Abstract
Dynamic changes in forest biomass are closely related to the carbon cycle, climate change, forest productivity and biodiversity. However, most previous studies mainly focused on the calculation of current forest biomass, and only a few studies attempted to predict future dynamic changes in forest biomass which obtained uncertain results. Therefore, this study comprehensively considered the effects of multi-stage continuous survey data of forest permanent sample plots, site condition factors and corresponding meteorological factors using Beijing as an example. The geographic detector method was used to screen the key interfering factors that affect the growth of forest biomass. Then, based on the back-propagation artificial neural network (BP-ANN) and support vector machine (SVM) learning methods, 80% of the sample data were extracted to train the model, and thereby verify the prediction accuracy of different modeling methods using different training samples. The results showed that the forest biomass prediction models based on both the machine learning algorithms had good fitting accuracy, and there was no significant difference in the prediction results between the two models. However, the SVM model was better than the BP-ANN. While the BP-ANN model provided more volatile predictions, and the accuracy was above 80%, the prediction results of the SVM model were relatively stable, and the accuracy was above 90%. This study not only provides good technical support for the scientific estimation of regional forest biomass in the future, but also offers reliable basic data for sustainable forest management, planning decisions, forest carbon sequestration and sustainable development.
1. Introduction
Forests are the dominant terrestrial plant communities, which not only maintain the balance of the global ecosystem, but also play an important role in the global carbon balance [1,2,3]. As the most basic quantitative characteristic of forest ecosystems, forest biomass and its dynamic changes are closely related to the carbon cycle, climate change, forest productivity and biodiversity, which makes them suitable to monitor the dynamic changes in forest resources at all levels as well as to measure the quality and productivity of the forest ecosystem [4,5,6]. In addition, forest biomass can also serve as an important indicator for evaluating forest productivity and judging the development status of forestry economy, playing an important role in promoting forest economy development and sustainable development [5,7,8]. Therefore, studying forest biomass can help us understand the ecological environment and ecosystem functions of the region, evaluate the contribution of forests to carbon cycling, the capacity of ecosystem service provision, and the stability of the ecosystem, which is of great significance for formulating science-based forest protection and management strategies.
In the past decade, research on forest biomass has increased significantly, especially in the development of biomass models using field data [9,10,11,12,13,14,15], but there is less research on estimating or predicting biomass in future stages. In forest biomass estimation, the vast majority of studies are based on individual tree scales, whereas this study estimates forest biomass based on fixed plots, which is important for calibrating and validating large-scale forest biomass and reflecting the relationship between various factors and forest biomass at a macro level [16,17,18]. Currently, the estimation methods of forest biomass include the clear-cutting method, mean wood method, relative growth method, and remote sensing estimation method [17,19,20,21,22,23,24,25]. The clear-cutting method is very accurate, and although some scholars later proposed a non-destructive method for measuring tree biomass, it appears to be less accurate in the calculation of forest biomass in large areas [26]. The mean wood method is relatively simple but offers low reliability. The relative growth method and remote sensing estimation method have higher accuracy and cause less damage to forest vegetation. Remote sensing data has certain advantages in forest biomass estimation, but there are still considerable uncertainties [27]. First, the spatial resolution is not fine enough to observe biomass changes in various corners of the forest and at different scales. Second, the cost of obtaining high-resolution data is high, which limits the application scope of remote sensing data in forest biomass monitoring and prediction. Third, due to the lack of future remote sensing images, existing forest biomass models often cannot make accurate predictions. Therefore, there is an urgent need for more reliable and low-cost model development methods to predict and analyze the spatiotemporal evolution of forest biomass.
Machine learning methods provide a workable solution for these problems, with great potential and advantages in predicting forest biomass [28]. Recently, machine learning methods have been widely applied in forest biomass prediction models [22,23,24,25,29]. Machine learning is an artificial intelligence technique that can automatically deduce patterns from data [30]. By analyzing large amounts of forest ecological data, such as terrain features, meteorological data, etc., features and patterns related to forest biomass are identified and used to establish a model for predicting forest biomass [15,31,32,33]. Lin et al. [34] established a stand harvesting model for Cunninghamia lanceolata plantations in northwest Fujian province using machine learning methods based on the back-propagation artificial neural network (BP-ANN) and support vector machine (SVM) algorithms, and the training sample precision of the model was above 0.93, wherein the fitting accuracy and generalization ability of the SVM model were better than those of the BP-ANN model. López-Serrano et al. [23] used remote sensing data to estimate forest biomass using three methods, KNN, RF and SVM, and the results indicated that SVM was the best choice. Pham et al. [24] established an aboveground biomass model of mangrove forests using an SVM regression method, and concluded that an integration of ALOS-2 PALSAR-2 and Sentinel-2A data with the SVR model can improve the AGB accuracy estimation of mangrove plantations in tropical areas. Li et al. [35] combined assimilation technology of the MODIS LAI time series with the random forest model to more accurately estimate bamboo forest aboveground biomass (AGB) in Zhejiang province. They provided a new method for estimating large-scale forest AGB based on low-resolution time series data. Wu et al. [36] used two nonparametric modeling approaches, random forest (RF) and support vector machine, to estimate AGB based on widely used Landsat images of the region. Rakesh et al. [37] used multi-source datasets based on machine learning algorithms for spatial estimation of forest biomass in India. The inclusion of multisource data using a random forest regression model increased the saturation range to 350 Mg ha−1, which is a significant improvement applicable to 94.7% of Indian forests. The model estimation error was reduced to 25.6% in the AGB range up to 350 Mg ha−1. Among many different machine learning methods [34,38,39,40,41], BP-ANN and SVM are considered to offer the best predictability and stability, with significant advantages in model construction. Therefore, using machine learning methods to accurately predict forest plot biomass is of great significance for promoting the sustainable utilization of forest resources and evaluation of carbon accounting.
Currently, most current studies focus on estimating forest biomass, with few studies focusing on predicting future forest biomass. Additionally, previous studies did not fully consider the impact of various factors in the selection of modeling factors. This study comprehensively considered the contribution of forest factors, site conditions, and meteorological factors in modeling, which improved the reliability of the prediction results. Moreover, there is no previous research on predicting future forest biomass based on sample plots in Beijing. Therefore, to address the practical need for sufficient predicted data on forest biomass in a specific region, using Beijing as a case study, this study aims to fill this research gap and provide a model for forest biomass prediction in other regions and help achieve the dual carbon targets, and enhance the forest ecosystem function. In this study, BP-ANN and SVM were utilized to establish a forest biomass prediction model for Beijing plots, followed by a comparison of the accuracy of the two models. The research findings can provide a scientific basis and decision support for promoting sustainable forestry development in Beijing, nationwide, and globally. This approach can help forest managers plan and manage forest resources more effectively, promoting a stable and healthy ecosystem development.
