Abstract
During the past two decades, three zoonotic coronaviruses have been identified as the cause of large-scale disease outbreaks–Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and Swine Acute Diarrhea Syndrome (SADS). SARS and MERS emerged in 2003 and 2012, respectively, and caused a worldwide pandemic that claimed thousands of human lives, while SADS struck the swine industry in 2017. They have common characteristics, such as they are all highly pathogenic to humans or livestock, their agents originated from bats, and two of them originated in China. Thus, it is highly likely that future SARS- or MERS-like coronavirus outbreaks will originate from bats, and there is an increased probability that this will occur in China. Therefore, the investigation of bat coronaviruses becomes an urgent issue for the detection of early warning signs, which in turn minimizes the impact of such future outbreaks in China. The purpose of the review is to summarize the current knowledge on viral diversity, reservoir hosts, and the geographical distributions of bat coronaviruses in China, and eventually we aim to predict virus hotspots and their cross-species transmission potential.
1. Introduction
Fifteen years after the first highly pathogenic human coronavirus caused the severe acute respiratory syndrome coronavirus (SARS-CoV) outbreak, another severe acute diarrhea syndrome coronavirus (SADS-CoV) devastated livestock production by causing fatal diseases in pigs. Both outbreaks began in China and were caused by coronaviruses of bat origin [1,2]. This increased the urgency to study bat coronaviruses in China to understand their potential of causing another virus outbreak.
In this review, we collected information from past epidemiology studies on bat coronaviruses in China, including the virus species identified, their host species, and their geographical distributions. We also discuss the future prospects of bat coronaviruses cross-species transmission and spread in China.
4. A SADS-CoV Model of Prediction and Other Hotspot Viruses
To predict the next CoV that will cause a virus outbreak in future, we list the general factors that may contribute to this outbreak. Firstly, bats host a large number of highly diverse CoVs. It is known that CoV genomes regularly undergo recombination during infection, and a rich gene pool can facilitate this process. Secondly, bat species are widely distributed and live close to humans. Thirdly, the viruses are pathogenic and transmissible. In this context, SADS-CoV and SARS-CoV outbreaks in China are not unexpected. By this model, there are other CoVs that have not yet caused virus outbreaks but should be monitored.
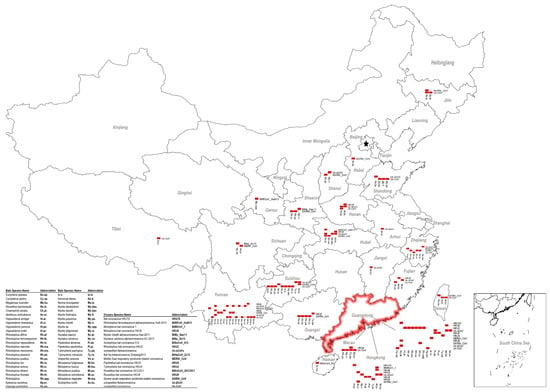
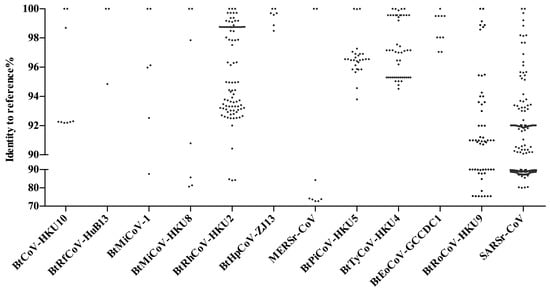
Within the family Vespertilionidae, the mouse-eared bats (Myotis) which favor roosting in abandoned human facilities are also a widespread genus of bats besides Pipistrellus bats. They carry a large number and genetically diversified HKU6-CoVs that are closely related to Myotis ricketti α-CoV Sax-2011 [36,38]. Moreover, bent-winged bats (Miniopterus spp.) carry a large variety of α-CoVs. One of the most frequently detected viruses is HKU8-CoV, which was first described circulating in M. pusillus in Hong Kong in 2005. Later, it was also found in M. magnate, M. fuliginosus, and M. schreibersii in Hong Kong, Guangdong, Yunnan, Fujian, and Hubei provinces, showing a great genetic diversity [32,33,34,35,37,41,60] (Figure 1). Besides HKU8-CoVs, bent-winged bats (Miniopterus spp.) also harbor a large amount of Miniopterus bat CoV 1 (BtMiCoV-1), which were called CoV1A or CoV1B previously. This viral species was found almost as frequently as HKU8-CoV in multiple provinces in China in Miniopterus bats, although these viruses showed a relatively small sequence variation between each other [32,33,34,35,37,41,60]. Genetic analysis indicates that BtMiCoV-1, HKU8-CoV, and HKU7-CoV (previous name) are different but closely related CoVs circulating in bent-winged bats and may have descended from a common