Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA-Extraction and Sequencing
2.2. N. vectensis Transcriptome Datasets
2.3. Raw Reads Processing and Filtering
2.4. Sequence Assembly and Viral Sequence Identification
2.5. Taxonomic Annotation
2.6. Reads Quantification
2.7. Validation of Candidate Viruses
2.8. Statistical Analysis
3. Results
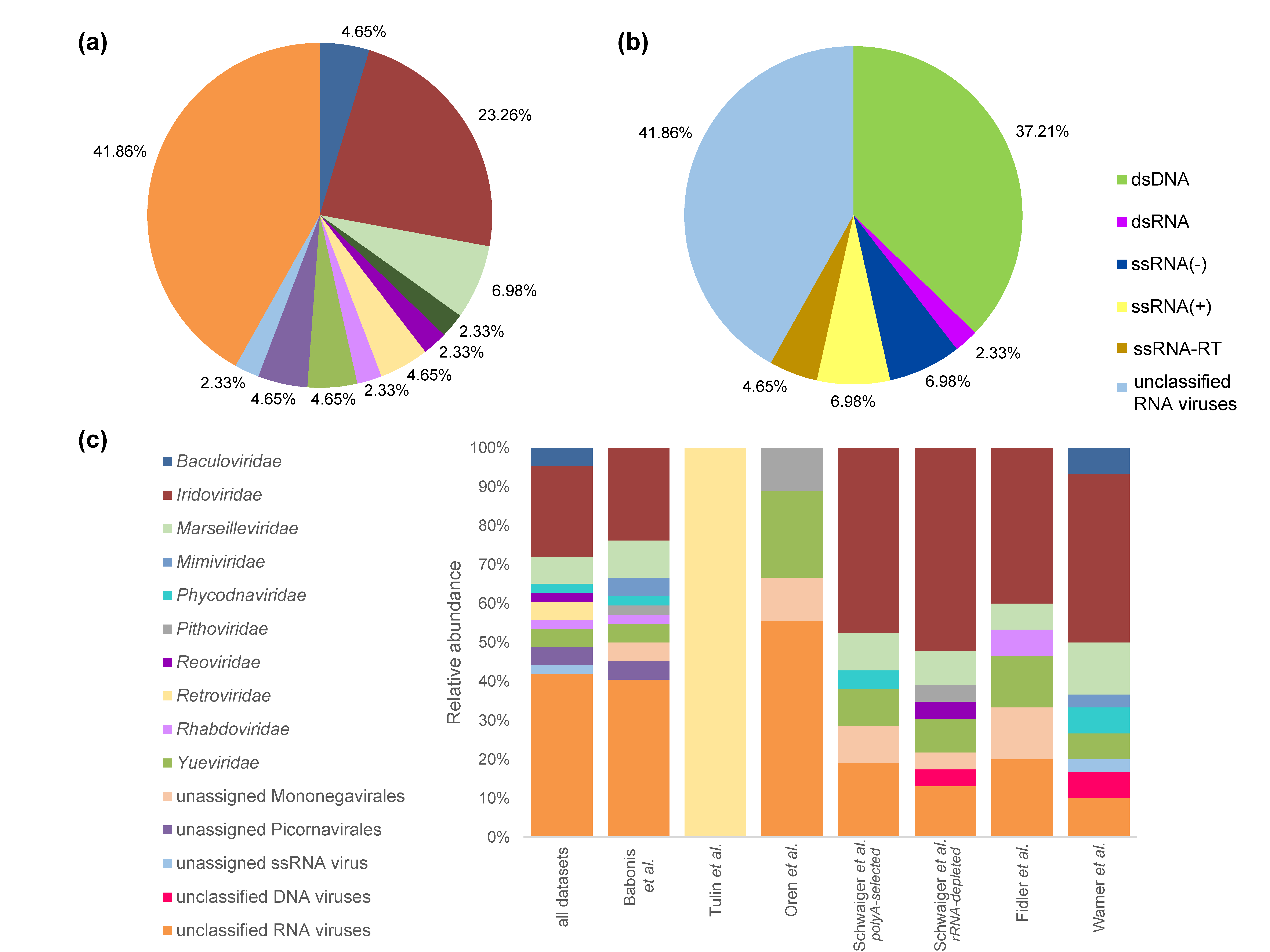
3.1. Viral Community Classification
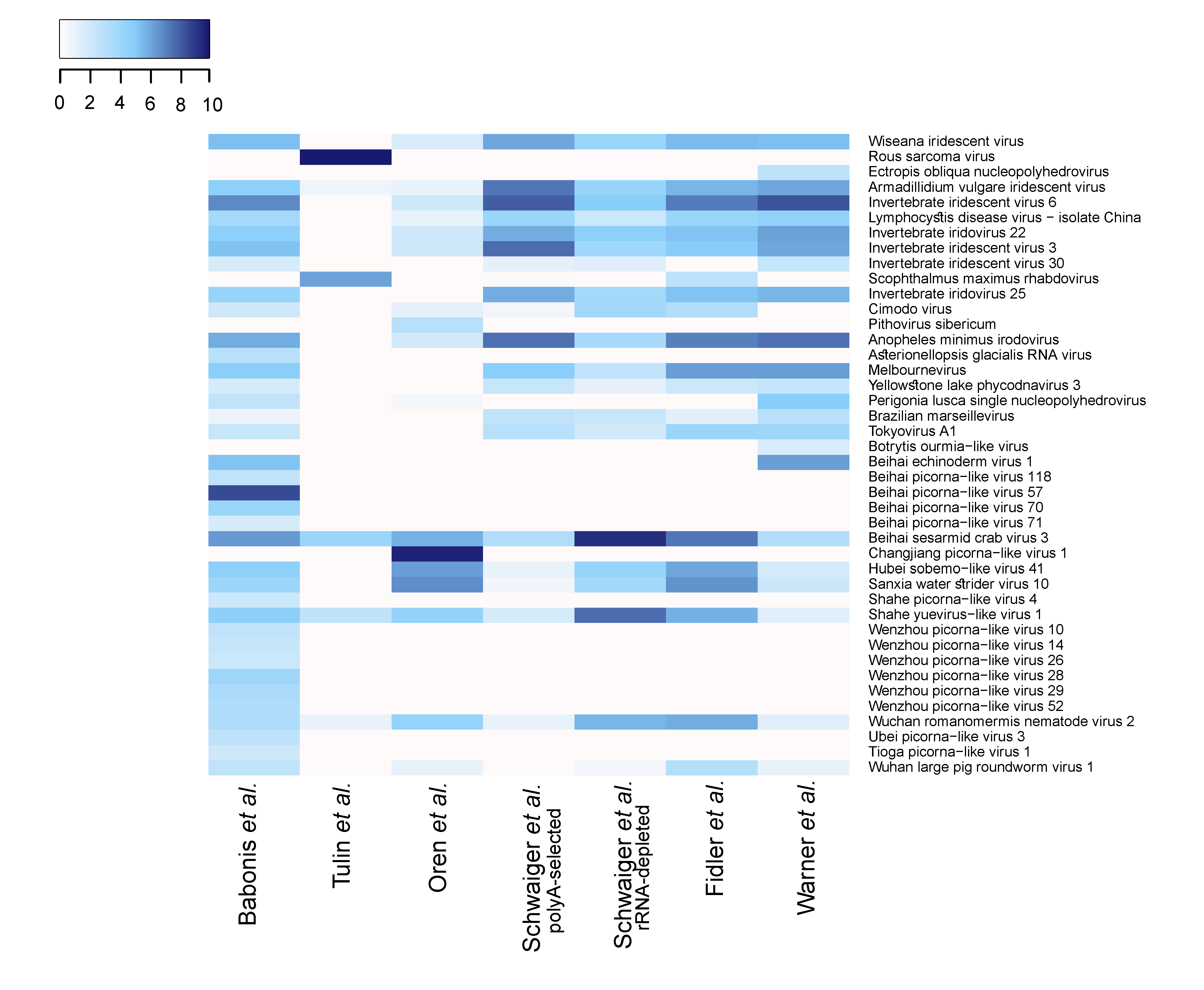
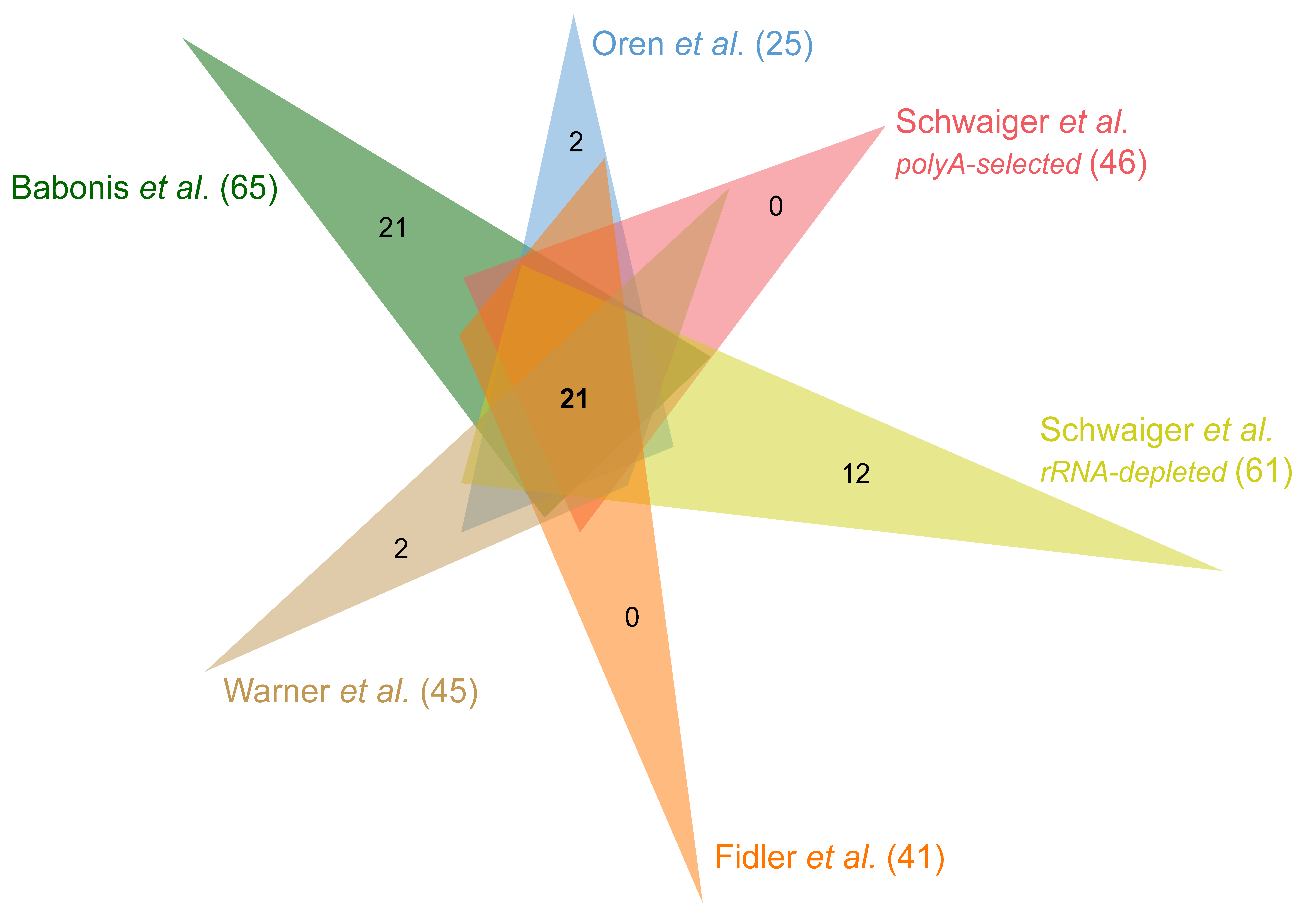
3.2. N. vectensis Core Virome
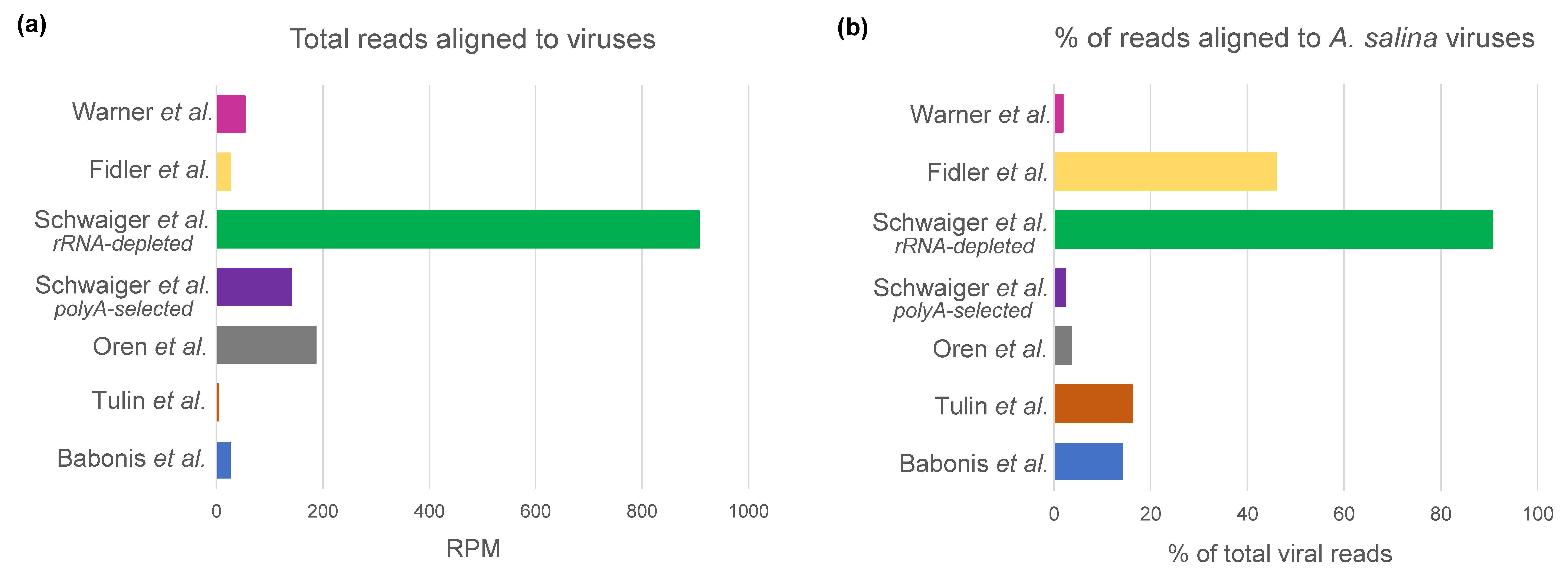
3.3. Interpopulation Comparison
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Grasis, J.A.; Lachnit, T.; Anton-Erxleben, F.; Lim, Y.W.; Schmieder, R.; Fraune, S.; Franzenburg, S.; Insua, S.; Machado, G.; Haynes, M.; et al. Species-specific viromes in the ancestral holobiont Hydra. PLoS ONE 2014, 9, e109952. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Liu, L.; Gong, T.; Tao, W.; Lin, B.; Li, C.; Zheng, X.; Zhu, S.; Jiang, W.; Zhou, R. Commensal viruses maintain intestinal intraepithelial lymphocytes via noncanonical RIG-I signaling. Nat. Immunol. 2019, 20, 1681–1691. [Google Scholar] [CrossRef]
- Vu, D.L.; Kaiser, L. The concept of commensal viruses almost 20 years later: Redefining borders in clinical virology. Clin. Microbiol. Infect. 2017, 23, 688–690. [Google Scholar] [CrossRef]