2. Materials and Methods
2.1. Overview of the Study Area
Beijing is an important economic center and population agglomeration area in China, and the health status of forest ecosystems has a significant impact on the lives of residents and economic development. It is located on the northwestern edge of the North China Plain (115°25′~117°30′ E, 39°28′~41°05′ N), with its center at 116°25′29″ E and 39°54′20″ N. Beijing’s topography is mainly composed of the Zhongshan Mountains, Yan Mountains, and Taihang Mountains, gradually rising from northwest to southeast. The highest peak is Badaling with an elevation of 2303 m. The city’s main rivers include the Bohai, Baihe, Yongding, and Nanyunhe, which form the three major natural zones of “Western Mountains, Northern Plains, and Eastern Waters”. Beijing belongs to a warm temperate semi-humid and semi-arid monsoon climate zone, with distinct seasons. Spring is mild, summer is hot and rainy, autumn is cool and pleasant, and winter is cold and dry. The average annual temperature is around 12 °C, and the annual precipitation is 600–700 mm. Due to differences in elevation, the climate factors, such as temperature and precipitation, also vary within the city. In addition, the vegetation coverage in different areas of the city varies. The vegetation growth and spatial distribution in Beijing are shown in Figure 1.
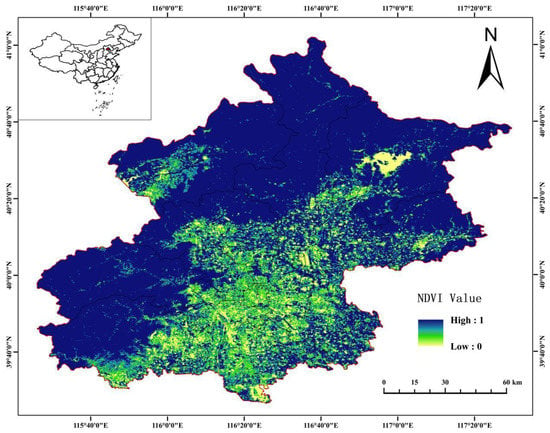
Figure 1.
Forest vegetation coverage map of Beijing.
Beijing’s total area is 16,807 square kilometers, with the urban area covering 1368 square kilometers. According to the latest forest resource inventory data, Beijing’s overall forest coverage has reached more than 40%. During the 15-year period from the sixth term to the eighth term (2004–2018), the forest area increased by 197,700 hectares, the forest coverage rate increased by 12.05%, and the forest stock increased by 13,987,800 cubic meters. The corresponding datapoints are listed in Table 1.

Table 1.
Forest area, forest cover ratio and forest accumulation from 2006 to 2016.
2.2. Data Source
The dataset mainly used in this study includes the National Forest Resource Continuous Inventory data and Meteorological data.
2.2.1. National Forest Resource Continuous Inventory Data
This study selected the forest inventory data of the 6th, 7th, and 8th periods in Beijing, including 214 permanent sample plots and 428 plot data records, all of which were forestland data. The permanent sample plot survey data mainly includes plot number, slope, aspect, GPS plane coordinates, vegetation cover, shrub cover, herb cover, average age, tree density (trees/ha), canopy density, average tree height (m), average diameter at breast height (cm), dominant tree species, breast height area G (m2/ha), and stock volume (m3/ha).
2.2.2. Calculating the Basic Biomass of Permanent Plots
In this study, the future forest biomass was predicted by modeling forest growth, so it was necessary to calculate the basic biomass of the forest permanent sample plot. The basic biomass of the permanent sample plot consists of the biomass of trees, shrubs and herbs. The tree biomass in the sample plot in this study was estimated using the linear fitting equation of biomass and storage volume by forest type and age group proposed by Xu et al. [42]. The biomass of shrubs and herbs was estimated using the method established by Wang et al. [43]. Based on the basic biomass of the permanent sample plot, this study calculated the forest growth for two consecutive periods.
2.2.3. Meteorological Data
The meteorological data used in this study were obtained from the China Meteorological Science Data Sharing Service Network and the ClimateAP software development kit. This study used two dominant climate factors, mean annual temperature and mean annual precipitation, which have a significant impact on forest growth, as the primary meteorological factors for modeling [32,44,45,46].
The historical meteorological data were obtained from the China Meteorological Science Data Sharing Service Network (http://data.cma.cn, accessed on 14 July 2021). Daily meteorological data from 21 meteorological stations in Beijing from 2004 to 2018 were selected, including daily mean temperature and mean precipitation (Figure 2). The missing data and outliers in some meteorological stations were supplemented using the inverse distance weighting method. To correspond the forest growth data for the two consecutive periods, a pivot table was used to organize the meteorological data for every five years, and the average values of mean annual precipitation and mean annual temperature for the corresponding period were calculated for modeling and validation.
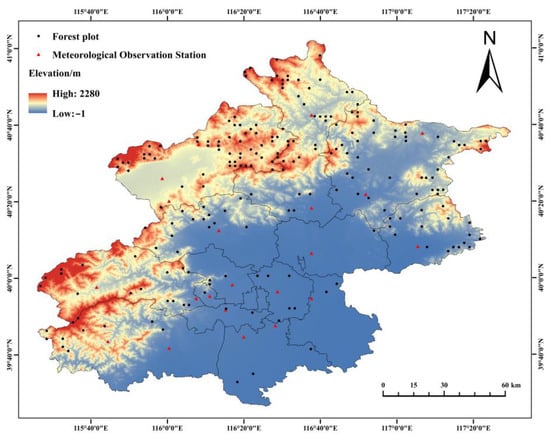
Figure 2.
Distribution map of continuous forest inventory data samples and meteorological stations in Beijing.
The future meteorological data were obtained from Climate AP [47,48]. This software covers future climate data predicted based on the IPCC Fifth Assessment Report (AR5). Climate AP can automatically extract corresponding climate data based on the query point location and the corresponding time series. Based on the coordinates of the inventory plots, annual climate data for five consecutive years from 2019 to 2023 (precipitation, temperature) were extracted and used to calculate the annual average of mean annual precipitation and mean annual temperature from 2019 to 2023 for model prediction.