ancestor [34]. Additionally, Rousettus leschenaultii bats in the family of Pteropodidae harbor HKU9-CoVs. As a fruit bat, Rousettus leschenaultii has a wider flying range than most of the insectivorous bats in China, thus it may carry viruses over long distances. A comparison of the reported HKU9-CoV sequences showed a high genetic diversity within this viral species [55,56,57] (Figure 2). The last CoV that should be mentioned is HKU10-CoV. HKU10-CoVs can be found in bats from different genera (Rousettus leschenaultii and Hipposideros pomona), suggesting interspecies transmission between bats [7,26,27,39]. A genetic difference can also be observed for this virus species (Figure 2). Above all, these viruses fit well in our SADS prediction model and should be monitored in our future studies.
5. Other Bat CoVs in China
In 2016, a novel β-CoV, Ro-BatCoVGCCDC1, was identified from the Rousettus leschenaultii bat. However, we confirmed the host was a closely related Eonycteris spelaea bat upon species identification and then renamed the virus as BtEoCoV-GCCDC1 (Table 1). The uniqueness of this virus is that it contains a gene that most likely originated from the p10 gene of a bat orthoreovirus [53]. A two-year follow-up study also illustrated that BtEoCoV-GCCDC1 persistently circulates among bats. Different to the genetically diverged HKU9-CoV, this virus is highly conserved (Figure 2). BtEoCoV-GCCDC1 has only been found in south Yunnan Province so far [54,55]. In addition, there are other bat CoVs that have been identified in China: Rhinolophus ferrumequinum α-CoV HuB-2013 [8], Myotis ricketti α-CoV Sax-2011 [8,37], Nyctalus velutinus α-CoV SC-2013 [8], Scotophilus bat CoV 512 [37], Hipposideros bat β-CoV Zhejiang2013, and a Murina leucogaster bat CoV, which has been described as the evolutionary ancestor of PEDV [37]. Notably, there are still many unclassified bat CoVs circulating in China, particularly in the northern part of the nation where bat viruses were rarely studied (Figure 1). According to the criteria defined by the ICTV, the CoV family will most likely expand following further investigation of bat CoVs in China.
7. Conclusions
Two bat origin CoVs caused large-scale epidemics in China over fourteen years, highlighting the risk of a future bat CoV outbreak in this nation. In this review, we have summarized the current findings related to bat CoV epidemiology in China, aiming to explore the associations between CoV species, bat species, and geographical locations, and eventually we aim to predict the cross-species transmission potential of these bat CoVs. Admittedly, the analysis may be affected by inaccurate or incomplete data. For example, not all research groups performed bat species identification or used Global Positioning System (GPS) during bat sampling. Bats in the north or west provinces were not surveyed either. Nonetheless, we believe this analysis is a good starting point for further research. Moreover, there are other outstanding questions that should be addressed in future studies: (1) given that most of the ICTV classified CoV species are from bats, why there are so many genetically divergent CoVs in bats, (2) the pathogenesis of most bat CoVs in humans remains unknown as the viruses have never been isolated or rescued—apart from the viruses identified during the outbreaks, many viruses pose a threat to human health, (3) although SARS-CoV and SADS-CoV were known to be transmitted from bats to human or swine, their exact transmission routes are unknown, and (4) why bats can maintain CoVs long-term without showing clinical symptoms of diseases. A unique bat immunity model has been proposed. The authors have shown that constitutively expressed bat interferon α may protect bats from infection [76], while some particularly dampened immune pathways may allow bats to have a higher tolerance against viral diseases [77]. While we start to unveil the mystery of unique bat immunity, there is still long way to go before we can fully understand the relationship between bats and coronaviruses.