- Barton, E.S.; White, D.W.; Cathelyn, J.S.; Brett-McClellan, K.A.; Engle, M.; Diamond, M.S.; Miller, V.L.; Virgin, H.W. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 2007, 447, 326–329. [Google Scholar] [CrossRef]
- Margulis, L. Words as battle cries—Symbiogenesis and the new field of endocytobiology. Bioscience 1990, 40, 673–677. [Google Scholar] [CrossRef]
- Grasis, J.A. The Intra-Dependence of Viruses and the Holobiont. Front. Immunol. 2017, 8, 1501. [Google Scholar] [CrossRef]
- Layden, M.J.; Rentzsch, F.; Rottinger, E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 408–428. [Google Scholar] [CrossRef]
- Technau, U.; Steele, R.E. Evolutionary crossroads in developmental biology: Cnidaria. Development 2011, 138, 1447–1458. [Google Scholar] [CrossRef]
- Zapata, F.; Goetz, F.E.; Smith, S.A.; Howison, M.; Siebert, S.; Church, S.H.; Sanders, S.M.; Ames, C.L.; McFadden, C.S.; France, S.C.; et al. Phylogenomic Analyses Support Traditional Relationships within Cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef]
- Thurber, R.V.; Payet, J.P.; Thurber, A.R.; Correa, A.M.S. Virus–host interactions and their roles in coral reef health and disease. Nat. Rev. Microbiol. 2017, 15, 205–216. [Google Scholar] [CrossRef]
- Bruwer, J.D.; Agrawal, S.; Liew, Y.J.; Aranda, M.; Voolstra, C.R. Association of coral algal symbionts with a diverse viral community responsive to heat shock. BMC Microbiol. 2017, 17, 174. [Google Scholar] [CrossRef]