2.3. Research and Construction Methods
Before conducting the modeling study, it is necessary to organize the above sample plot data. As the sample plot data is a continuous survey data of multiple periods, the correlation between the data is poor, and there is a lack of intuitive continuous variability. In order to better correlate the sample plot data, we use the following methods to conduct correlation analysis on the sample plot data separately. The continuous survey plots use a “point sampling” system, which can be directly correlated based on the fixed plot coordinates. This study selected the relational database management software MySQL developed by the Swedish MySQLAB company to calculate and match the correlation between different period plot data. The data of the sixth, seventh, and eighth periods were imported into the software, and then matched according to the coordinate of each two-period data, realizing the one-to-one correlation of the three-period plot data. At the same time, in order to realize the prediction function for the fixed plots in Beijing, the latest continuous survey plot data was classified, organized, and correspondingly correlated.
2.3.1. Correlation Analysis
Geographic Detector is a tool used to analyze geographic phenomena and explore relationships between geographic factors [49]. It combines GIS, remote sensing, and statistical methods to help researchers better understand geographic phenomena and explore relationships between variables. Geographic Detector can analyze the relationships between multiple variables to find their mutual influence and spatial relationships, including linear and nonlinear relationships. By comparing the contribution of variables to the target variable, Geographic Detector can select variables that are important to the target variable, reduce the complexity of the model, and improve the accuracy and reliability of the model. Geographic Detector can consider spatial autocorrelation, which helps to select variables with regional differences in factor selection, and improve the adaptability and generalizability of the model.
The stand factors, site conditions, and meteorological factors are all important factors affecting forest biomass, and the influence of each factor on forest biomass varies depending on the region, environment, vegetation type, and other conditions [33,50,51,52,53,54,55]. According to extensive research, this study selected forest stand factors (mean stand age, mean stand density, mean diameter at breast height, mean stand height, and mean cross-sectional area at breast height), site conditions (slope, and aspect), and meteorological factors (mean annual precipitation, and mean annual temperature) as variables. This study used Geographic Detector to analyze the correlation among variable factors and plotted the following factor interaction diagram using the R software environment (Figure 3). It can be seen that the impact of any two independent factors was enhanced after interaction. However, the correlation between terrain factors, such as slope and forest growth, was weak, while the correlation between meteorological factors was strong. The main reason is that the impact of terrain factors on forest growth can ultimately be reflected in temperature and precipitation [32,44,45,46].
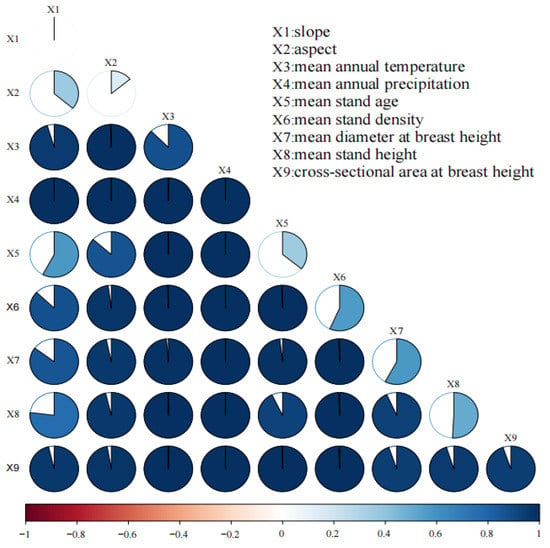
Figure 3.
The factor interaction plot.
Therefore, this study combined the site conditions and meteorological factors with forest survey factors for model training and testing, and subsequently analyzed the forest sample biomass model for Beijing.
2.3.2. BP-ANN Model
Back-propagation artificial neural network (BP-ANN) is a machine learning algorithm based on neural networks that can be used for prediction and classification of problems [56]. In predicting forest biomass, BP-ANN has many advantages, as this model can adaptively learn the nonlinear relationship between input and output through training, which can better capture the complex relationships and nonlinear features of forest biomass, thus improving the estimation accuracy [57]. Compared to traditional linear regression models, BP-ANN can better handle nonlinear relationships and high-dimensional data. BP-ANN is able to handle missing data and outliers because its training process is based on a large number of data samples, which can be used to infer missing values or filter out outliers based on multiple sample features. In addition, BP-ANN can perform iterative training and tuning to improve the accuracy of model prediction by continuously adjusting network structure and hyperparameters [58]. In summary, BP-ANN is a powerful machine learning algorithm that can be used to predict forest biomass, with good prediction accuracy and scalability.
BP-ANN is a multilayer feedforward neural network trained using an error backward-propagation algorithm, which has a single hidden layer or multiple hidden layers [59]. In 1989, Funahashi proved that a Rumelhart–Hinton–Williams multi-layer neural network could approximately realize arbitrary continuous mapping, and its output was an s-type function [60,61]. The accuracy is higher than that of a network with a single hidden layer. In the process of model establishment, the topology of BP-ANN is composed of an input layer, a hidden layer, and an output layer. Neurons in the same layer do not affect each other, and the neuron state of each layer only affects the neuron state of the next layer [59]. However, the number of parameters will increase exponentially with the number of hidden layers. When it reaches a certain number of layers, the classification effect will become less and less obvious when further hidden layers are added.
In this study, we used a single hidden layer to construct the model. The sorted data were randomly divided into test sets and training sets, while the input and output vectors were normalized to a range of [0, 1] using the Max and Min method. The topological structure of BP-ANN was determined to be 9-14-1, and the empirical range of weight and threshold value was [−1, 1], so that the range of optimization can be widened appropriately. The number of learning iterations was set to 1000, with a minimum training target error of 0.00001 and a learning rate of 0.1. To improve the learning ability of BP-ANN, the algorithm was optimized. To pass in front of the signal process, it imports data from the input layer, constantly training the model in the hidden layer, and then reaches the output layer; if the output of and comparison between the predicted values and the real value if it do not meet expectations, it goes into the reverse transmission, and uninterruptedly adjusts the weights and thresholds, until the minimum error output value is close to the real value. Finally, the prediction results are inversely normalized and visualized, while the model error evaluation is given as the output (Figure 4).