Author Contributions
P.Z. and Z.-L.S. designed the study. Y.F. and K.Z. analyzed the data. P.Z. and Y.F. prepared the manuscript.
Funding
The work was supported by China’s National Science and Technology Major Project on Infectious Diseases (2018ZX10101004) and the National Natural Science Foundation of China (Excellent Young Scholars to PZ 81822028 and 81661148058).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255. [Google Scholar] [CrossRef] [PubMed]
- Coronavirinae in ViralZone. Available online: https://viralzone.expasy.org/785 (accessed on 28 January 2019).
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- ICTV Virus Taxonomy: 2018 Release. 2018. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 28 January 2019).
- Ge, X.Y.; Wang, N.; Zhang, W.; Hu, B.; Li, B.; Zhang, Y.Z.; Zhou, J.H.; Luo, C.M.; Yang, X.L.; Wu, L.J.; et al. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016, 31, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; Ren, X.; He, G.; Zhang, J.; Yang, J.; Qian, Z.; Dong, J.; Sun, L.; Zhu, Y.; et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016, 10, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yoo, D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016, 226, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, H.; Zhao, J.; Zhong, Q.; Jin, J.H.; Zhang, G.Z. Evolution of infectious bronchitis virus in China over the past two decades. J. Gen. Virol. 2016, 97, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.A.; Wu, G.; St Leger, J.; Nordhausen, R.W.; Wang, D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Halpin, R.; Wang, S.; Ghedin, E.; Spiro, D.J.; Saif, L.J. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J. Gen. Virol. 2011, 92, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Cowled, C. Bats and Viruses: A New Frontior of Emerging Infectious Diseases; John Wiley Sons Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Yang, X.L.; Tan, C.W.; Anderson, D.E.; Jiang, R.D.; Li, B.; Zhang, W.; Zhu, Y.; Lim, X.F.; Zhou, P.; Liu, X.L.; et al. Characterization of a filovirus (Mengla virus) from Rousettus bats in China. Nat. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Hosseini, P.R.; Zambrana-Torrelio, C.; Ross, N.; Bogich, T.L.; Daszak, P. Host and viral traits predict zoonotic spillover from mammals. Nature 2017, 546, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Li, K.S.M.; Huang, Y.; Tsoi, H.W.; Wong, B.H.L.; Wong, S.S.Y.; Leung, S.Y.; Chan, K.H.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef] [PubMed]
- Li, W.D.; Shi, Z.L.; Yu, M.; Ren, W.Z.; Smith, C.; Epstein, J.H.; Wang, H.Z.; Crameri, G.; Hu, Z.H.; Zhang, H.J.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.J.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Duong, V.; Hul, V.; San, S.; Davun, H.; Omaliss, K.; Chea, S.; Hassanin, A.; Theppangna, W.; Silithammavong, S.; et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017, 48, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef] [PubMed]
- Leopardi, S.; Holmes, E.C.; Gastaldelli, M.; Tassoni, L.; Priori, P.; Scaravelli, D.; Zamperin, G.; De Benedictis, P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018, 58, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Li, K.S.M.; Tsang, A.K.L.; Shek, C.T.; Wang, M.; Choi, G.K.Y.; Guo, R.T.; Wong, B.H.L.; Poon, R.W.S.; Lam, C.S.F.; et al. Recent Transmission of a Novel Alphacoronavirus, Bat Coronavirus HKU10, from Leschenault’s Rousettes to Pomona Leaf-Nosed Bats: First Evidence of Interspecies Transmission of Coronavirus between Bats of Different Suborders. J. Virol. 2012, 86, 11906–11918. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhang, Y.; Xu, L.; Yang, W.; Yang, F.; Feng, Y.; Xia, L.; Zhou, J.; Zhen, W.; Feng, Y.; et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014, 88, 7070–7082. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.Q.; Chen, D.H.; Tan, W.P.; Qiu, S.Y.; Xu, D.; Liang, H.X.; Chen, M.X.; Li, X.; Lin, Z.S.; Liu, W.K.; et al. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: A study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.C.; Lam, C.S.; Luk, H.K.; Wong, E.Y.; Lee, R.A.; So, L.Y.; Chan, K.H.; Cheng, V.C.; Yuen, K.Y.; Woo, P.C.; et al. A six-year descriptive epidemiological study of human coronavirus infections in hospitalized patients in Hong Kong. Virol. Sin. 2016, 31, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, X.D.; Guo, W.P.; Zhou, R.H.; Wang, M.R.; Wang, C.Q.; Ge, S.; Mei, S.H.; Li, M.H.; Shi, M.; et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology 2015, 474, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Provacia, L.B.; Smits, S.L.; Martina, B.E.; Raj, V.S.; Doel, P.V.; Amerongen, G.V.; Moorman-Roest, H.; Osterhaus, A.D.; Haagmans, B.L. Enteric coronavirus in ferrets, The Netherlands. Emerg. Infect. Dis. 2011, 17, 1570–1571. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.; Chu, D.K.; Chan, K.H.; Wong, O.K.; Ellis, T.M.; Leung, Y.H.; Lau, S.K.; Woo, P.C.; Suen, K.Y.; Yuen, K.Y.; et al. Identification of a novel coronavirus in bats. J. Virol. 2005, 79, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Poon, L.L.; Chan, K.H.; Chen, H.; Guan, Y.; Yuen, K.Y.; Peiris, J.S. Coronaviruses in bent-winged bats (Miniopterus spp.). J. Gen. Virol. 2006, 87, 2461–2466. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Peiris, J.S.; Chen, H.; Guan, Y.; Poon, L.L. Genomic characterizations of bat coronaviruses (1A, 1B and HKU8) and evidence for co-infections in Miniopterus bats. J. Gen. Virol. 2008, 89, 1282–1287. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yang, L.; Ren, X.; Zhang, J.; Dong, J.; Sun, L.; Zhu, Y.; Yang, F.; Zhang, S.; Wu, Z.; et al. Genetic diversity of coronaviruses in Miniopterus fuliginosus bats. Sci. China Life Sci. 2016, 59, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yang, X.L.; Li, B.; Liu, Q.; Zhang, Q.; Liu, H.; Kan, H.P.; Wong, K.C.; Chek, S.N.; He, X.; et al. Detection of diverse viruses in alimentary specimens of bats in Macau. Virol. Sin. 2017, 32, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.D.; Wang, W.; Hao, Z.Y.; Wang, Z.X.; Guo, W.P.; Guan, X.Q.; Wang, M.R.; Wang, H.W.; Zhou, R.H.; Li, M.H.; et al. Extensive diversity of coronaviruses in bats from China. Virology 2017, 507, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Li, K.S.; Poon, R.W.; Wong, B.H.; Tsoi, H.W.; Yip, B.C.; Huang, Y.; Chan, K.H.; Yuen, K.Y. Molecular diversity of coronaviruses in bats. Virology 2006, 351, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Wang, M.; Lau, S.K.; Xu, H.; Poon, R.W.; Guo, R.; Wong, B.H.; Gao, K.; Tsoi, H.W.; Huang, Y.; et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 2007, 81, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Feng, Y.; Chen, H.; Luk, H.K.; Yang, W.H.; Li, K.S.; Zhang, Y.Z.; Huang, Y.; Song, Z.Z.; Chow, W.N.; et al. Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination. J. Virol. 2015, 89, 10532–10547. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, F.; Yang, W.; Jiang, T.; Lu, G.; He, B.; Li, X.; Hu, T.; Chen, G.; Feng, Y.; et al. Detection and characterization of diverse alpha- and betacoronaviruses from bats in China. Virol. Sin. 2016, 31, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch. Virol. 2013, 158, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.C.; Zhang, J.X.; Zhang, S.Y.; Wang, P.; Fan, X.H.; Li, L.F.; Li, G.; Dong, B.Q.; Liu, W.; Cheung, C.L.; et al. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006, 80, 7481–7490. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Wang, M.; Lam, C.S.; Xu, H.; Guo, R.; Chan, K.H.; Zheng, B.J.; et al. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 2007, 367, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Li, K.S.; Huang, Y.; Shek, C.T.; Tse, H.; Wang, M.; Choi, G.K.; Xu, H.; Lam, C.S.; Guo, R.; et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010, 84, 2808–2819. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Tsang, A.K.; Fan, R.Y.; Luk, H.K.; Cai, J.P.; Chan, K.H.; Zheng, B.J.; Wang, M.; et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J. Virol. 2015, 89, 3076–3092. [Google Scholar] [CrossRef] [PubMed]