- Levin, R.A.; Voolstra, C.R.; Weynberg, K.D.; van Oppen, M.J. Evidence for a role of viruses in the thermal sensitivity of coral photosymbionts. ISME J. 2017, 11, 808–812. [Google Scholar] [CrossRef]
- Bruwer, J.D.; Voolstra, C.R. First insight into the viral community of the cnidarian model metaorganism Aiptasia using RNA-Seq data. PeerJ 2018, 6, e4449. [Google Scholar] [CrossRef]
- Soffer, N.; Brandt, M.E.; Correa, A.M.; Smith, T.B.; Thurber, R.V. Potential role of viruses in white plague coral disease. ISME J. 2014, 8, 271–283. [Google Scholar] [CrossRef]
- Weynberg, K.D.; Voolstra, C.R.; Neave, M.J.; Buerger, P.; van Oppen, M.J. From cholera to corals: Viruses as drivers of virulence in a major coral bacterial pathogen. Sci. Rep. 2015, 5, 17889. [Google Scholar] [CrossRef]
- Marhaver, K.L.; Edwards, R.A.; Rohwer, F. Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 2008, 10, 2277–2286. [Google Scholar] [CrossRef]
- Vega Thurber, R.L.; Barott, K.L.; Hall, D.; Liu, H.; Rodriguez-Mueller, B.; Desnues, C.; Edwards, R.A.; Haynes, M.; Angly, F.E.; Wegley, L.; et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. USA 2008, 105, 18413–18418. [Google Scholar] [CrossRef]