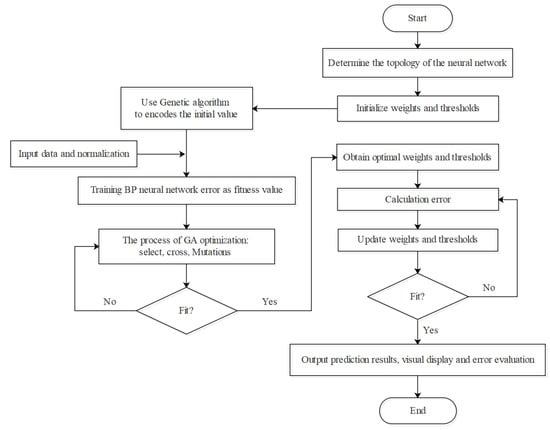
Figure 4.
Modeling flowchart using BP-ANN.
2.3.3. SVM Modeling
The support vector machine (SVM) is also a commonly used machine learning algorithm that can be used for classification and regression problems [62]. In predicting forest biomass, SVM has many advantages, as it can handle non-linear relationships and high-dimensional data by selecting appropriate kernel functions to map the data to a high-dimensional space, thus more accurately capturing the key factors that affect forest biomass [63]. SVM can undergo iterative training and parameter tuning, improving the model prediction accuracy by continuously adjusting hyperparameters and kernel functions. Its learning algorithm seeks the optimal compromise between model complexity (namely, the learning accuracy of training samples) and learning ability for limited sample information [64,65]. In summary, SVM is a powerful machine learning algorithm that can be used to predict forest biomass with good predictive accuracy and scalability. However, training and prediction on large-scale datasets may consume a longer time and more computing resources. Therefore, when selecting an algorithm, the size and complexity of the dataset should be considered.
This study was carried out using the libsvm toolbox. According to the requirements of the SVM model, the input and output vectors were normalized, and the encapsulated svmrp was used to predict the number growth of svm, and the optimal parameters C (penalty parameter) and G (kernel parameter) were found. In order to improve the learning ability of the SVM network, the particle swarm optimization algorithm was used for parameter optimization, whereby the penalty parameter C and kernel parameter G were used as optimization variables with cross-validation. The initial range of the penalty parameter C was 0~100, the initial range of the kernel parameter G was 0~1000, the termination algebra was 100, the population number was 20, the variation probability was 0.01, and the functional error precision was set to 0.0001. After optimization, the optimal value of the parameter C was 11.3137 and that of G was 0.7071, which were used in the SVM. The optimal parameters were used for SVM network training, simulation prediction and inverse normalization, and the prediction results were obtained and visualized. At the same time, the model error evaluation was generated as the output (Figure 5).
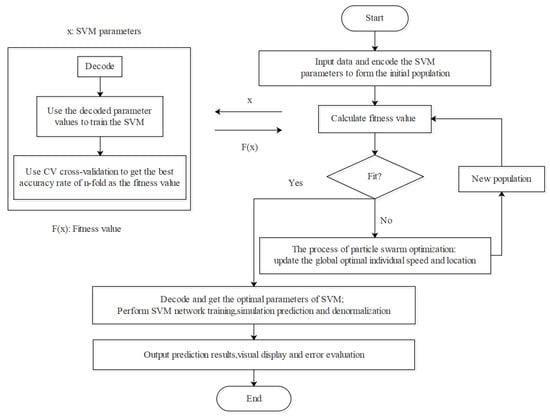
Figure 5.
Modeling flowchart using SVM.
2.4. Model Assessment
In order to evaluate the prediction performance of the model and further test its applicability, the determination coefficient (R2), mean square error (MSE), root mean square error (RMSE) and mean absolute percentage error (MAPE) were used as evaluation indexes to explain the stability of model fitting, the criterion for judging the quality of the model and the accuracy of the prediction results. The corresponding equations are as follows:
Determination coefficient (R2):
Mean square error (MSE):
Root Mean Square Error (RMSE):
Mean Absolute Percentage Error (MAPE)
where, n is the number of samples, is the predicted value, is the actual value, and is the average value.
3. Results
This study is based on 428 sorted data samples. During the training phase, the original data was automatically divided into two non-overlapping parts, one for model training and the other for validation. The training data was used to train the model, and the validation data was used to verify the accuracy and generalizability of the model, to ensure that the model can effectively predict new data. By adjusting the parameters of the model and optimizing the algorithm, the predictive performance of the model was continuously improved to achieve higher accuracy and stability. We randomly sampled 80% of the data for training. This data selection method and ratio have been widely used by many scholars, such as Zeng et al., who established a larch forest biomass equation [66], or Zhu et al., who used this model to accurately estimate the biomass of heterogeneous and dense mangroves in worldview-2 images [67].
After developing and constructing the BP-ANN model, we obtained the prediction results shown in Figure 6. As can be seen in the figure, when the training samples accounted for 80% of the total dataset, the forest biomass growth predicted by the model was basically consistent with the actual value, indicating a good training result.
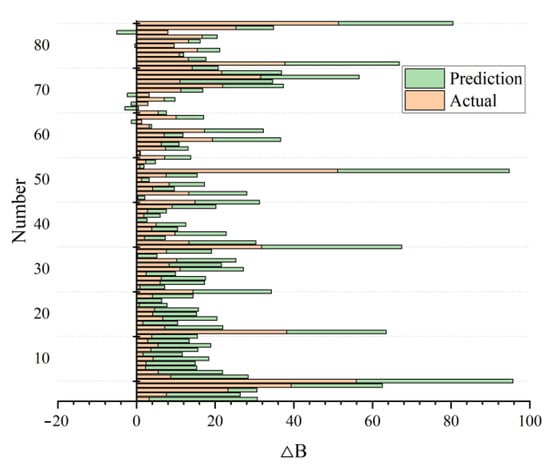
Figure 6.
Predicted and actual values of forest biomass growth (∆B) based on the BP-ANN prediction model.
At the same time, we found that the prediction model based on BP-ANN was slightly unstable with different training times, and the R2 fluctuated within a certain range. We randomly selected the results three times (Table 2), and the R2 of the three operation results basically remained above 0.85, which reflected the prediction results and illustrated the feasibility of the model.

Table 2.
The fitting results based on BP-ANN.
After developing and constructing the SVM model, we obtained the prediction results shown in Figure 7. When the training samples accounted for 80% of the total dataset, the forest biomass growth predicted by the model was in good agreement with the actual value, indicating a good training result.