- Bardos, V.; Schwanzer, V.; Pesko, J. Identification of Tettnang virus (‘possible arbovirus’) as mouse hepatitis virus. Intervirology 1980, 13, 275–283. [Google Scholar] [PubMed]
- Corman, V.M.; Kallies, R.; Philipps, H.; Gopner, G.; Muller, M.A.; Eckerle, I.; Brunink, S.; Drosten, C.; Drexler, J.F. Characterization of a novel betacoronavirus related to middle East respiratory syndrome coronavirus in European hedgehogs. J. Virol. 2014, 88, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.M.; Wang, N.; Yang, X.L.; Liu, H.Z.; Zhang, W.; Li, B.; Hu, B.; Peng, C.; Geng, Q.B.; Zhu, G.J.; et al. Discovery of Novel Bat Coronaviruses in South China That Use the Same Receptor as Middle East Respiratory Syndrome Coronavirus. J. Virol. 2018, 92, e00116-18. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Zhang, L.B.; Luk, H.K.H.; Xiong, L.F.; Peng, X.W.; Li, K.S.M.; He, X.Y.; Zhao, P.S.H.; Fan, R.Y.Y.; Wong, A.C.P.; et al. Receptor Usage of a Novel Bat Lineage C Betacoronavirus Reveals Evolution of Middle East Respiratory Syndrome-Related Coronavirus Spike Proteins for Human Dipeptidyl Peptidase 4 Binding. J. Infect. Dis. 2018, 218, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Li, K.S.M.; Tsang, A.K.L.; Lam, C.S.F.; Ahmed, S.; Chen, H.L.; Chan, K.H.; Woo, P.C.Y.; Yuen, K.Y. Genetic Characterization of Betacoronavirus Lineage C Viruses in Bats Reveals Marked Sequence Divergence in the Spike Protein of Pipistrellus Bat Coronavirus HKU5 in Japanese Pipistrelle: Implications for the Origin of the Novel Middle East Respiratory Syndrome Coronavirus. J. Virol. 2013, 87, 8638–8650. [Google Scholar] [PubMed]
- Yang, L.; Wu, Z.Q.; Ren, X.W.; Yang, F.; Zhang, J.P.; He, G.M.; Dong, J.; Sun, L.L.; Zhu, Y.F.; Zhang, S.Y.; et al. MERS-Related Betacoronavirus in Vespertilio superans Bats, China. Emerg. Infect. Dis. 2014, 20, 1260–1262. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, W.J.; Xu, W.; Jin, T.; Zhao, Y.; Song, J.; Shi, Y.; Ji, W.; Jia, H.; Zhou, Y.; et al. A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene. PLoS Pathog. 2016, 12, e1005883. [Google Scholar] [CrossRef] [PubMed]
- Obameso, J.O.; Li, H.; Jia, H.; Han, M.; Zhu, S.; Huang, C.; Zhao, Y.; Zhao, M.; Bai, Y.; Yuan, F.; et al. The persistent prevalence and evolution of cross-family recombinant coronavirus GCCDC1 among a bat population: A two-year follow-up. Sci. China Life Sci. 2017, 60, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Li, B.; Jiang, R.D.; Hu, B.J.; Luo, D.S.; Zhu, G.J.; Hu, B.; Liu, H.Z.; Zhang, Y.Z.; Yang, X.L.; et al. Longitudinal Surveillance of Betacoronaviruses in Fruit Bats in Yunnan Province, China During 2009–2016. Virol. Sin. 2018, 33, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Poon, R.W.; Wong, B.H.; Wang, M.; Huang, Y.; Xu, H.; Guo, R.; Li, K.S.; Gao, K.; Chan, K.H.; et al. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 2010, 84, 11385–11394. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Li, Y.; Yang, X.; Zhang, H.; Zhou, P.; Zhang, Y.; Shi, Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012, 86, 4620–4630. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Li, W.D.; Yu, M.; Hao, P.; Zhang, Y.; Zhou, P.; Zhang, S.Y.; Zhao, G.P.; Zhong, Y.; Wang, S.Y.; et al. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 2006, 87, 3355–3359. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.F.; Hon, C.C.; Li, Y.; Wang, D.M.; Xu, G.L.; Zhang, H.J.; Zhou, P.; Poon, L.L.M.; Lam, T.T.Y.; Leung, F.C.C.; et al. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J. Gen. Virol. 2010, 91, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.G.; Yang, L.; Ren, X.W.; Zhang, J.P.; Yang, F.; Zhang, S.Y.; Jin, Q. ORF8-Related Genetic Evidence for Chinese Horseshoe Bats as the Source of Human Severe Acute Respiratory Syndrome Coronavirus. J. Infect. Dis. 2016, 213, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Hu, B.; Wang, B.; Wang, M.N.; Zhang, Q.; Zhang, W.; Wu, L.J.; Ge, X.Y.; Zhang, Y.Z.; Daszak, P.; et al. Isolation and Characterization of a Novel Bat Coronavirus Closely Related to the Direct Progenitor of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2016, 90, 3253–3256. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zeng, L.P.; Yang, X.L.; Ge, X.Y.; Zhang, W.; Li, B.; Xie, J.Z.; Shen, X.R.; Zhang, Y.Z.; Wang, N.; et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017, 13, e1006698. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zhu, C.Q.; Ai, L.L.; He, T.; Wang, Y.; Ye, F.Q.; Yang, L.; Ding, C.X.; Zhu, X.H.; Lv, R.C.; et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 2018, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Zhu, C.; Wang, Y.; Ai, L.; Yang, L.; Ye, F.; Ding, C.; Chen, J.; He, B.; Zhu, J.; et al. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci. Rep. 2017, 7, 10917. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [PubMed]
- Woo, P.C.; Lau, S.K.; Tsang, C.C.; Lau, C.C.; Wong, P.C.; Chow, F.W.; Fong, J.Y.; Yuen, K.Y. Coronavirus HKU15 in respiratory tract of pigs and first discovery of coronavirus quasispecies in 5′-untranslated region. Emerg. Microbes Infect. 2017, 6, e53. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Qu, X.; Li, W.; Han, Z.; Yu, M.; Zhou, P.; Zhang, S.Y.; Wang, L.F.; Deng, H.; Shi, Z. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008, 82, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wu, Z.Q.; Ren, X.W.; Yang, F.; He, G.M.; Zhang, J.P.; Dong, J.; Sun, L.L.; Zhu, Y.F.; Du, J.; et al. Novel SARS-like Betacoronaviruses in Bats, China, 2011. Emerg. Infect. Dis. 2013, 19, 989–991. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Yount, B.L., Jr.; Debbink, K.; Agnihothram, S.; Gralinski, L.E.; Plante, J.A.; Graham, R.L.; Scobey, T.; Ge, X.Y.; Donaldson, E.F.; et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015, 21, 1508–1513. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, S.Y.; Yang, X.L.; Huang, H.M.; Zhang, Y.J.; Guo, H.; Luo, C.M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Reusken, C.B.; Raj, V.S.; Koopmans, M.P.; Haagmans, B.L. Cross host transmission in the emergence of MERS coronavirus. Curr. Opin. Virol. 2016, 16, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. MBio 2017, 8, e00373-17. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Du, L.; Liu, C.; Wang, L.; Ma, C.; Tang, J.; Baric, R.S.; Jiang, S.; Li, F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA 2014, 111, 12516–12521. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Su, S.; Bi, Y.; Wong, G.; Gao, G.F. Bat-Origin Coronaviruses Expand Their Host Range to Pigs. Trends Microbiol. 2018, 26, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Tachedjian, M.; Wynne, J.W.; Boyd, V.; Cui, J.; Smith, I.; Cowled, C.; Ng, J.H.; Mok, L.; Michalski, W.P.; et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. Proc. Natl. Acad. Sci. USA 2016, 113, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Li, Y.; Shen, X.; Goh, G.; Zhu, Y.; Cui, J.; Wang, L.F.; Shi, Z.L.; Zhou, P. Dampened STING-Dependent Interferon Activation in Bats. Cell Host Microbe 2018, 23, 297–301. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).