- Correa, A.M.; Ainsworth, T.D.; Rosales, S.M.; Thurber, A.R.; Butler, C.R.; Vega Thurber, R.L. Viral Outbreak in Corals Associated with an In Situ Bleaching Event: Atypical Herpes-Like Viruses and a New Megavirus Infecting Symbiodinium. Front. Microbiol. 2016, 7, 127. [Google Scholar] [CrossRef]
- Leach, W.B.; Carrier, T.J.; Reitzel, A.M. Diel patterning in the bacterial community associated with the sea anemone Nematostella vectensis. Ecol. Evol. 2019, 9, 9935–9947. [Google Scholar] [CrossRef]
- Mortzfeld, B.M.; Urbanski, S.; Reitzel, A.M.; Kunzel, S.; Technau, U.; Fraune, S. Response of bacterial colonization in Nematostella vectensis to development, environment and biogeography. Environ. Microbiol. 2016, 18, 1764–1781. [Google Scholar] [CrossRef]
- Har, J.Y.; Helbig, T.; Lim, J.H.; Fernando, S.C.; Reitzel, A.M.; Penn, K.; Thompson, J.R. Microbial diversity and activity in the Nematostella vectensis holobiont: Insights from 16S rRNA gene sequencing, isolate genomes, and a pilot-scale survey of gene expression. Front. Microbiol. 2015, 6, 818. [Google Scholar] [CrossRef]
- Felix, M.A.; Ashe, A.; Piffaretti, J.; Wu, G.; Nuez, I.; Belicard, T.; Jiang, Y.; Zhao, G.; Franz, C.J.; Goldstein, L.D.; et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011, 9, e1000586. [Google Scholar] [CrossRef]
- Carbonell, A.; Carrington, J.C. Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef]
- Hand, C.; Uhlinger, K.R. The Culture, Sexual and Asexual Reproduction, and Growth of the Sea Anemone Nematostella vectensis. Biol. Bull. 1992, 182, 169–176. [Google Scholar] [CrossRef]