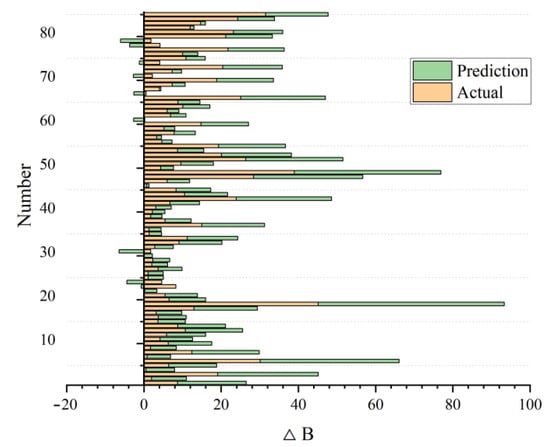
Figure 7.
Predicted and actual values of forest biomass growth (∆B) based on the SVM prediction model.
At the same time, we randomly selected the results three times. Table 3 shows that with different training times, the prediction model based on SVM had good stability. The R2 fluctuated only slightly, and the R2 of the three operation results basically remained above 0.9. This indicated high precision, which can well reflect the prediction results, confirming the feasibility and superiority of the model.

Table 3.
The fitting results based on the SVM prediction model.
By using the two models established above and taking the model built with 80% of the training samples as the standard, the forest growth of the next period (2019–2023) of the sample plot can be predicted. Based on this, the forest biomass of the ninth period can be obtained by adding the basic biomass of the eighth period permanent sample plot (Figure 8).
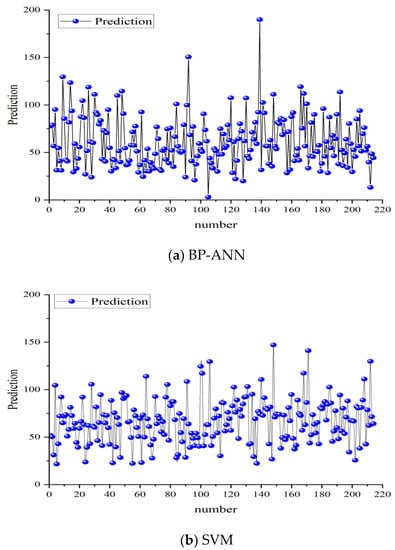
Figure 8.
Prediction results of the BP-ANN and SVM models developed in this study.
In order to express the predictive performance of the two models more intuitively, the established models were used to predict all data. After plotting the predicted values and standard values on the same coordinate system (Figure 9) and the error graphs of two models (Figure 10), we can observe the distribution of the scatterplot to compare the differences and correlations between the predicted values and standard values. Overall, the scatterplot showed that the points are dense and concentrated near a straight line, indicating relatively accurate predictions. In addition, some points were relatively scattered, which may be due to additional factors. In this case, we can continue to analyze these outliers to understand their relationship with other factors. Compared to the sub-models, the predictions of the SVM model were more concentrated near the straight line than those of the BP-ANN model (R2SVM > R2BP-ANN), indicating higher accuracy. These results indicate a strong correlation between predicted and standard values, demonstrating that the model can provide a valuable reference for forest resource management and carbon cycle research.
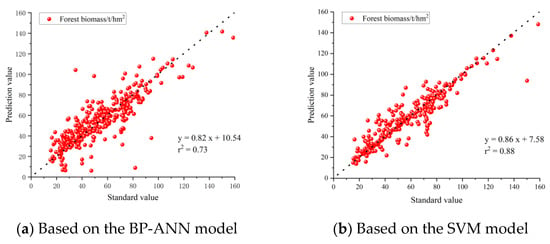
Figure 9.
Correlation analysis of the predicted and standard values.
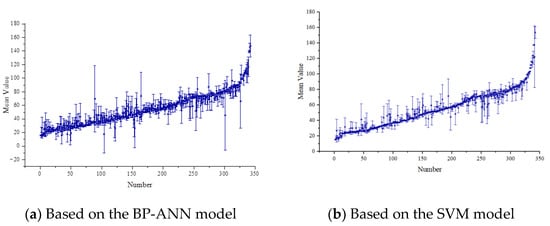
Figure 10.
Mean error map of the BP-ANN and SVM models.
In this study, we found that the prediction accuracy of the models based on the BP-ANN and SVM methods can reach more than 80%. As can be seen from Table 2 and Table 3, when the training samples of the BP-ANN and SVM prediction models included 80% of the data, the results of multiple runs fluctuated differently and the influence on the accuracy was different. Therefore, in order to further compare the stability of the two prediction models with different training samples, we reduced the number of training samples to determine the impact of the number of training samples on each model. Because the training samples generally contain more than 50% of the data, in order to highlight the comparison between research and construction models, we selected numbers of samples with an interval of 10% for training, and we obtained the model prediction fitting results when the number of training samples was 70% (Figure 11).
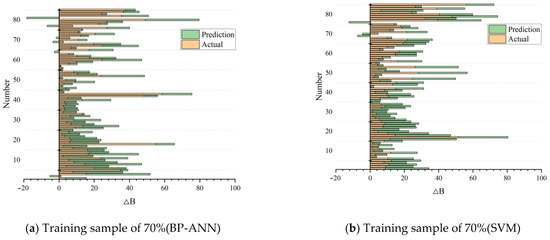
Figure 11.
Predicted and actual values of forest biomass growth (∆B) based on the BP-ANN model and the SVM model.
By comparing the two models separately (Table 4), it can be observed that the variation of training sample size has a certain impact on both prediction models. Both the BP-ANN and SVM prediction models showed a downward trend in R2 as the number of samples decreased (Table 4).

Table 4.
The fitting results of the BP-ANN and SVM models.
4. Discussion
Establishing forest biomass models based on machine learning methods opens great possibilities for more reliable estimation and prediction [30]. This study accurately estimated the forest biomass of the permanent sample plots in Beijing using two machine learning methods, providing a reference for solving regional forest biomass estimation problems. The findings of this study can enhance our comprehension of the productivity and carbon stocks of forest ecosystems, and evaluate the response and adaptability of forests to climate change in Beijing.
