral replication in vitro. J. Virol. 2010, 84, 8986–8989. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fenaux, M.; Opriessnig, T.; Halbur, P.G.; Elvinger, F.; Meng, X.J. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J. Virol. 2004, 78, 13440–13446. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, J.; Melnick, J.L.; Rawls, W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 1968, 2, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Young, D.F.; Andrejeva, L.; Livingstone, A.; Goodbourn, S.; Lamb, R.A.; Collins, P.L.; Elliott, R.M.; Randall, R.E. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 2003, 77, 2174–2181. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.E.; Randall, R.E.; Adamson, C.S. Inhibitors of the interferon response enhance virus replication in vitro. PLoS ONE 2014, 9, e112014. [Google Scholar] [CrossRef] [PubMed]
- Walia, R.; Dardari, R.; Chaiyakul, M.; Czub, M. Porcine circovirus-2 capsid protein induces cell death in PK15 cells. Virology 2014, 468–470, 126–132. [Google Scholar] [CrossRef]
- Ma, T.; Ouyang, T.; Ouyang, H.; Chen, F.; Peng, Z.; Chen, X.; Pang, D.; Ren, L. Porcine circovirus 2 proliferation can be enhanced by stably expressing porcine IL-2 gene in PK-15 cell. Virus Res. 2017, 227, 143–149. [Google Scholar] [CrossRef]
- Franzo, G.; He, W.; Correa-Fiz, F.; Li, G.; Legnardi, M.; Su, S.; Segales, J. A Shift in Porcine Circovirus 3 (PCV-3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv. Sci. (Weinh) 2019, 6, 1901004. [Google Scholar] [CrossRef]
- Ladekjaer-Mikkelsen, A.S.; Nielsen, J.; Stadejek, T.; Storgaard, T.; Krakowka, S.; Ellis, J.; McNeilly, F.; Allan, G.; Botner, A. Reproduction of postweaning multisystemic wasting syndrome (PMWS) in immunostimulated and non-immunostimulated 3-week-old piglets experimentally infected with porcine circovirus type 2 (PCV2). Vet. Microbiol. 2002, 89, 97–114. [Google Scholar] [CrossRef]
- Opriessnig, T.; Patterson, A.R.; Elsener, J.; Meng, X.J.; Halbur, P.G. Influence of maternal antibodies on efficacy of porcine circovirus type 2 (PCV2) vaccination to protect pigs from experimental infection with PCV2. Clin. Vaccine Immunol. 2008, 15, 397–401. [Google Scholar] [CrossRef]
- Meerts, P.; Misinzo, G.; Lefebvre, D.; Nielsen, J.; Botner, A.; Kristensen, C.S.; Nauwynck, H.J. Correlation between the presence of neutralizing antibodies against porcine circovirus 2 (PCV2) and protection against replication of the virus and development of PCV2-associated disease. BMC Vet. Res. 2006, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Thacker, E.L.; Yu, S.; Fenaux, M.; Meng, X.J.; Halbur, P.G. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet. Pathol. 2004, 41, 624–640. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Meng, X.J.; Halbur, P.G. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007, 19, 591–615. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Halbur, P.G. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012, 164, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, M.; Halbur, P.G.; Haqshenas, G.; Royer, R.; Thomas, P.; Nawagitgul, P.; Gill, M.; Toth, T.E.; Meng, X.J. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: Characterization of clinical disease, virus distribution, and pathologic lesions. J. Virol. 2002, 76, 541–551. [Google Scholar] [CrossRef]


| Isolate and Original Tissue | Case | GenBank Accession # | PCV Differential qPCR from Original Tissue | PCV3 qPCR Results 4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCV1 | PCV2 | PCV3 | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | |||
| ISU58312 1 | 1 | MK568469 | ||||||||||||
| Heart, liver, kidney homogenate | ≥37.0 | ≥37.0 | 14.4 | 19.1 | 22.3 | 23.7 | 27.1 5 | NP | NP | NP | NP | NP | ||
| Brain | ≥37.0 | ≥37.0 | 7.5 | 12.3 | 15.5 | 17.5 | 20.9 5 | NP | NP | NP | NP | NP | ||
| ISU44806 2 | 2 | MK568470 | ||||||||||||
| Lung, heart homogenate | ≥37.0 | ≥37.0 | 9.4 | 14.1 | 16.8 | 18.5 | 21.5 5 | NP | NP | NP | NP | NP | ||
| Kidney | ≥37.0 | ≥37.0 | 12.5 | 16.7 | 19.6 | 21.1 | 24.1 5 | NP | NP | NP | NP | NP | ||
| ISU27734 3 | 3 | MK058528, MK058429 | ||||||||||||
| Lung | ≥37.0 | ≥37.0 | 13.5 | 14.1 5 | 16.1 | 17.8 5 | 19.1 | 22.2 | 24.1 5 | 25.4 | 27.7 | 29.9 5 | ||
| Brain | ≥37.0 | ≥37.0 | 13.3 | 19.8 | 22.9 | 25.3 | 28.6 | 33.5 | ≥37.0 | ≥37.0 | - | - | ||
| Liver | ≥37.0 | ≥37.0 | 19.3 | 26.8 | 30.7 | ≥37.0 | ≥37.0 | - | - | - | - | - | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora-Díaz, J.; Piñeyro, P.; Shen, H.; Schwartz, K.; Vannucci, F.; Li, G.; Arruda, B.; Giménez-Lirola, L. Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs. Viruses 2020, 12, 219. https://doi.org/10.3390/v12020219
Mora-Díaz J, Piñeyro P, Shen H, Schwartz K, Vannucci F, Li G, Arruda B, Giménez-Lirola L. Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs. Viruses. 2020; 12(2):219. https://doi.org/10.3390/v12020219
Chicago/Turabian StyleMora-Díaz, Juan, Pablo Piñeyro, Huigang Shen, Kent Schwartz, Fabio Vannucci, Ganwu Li, Bailey Arruda, and Luis Giménez-Lirola. 2020. "Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs" Viruses 12, no. 2: 219. https://doi.org/10.3390/v12020219
APA StyleMora-Díaz, J., Piñeyro, P., Shen, H., Schwartz, K., Vannucci, F., Li, G., Arruda, B., & Giménez-Lirola, L. (2020). Isolation of PCV3 from Perinatal and Reproductive Cases of PCV3-Associated Disease and In Vivo Characterization of PCV3 Replication in CD/CD Growing Pigs. Viruses, 12(2), 219. https://doi.org/10.3390/v12020219