- Stefanik, D.J.; Friedman, L.E.; Finnerty, J.R. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 2013, 8, 916–923. [Google Scholar] [CrossRef]
- McCarthy, S.D.; Dugon, M.M.; Power, A.M. ‘Degraded’ RNA profiles in Arthropoda and beyond. PeerJ 2015, 3, e1436. [Google Scholar] [CrossRef]
- Babonis, L.S.; Martindale, M.Q.; Ryan, J.F. Do novel genes drive morphological novelty? An investigation of the nematosomes in the sea anemone Nematostella vectensis. BMC Evol. Biol. 2016, 16, 114. [Google Scholar] [CrossRef]
- Tulin, S.; Aguiar, D.; Istrail, S.; Smith, J. A quantitative reference transcriptome for Nematostella vectensis early embryonic development: A pipeline for de novo assembly in emerging model systems. EvoDevo 2013, 4, 16. [Google Scholar] [CrossRef]
- Oren, M.; Tarrant, A.M.; Alon, S.; Simon-Blecher, N.; Elbaz, I.; Appelbaum, L.; Levy, O. Profiling molecular and behavioral circadian rhythms in the non-symbiotic sea anemone Nematostella vectensis. Sci. Rep. 2015, 5, 11418. [Google Scholar] [CrossRef]
- Schwaiger, M.; Schonauer, A.; Rendeiro, A.F.; Pribitzer, C.; Schauer, A.; Gilles, A.F.; Schinko, J.B.; Renfer, E.; Fredman, D.; Technau, U. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 2014, 24, 639–650. [Google Scholar] [CrossRef]