In accuracy evaluation, R2 is mainly used to measure the degree of fit of a model, which reflects its quality, while other indicators are mainly used to evaluate the accuracy of the predicted values [68]. MAPE measures the relative size of deviations, which is not easily affected by extreme values. MSE and RMSE measure the absolute size of deviations between measured and predicted values, which are more sensitive to outliers. In the model constructed in this study, R2 was relatively high, but RMSE was also high, indicating that the overall fitting effect was good in the prediction results, but there were some outliers. This is mainly because different types and ages of forests have different rates of forest biomass growth [5,18,69]. Therefore, evaluating the quality of a model cannot be separated from specific application scenarios and datasets. It is basically meaningless to simply judge which model is good or bad. Overall, both models were relatively stable, which was mainly attributed to the reliability, representativeness and accuracy of the data used in modeling, the use of geographic detectors for correlation analysis of variables to identify important explanatory variables, and the combination of variables to optimize the explanatory power and prediction accuracy of the models. However, this also indicates that higher requirements are placed on the data sources when using machine learning methods for modeling. Attention should be paid to the processing of missing values and outliers, variable selection, as well as other aspects of data processing and screening.
Using machine learning methods can effectively improve the accuracy of model prediction, but due to the complexity and uncertainty of the prediction object, the results of a single modeling method may be biased [24,34,37]. In this case, the optimal integration of various modeling algorithms provides another alternative to reduce this uncertainty [70]. Dai et al. [50] pointed out that using combination models can greatly improve the accuracy of estimation results. Che et al. [71] demonstrated that the output of the combination prediction model is more accurate. Smuga-Kogut et al. [30] used artificial neural networks and random forest algorithms to generate mixed models, and the fitting degree was improved to R2 = 0.961. This suggests that the use of combined prediction models can comprehensively improve the accuracy and overall effect of predictions, thereby achieving a better performance [72]. However, the universality of combination models still needs to be explored and verified. Therefore, the role of combination models in forest biomass models should be studied in depth in the future.
Forest diversity (in terms of types, climates, site conditions, etc.), long-term temporal scales, and different model construction methods can all lead to differences in forest biomass estimation at different scales [6,8,73]. Currently, research on forest biomass models mostly focuses on vegetation factors, while ignoring the influence of topography and weather on the models. In this study, site conditions and meteorological factors were added as independent variables to the biomass model based on stand factors. The increase in the number of independent variables brings the estimate of biomass closer to the true value [74], but it also reduces the general effectiveness of the biomass model. Therefore, theories from other disciplines (ecology, biology, meteorology, etc.) should be cross-referenced when constructing a biomass model, and the balance between statistical standards and practical application requirements should be considered [75]. Additionally, the prediction in this study is primarily based on fixed sample sites and has not yet been extended to the entire region (surface). Therefore, the next step will focus on predicting forest biomass for the entire region (surface). At the same time, improving the practicality and representativeness of the model, carrying out regional and national forest biomass estimation and evaluation, as well as establishing a universal biomass model suitable for large regions are all worthwhile goals for future studies.
In the context of global carbon cycling, forest biomass prediction plays a critical role in carbon sequestration, sustainable forest management, and response to global climate change [76]. In the long run, the high population density of Beijing and worldwide, the issuance of relevant policies, and the willingness of the public to participate in protection are all important for the growth of forest biomass. The research results of this study are significant for promoting sustainable forestry development in Beijing, as it can aid forest managers in developing more scientific forestry management strategies and evaluating the ecological benefits of forest ecosystems. However, forests are subject to dynamic changes in biomass due to natural succession and air pollution [77,78]. In addition, human activities, such as changes in forest landscape patterns, land use types, and fires, also have significant impacts on forest biomass [79,80,81,82]. Therefore, future studies should pay more attention to combining models and adding human activities as variables to obtain more accurate forest biomass estimates and understanding potential dynamic changes. These methods will provide a valuable reference for research on forest carbon storage and carbon balance.
Although this study made a good attempt at predicting regional forest biomass, there are still two areas where there is room for improvement in future research. The first area of potential improvement is that the prediction in this study is based on fixed sample locations, primarily used for point prediction, and has not yet been extended to the entire region (surface). The focus of future work will be expanding from point to surface prediction. Another potential area for improvement is the significant influence of human activities. In future research, changes in land cover and landscape patterns should be included as driving factors in modeling.
5. Conclusions
The results showed that the forest biomass prediction models based on both machine learning algorithms had good fitting accuracy, and there was no significant difference in the prediction results between the two models. However, the SVM model was better than BP-ANN. While the BP-ANN model provided more volatile predictions, and the accuracy was above 80%, the prediction results of the SVM model were relatively stable, and the accuracy was above 90%. We believe that machine learning models can better predict the potential of forest carbon sequestration and sequestration enhancement, which can provide a valuable reference for the improvement of forest quality and sustainable forest management in Beijing, nationwide, and globally.
Author Contributions
Conceptualization, J.L. and Q.Z.; methodology, J.L.; software, C.Y.; validation, J.L., C.P. and X.L.; formal analysis, C.Y.; investigation, J.L. and X.L.; resources, J.L.; data curation, J.L. and C.P.; writing—original draft preparation, J.L.; writing—review and editing, J.L. and C.Y.; visualization, J.L.; supervision, Q.Z.; project administration, Q.Z.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China under grant 32001249 and the State Key Laboratory Foundation of China under Grant A314021402-202220.
Data Availability Statement
Not applicable.
Acknowledgments
We are particularly grateful to the staff who collected data for forest resource inventory, whose work has provided data support for our research. Meanwhile, we are grateful to the staff at the department of Geographical Sciences of Northwest A&F University, and other people who have contributed to this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, A.; Kushwaha, S.K.P.; Nandy, S.; Padalia, H.; Ghosh, S.; Srivastava, A.; Kumari, N. Aboveground Forest Biomass Estimation by the Integration of TLS and ALOS PALSAR Data Using Machine Learning. Remote Sens. 2023, 15, 1143. [Google Scholar] [CrossRef]
- Chi, J.; Zhao, P.; Klosterhalfen, A.; Jocher, G.; Kljun, N.; Nilsson, M.B.; Peichl, M. Forest Floor Fluxes Drive Differences in the Carbon Balance of Contrasting Boreal Forest Stands. Agric. For. Meteorol. 2021, 306, 108454. [Google Scholar] [CrossRef]
- Buchholz, T.; Friedland, A.J.; Hornig, C.E.; Keeton, W.S.; Zanchi, G.; Nunery, J. Mineral Soil Carbon Fluxes in Forests and Implications for Carbon Balance Assessments. GCB Bioenergy 2014, 6, 305–311. [Google Scholar] [CrossRef]
- Primary Productivity of the Biosphere|SpringerLink. Available online: https://link.springer.com/book/10.1007/978-3-642-80913-2 (accessed on 20 March 2023).