- Fidler, A.L.; Vanacore, R.M.; Chetyrkin, S.V.; Pedchenko, V.K.; Bhave, G.; Yin, V.P.; Stothers, C.L.; Rose, K.L.; McDonald, W.H.; Clark, T.A.; et al. A unique covalent bond in basement membrane is a primordial innovation for tissue evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 331–336. [Google Scholar] [CrossRef]
- Warner, J.F.; Guerlais, V.; Amiel, A.R.; Johnston, H.; Nedoncelle, K.; Rottinger, E. NvERTx: A gene expression database to compare embryogenesis and regeneration in the sea anemone Nematostella vectensis. Development 2018, 145. [Google Scholar] [CrossRef]
- Andrew, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 July 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef]
- RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019, 47, D221–D229. [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, N.; Aljanahi, A.; Nandakumar, S.; Mikailov, M.; Khan, A.S. A Reference Viral Database (RVDB) To Enhance Bioinformatics Analysis of High-Throughput Sequencing for Novel Virus Detection. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Romiguier, J.; Gayral, P.; Ballenghien, M.; Bernard, A.; Cahais, V.; Chenuil, A.; Chiari, Y.; Dernat, R.; Duret, L.; Faivre, N.; et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 2014, 515, 261–263. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- RStudio: Integrated Development Environment for R; RStudio: Boston, MA, USA, 2016.
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV Virus Taxonomy Profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 2008, 153, 715–727. [Google Scholar] [CrossRef]
- Majerciak, V.; Ni, T.; Yang, W.; Meng, B.; Zhu, J.; Zheng, Z.M. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013, 9, e1003749. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef]
- He, M.; Jiang, Z.; Li, S.; He, P. Presence of poly(A) tails at the 3’-termini of some mRNAs of a double-stranded RNA virus, southern rice black-streaked dwarf virus. Viruses 2015, 7, 1642–1650. [Google Scholar] [CrossRef]
- Slomovic, S.; Fremder, E.; Staals, R.H.G.; Pruijn, G.J.M.; Schuster, G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc. Natl. Acad. Sci. USA 2010, 107, 7407–7412. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Zhang, C.; Pei, X.; Wang, Z.; Jia, S. Presence of poly(A) and poly(A)-rich tails in a positive-strand RNA virus known to lack 3 poly(A) tails. Virology 2014, 454–455, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vierna, J.; Wehner, S.; Honer zu Siederdissen, C.; Martinez-Lage, A.; Marz, M. Systematic analysis and evolution of 5S ribosomal DNA in metazoans. Heredity 2013, 111, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Waldron, F.M.; Stone, G.N.; Obbard, D.J. Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PLoS Genet. 2018, 14, e1007533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, Y.-H.; Wang, X.; Zhang, H.-X.; Lin, Q. The mitochondrial genome of a sea anemone Bolocera sp. exhibits novel genetic structures potentially involved in adaptation to the deep-sea environment. Ecol. Evol. 2017, 7, 4951–4962. [Google Scholar] [CrossRef]
- Muller, E.M.; Fine, M.; Ritchie, K.B. The stable microbiome of inter and sub-tidal anemone species under increasing pCO(2). Sci. Rep. 2016, 6, 37387. [Google Scholar] [CrossRef]
- Hand, C.; Uhlinger, K.R. The Unique, Widely Distributed, Estuarine Sea-Anemone, Nematostella vectensis Stephenson—A Review, New Facts, and Questions. Estuaries 1994, 17, 501–508. [Google Scholar] [CrossRef]
- Heck, K.L.; Able, K.W.; Roman, C.T.; Fahay, M.P. Composition, abundance, biomass, and production of macrofauna in a New England estuary: Comparisons among eelgrass meadows and other nursery habitats. Estuaries 1995, 18, 379–389. [Google Scholar] [CrossRef]
- Tietjen, J.H. The ecology of shallow water meiofauna in two New England estuaries. Oecologia 1969, 2, 251–291. [Google Scholar] [CrossRef]
- Wood-Charlson, E.M.; Weynberg, K.D.; Suttle, C.A.; Roux, S.; van Oppen, M.J.H. Metagenomic characterization of viral communities in corals: Mining biological signal from methodological noise. Environ. Microbiol. 2015, 17, 3440–3449. [Google Scholar] [CrossRef]
- Weynberg, K.D.; Laffy, P.W.; Wood-Charlson, E.M.; Turaev, D.; Rattei, T.; Webster, N.S.; van Oppen, M.J.H. Coral-associated viral communities show high levels of diversity and host auxiliary functions. PeerJ 2017, 5, e4054. [Google Scholar] [CrossRef]
- Thornhill, D.J.; Xiang, Y.; Pettay, D.T.; Zhong, M.; Santos, S.R. Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 2013, 22, 4499–4515. [Google Scholar] [CrossRef] [PubMed]