- Dai, E.; Wu, Z.; Ge, Q.; Xi, W.; Wang, X. Predicting the Responses of Forest Distribution and Aboveground Biomass to Climate Change under RCP Scenarios in Southern China. Glob. Chang. Biol. 2016, 22, 3642–3661. [Google Scholar] [CrossRef] [PubMed]
- Remote Sensing|Free Full-Text|Estimating the Aboveground Biomass for Planted Forests Based on Stand Age and Environmental Variables. Available online: https://www.mdpi.com/2072-4292/11/19/2270/htm (accessed on 21 March 2023).
- Köhl, M.; Lasco, R.; Cifuentes, M.; Jonsson, Ö.; Korhonen, K.T.; Mundhenk, P.; de Jesus Navar, J.; Stinson, G. Changes in Forest Production, Biomass and Carbon: Results from the 2015 UN FAO Global Forest Resource Assessment. For. Ecol. Manag. 2015, 352, 21–34. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, D.; Chen, X.; Zhang, Y.; Maisupova, B.; Tao, Y. The Spatiotemporal Patterns of Vegetation Coverage and Biomass of the Temperate Deserts in Central Asia and Their Relationships with Climate Controls. Remote Sens. Environ. 2016, 175, 271–281. [Google Scholar] [CrossRef]
- Ribeiro, N.S.; Matos, C.N.; Moura, I.R.; Washington-Allen, R.A.; Ribeiro, A.I. Monitoring Vegetation Dynamics and Carbon Stock Density in Miombo Woodlands. Carbon Balance Manag. 2013, 8, 11. [Google Scholar] [CrossRef]
- Lu, C.; Xu, H.; Zhang, J.; Wang, A.; Wu, H.; Bao, R.; Ou, G. A Method for Estimating Forest Aboveground Biomass at the Plot Scale Combining the Horizontal Distribution Model of Biomass and Sampling Technique. Forests 2022, 13, 1612. [Google Scholar] [CrossRef]
- Ribeiro, N.S.; Saatchi, S.S.; Shugart, H.H.; Washington-Allen, R.A. Washington-Allen, Aboveground biomass and leaf area index(LAI) mapping for Niassa Reserve, northern Mozambique. J. Geophys. Res. 2008, 113, G02S02. [Google Scholar] [CrossRef]
- Saatchi, S.S.; Harris, N.L.; Brown, S.; Lefsky, M.; Mitchard, E.T.; Salas, W.; Zutta, B.R.; Buermann, W.; Lewis, S.L.; Hagen, S.; et al. Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 2011, 108, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, T.M. Live Above- and Belowground Biomass of a Mozambican Evergreen Forest: A Comparison of Estimates Based on Regression Equations and Biomass Expansion Factors. For. Ecosyst. 2016, 3, 28. [Google Scholar] [CrossRef]
- Guedes, B.S.; Sitoe, A.A.; Olsson, B.A. Allometric Models for Managing Lowland Miombo Woodlands of the Beira Corridor in Mozambique. Glob. Ecol. Conserv. 2018, 13, e00374. [Google Scholar] [CrossRef]
- Lisboa, S.N.; Guedes, B.S.; Ribeiro, N.; Sitoe, A. Biomass Allometric Equation and Expansion Factor for a Mountain Moist Evergreen Forest in Mozambique. Carbon Balance Manag. 2018, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Ou, Q.; Li, H.; Yang, Y. Factors Affecting the Biomass Conversion and Expansion Factor of Masson Pine in Fujian Province. Acta Ecol. Sin. 2017, 37, 5756–5764. [Google Scholar] [CrossRef]
- Liu, J.; Feng, Z.; Mannan, A.; Khan, T.U.; Cheng, Z. Comparing Non-Destructive Methods to Estimate Volume of Three Tree Taxa in Beijing, China. Forests 2019, 10, 92. [Google Scholar] [CrossRef]
- Fu, L.; Zeng, W.; Tang, S. Individual Tree Biomass Models to Estimate Forest Biomass for Large Spatial Regions Developed Using Four Pine Species in China. For. Sci. 2017, 63, 241–249. [Google Scholar] [CrossRef]
- Claus, A.; George, E. Effect of Stand Age on Fine-Root Biomass and Biomass Distribution in Three European Forest Chronosequences. Can. J. For. Res.-Rev. Can. Rech. For. 2005, 35, 1617–1625. [Google Scholar] [CrossRef]
- Van Den Berge, S.; Vangansbeke, P.; Calders, K.; Vanneste, T.; Baeten, L.; Verbeeck, H.; Krishna Moorthy, S.P.; Verheyen, K. Biomass Expansion Factors for Hedgerow-Grown Trees Derived from Terrestrial LiDAR. BioEnergy Res. 2021, 14, 561–574. [Google Scholar] [CrossRef]
- Li, L.; Zhou, B.; Liu, Y.; Wu, Y.; Tang, J.; Xu, W.; Wang, L.; Ou, G. Reduction in Uncertainty in Forest Aboveground Biomass Estimation Using Sentinel-2 Images: A Case Study of Pinus densata Forests in Shangri-La City, China. Remote Sens. 2023, 15, 559. [Google Scholar] [CrossRef]
- Hopman, H.J.; Chan, S.M.S.; Chu, W.C.W.; Lu, H.; Tse, C.-Y.; Chau, S.W.H.; Lam, L.C.W.; Mak, A.D.P.; Neggers, S.F.W. Personalized Prediction of Transcranial Magnetic Stimulation Clinical Response in Patients with Treatment-Refractory Depression Using Neuroimaging Biomarkers and Machine Learning. J. Affect. Disord. 2021, 290, 261–271. [Google Scholar] [CrossRef]
- Lopez-Serrano, P.M.; Lopez-Sanchez, C.A.; Alvarez-Gonzalez, J.G.; Garcia-Gutierrez, J. A Comparison of Machine Learning Techniques Applied to Landsat-5 TM Spectral Data for Biomass Estimation. Can. J. Remote Sens. 2016, 42, 690–705. [Google Scholar] [CrossRef]