- Habayeb, M.S.; Ekengren, S.K.; Hultmark, D. Nora virus, a persistent virus in Drosophila, defines a new picorna-like virus family. J. Gen. Virol. 2006, 87, 3045–3051. [Google Scholar] [CrossRef] [PubMed]
- Balla, K.M.; Rice, M.C.; Gagnon, J.A.; Elde, N.C. Discovery of a prevalent picornavirus by visualizing zebrafish immune responses. bioRxiv 2019. [Google Scholar] [CrossRef]
- Weiss, R.A.; Vogt, P.K. 100 years of Rous sarcoma virus. J. Exp. Med. 2011, 208, 2351–2355. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Hayward, D.C.; Cooke, I.; Wang, W.; Moya, A.; Siemering, K.R.; Sprungala, S.; Ball, E.E.; Foret, S.; Miller, D.J. The Whole-Genome Sequence of the Coral Acropora millepora. Genome Biol. Evol. 2019, 11, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Hwang, D.S.; Lee, J.S.; Song, J.I.; Seo, T.K.; Won, Y.J. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol. Phylogenet. Evol. 2012, 62, 329–345. [Google Scholar] [CrossRef] [PubMed]
| Reference | No. of Samples | Raw Read Pairs | Retained Reads | Total Number of Contigs | Identified Viral Sequences |
|---|---|---|---|---|---|
| Babonis et al. 2016 † [31] | 9 | 364,726,242 | 18,134,716 | 99,241 | 76 |
| Tulin et al. 2013 a [32] | 6 | 112,159,243 | 5,251,525 | 7631 | 6 |
| Oren et al. 2015 a [33] | 13 | 105,403,849 | 6,805,643 | 10,968 | 12 |
| Shwaiger et al. 2014 a polyA-selected [34] | 8 | 532,867,635 | 75,383,380 | 40,225 | 50 |
| Shwaiger et al. 2014 b rRNA-depleted [34] | 2 | 155,212,236 | 23,904,647 | 6470 | 62 |
| Fidler et al. 2014 a [35] | 1 | 95,331,053 | 6,988,326 | 9902 | 25 |
| Warner et al. 2018 a [36] | 15 | 542,474,332 | 35,925,820 | 24,934 | 73 |
| All datasets merged | 54 | 1,908,174,590 | 172,394,057 | 155,322 | 94 |
| Babonis et al. [31] | Tulin et al. [32] | Oren et al. [33] | Schwaiger et al. polyA-selected [34] | Schwaiger et al. rRNA-depleted [34] | Fidler et al. [35] | Warner et al. [36] | |
|---|---|---|---|---|---|---|---|
| All reads aligned to viruses | 9544 | 388 | 19,676 | 74,721 | 140,782 | 2429 | 28,732 |
| SDs from mean | −0.5133 | −0.5837 | −0.0156 | −0.1597 | 2.218 | −0.5154 | −0.4302 |
| Reads aligned to A. salina viruses | 1362 | 63 | 721 | 1783 | 127,633 | 1116 | 537 |
| SDs from mean | −0.3805 | −0.3908 | −0.3705 | −0.3818 | 2.2676 | −0.3547 | −0.3894 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewandowska, M.; Hazan, Y.; Moran, Y. Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis. Viruses 2020, 12, 218. https://doi.org/10.3390/v12020218
Lewandowska M, Hazan Y, Moran Y. Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis. Viruses. 2020; 12(2):218. https://doi.org/10.3390/v12020218
Chicago/Turabian StyleLewandowska, Magda, Yael Hazan, and Yehu Moran. 2020. "Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis" Viruses 12, no. 2: 218. https://doi.org/10.3390/v12020218
APA StyleLewandowska, M., Hazan, Y., & Moran, Y. (2020). Initial Virome Characterization of the Common Cnidarian Lab Model Nematostella vectensis. Viruses, 12(2), 218. https://doi.org/10.3390/v12020218