- Pham, T.D.; Yoshino, K.; Le, N.N.; Bui, D.T. Estimating Aboveground Biomass of a Mangrove Plantation on the Northern Coast of Vietnam Using Machine Learning Techniques with an Integration of ALOS-2 PALSAR-2 and Sentinel-2A Data. Int. J. Remote Sens. 2018, 39, 7761–7788. [Google Scholar] [CrossRef]
- Huang, W.; Li, W.; Xu, J.; Ma, X.; Li, C.; Liu, C. Hyperspectral Monitoring Driven by Machine Learning Methods for Grassland Above-Ground Biomass. Remote Sens. 2022, 14, 2086. [Google Scholar] [CrossRef]
- Lopez-Lopez, S.F.; Martinez-Trinidad, T.; Benavides-Meza, H.; Garcia-Nieto, M.; de los Santos-Posadas, H.M. Non-Destructive Method for above-Ground Biomass Estimation of Fraxinus Uhdei (Wenz.) Lingelsh in an Urban Forest. Urban For. Urban Green. 2017, 24, 62–70. [Google Scholar] [CrossRef]
- Urbazaev, M.; Thiel, C.; Cremer, F. Estimation of forest aboveground biomass and uncertainties by integration of field measurements, airborne LiDAR, and SAR and optical satellite data in Mexico. Carbon Balance Manag. 2018, 13, 5. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Li, C.; Liu, Z. Forest Aboveground Biomass Estimation Using Landsat 8 and Sentinel-1A Data with Machine Learning Algorithms. Sci. Rep. 2020, 10, 9952. [Google Scholar] [CrossRef]
- Su, H.; Shen, W.; Wang, J.; Ali, A.; Li, M. Machine Learning and Geostatistical Approaches for Estimating Aboveground Biomass in Chinese Subtropical Forests. For. Ecosyst. 2020, 7, 64. [Google Scholar] [CrossRef]
- Smuga-Kogut, M.; Kogut, T.; Markiewicz, R.; Slowik, A. Use of Machine Learning Methods for Predicting Amount of Bioethanol Obtained from Lignocellulosic Biomass with the Use of Ionic Liquids for Pretreatment. Energies 2021, 14, 243. [Google Scholar] [CrossRef]
- HU, Y.; MA, L.; LI, R.; KE, Z.; YANG, J.; LIU, Z. Factor Analysis of Underground Biomass in Forest Ecosystem on the Loess Plateau. Acta Ecol. Sin. 2021, 41, 8643–8653. [Google Scholar] [CrossRef]
- Dulamsuren, C.; Hauck, M.; Bader, M.; Osokhjargal, D.; Oyungerel, S.; Nyambayar, S.; Runge, M.; Leuschner, C. Water Relations and Photosynthetic Performance in Larix Sibirica Growing in the Forest-Steppe Ecotone of Northern Mongolia. Tree Physiol. 2009, 29, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Newton, P.F. Simulating the Potential Effects of a Changing Climate on Black Spruce and Jack Pine Plantation Productivity by Site Quality and Locale through Model Adaptation. Forests 2016, 7, 223. [Google Scholar] [CrossRef]
- Jiang, F.; Sun, H.; Ma, K.; Fu, L.; Tang, J. Improving Aboveground Biomass Estimation of Natural Forests on the Tibetan Plateau Using Spaceborne LiDAR and Machine Learning Algorithms. Ecol. Indic. 2022, 143, 109365. [Google Scholar] [CrossRef]
- Li, X.; Du, H.; Mao, F.; Zhou, G.; Chen, L.; Xing, L.; Fan, W.; Xu, X.; Liu, Y.; Cui, L.; et al. Estimating Bamboo Forest Aboveground Biomass Using EnKF-Assimilated MODIS LAI Spatiotemporal Data and Machine Learning Algorithms. Agric. For. Meteorol. 2018, 256, 445–457. [Google Scholar] [CrossRef]
- Wu, C.; Tao, H.; Zhai, M.; Lin, Y.; Wang, K.; Deng, J.; Shen, A.; Gan, M.; Li, J.; Yang, H. Using Nonparametric Modeling Approaches and Remote Sensing Imagery to Estimate Ecological Welfare Forest Biomass. J. For. Res. 2018, 29, 151–161. [Google Scholar] [CrossRef]
- Fararoda, R.; Reddy, R.S.; Rajashekar, G.; Chand, T.R.K.; Jha, C.S.; Dadhwal, V.K. Improving Forest above Ground Biomass Estimates over Indian Forests Using Multi Source Data Sets with Machine Learning Algorithm. Ecol. Inform. 2021, 65, 101392. [Google Scholar] [CrossRef]
- Mas, J.F.; Flores, J.J. The Application of Artificial Neural Networks to the Analysis of Remotely Sensed Data. Remote Sens. 2008, 29, 617–663. [Google Scholar] [CrossRef]
- Szantoi, Z.; Escobedo, F.J.; Abd-Elrahman, A.; Pearlstine, L.; Dewitt, B.; Smith, S. Classifying spatially heterogeneous wetland communities using machine learning algorithms and spectral and textural features. Environ. Monit. Assess. 2015, 187, 262. [Google Scholar] [CrossRef]
- Foody, G.M.; Cutler, M.E.; McMorrow, J.; Pelz, D.; Tangki, M.; Boyd, D.S. Mapping the biomass of Bornean tropical rain forest from remotely sensed data. Glob. Ecol. Biogeogr. 2001, 10, 379. [Google Scholar] [CrossRef]
- Zhang, C.; Denka Durgan, S.; Sirianni, H.; Mishra, D. Quantification of sawgrass marsh aboveground biomass in the coastal Everglades using object-based ensemble analysis and Landsat data. Remote Sens. Environ. 2017, 204, 366–379. [Google Scholar] [CrossRef]
- Xu, X.; Cao, M.; Li, K. Temporal-Spatial Dynamics of Carbon Storage of Forest Vegetation in China. Prog. Geogr. 2007, 26, 1–10. [Google Scholar]
- Wang, H.; Niu, S.; Shao, X.; Zhang, C. Study on biomass estimation methods of understory shrubs and herbs in forest ecosystem. Acta Pratacult. Sin. 2014, 23, 20–29. [Google Scholar]
- Shen, C.; Lei, X.; Liu, H.; Wang, L.; Liang, W. Potential impacts of regional climate change on site productivity of Larix olgensis plantations in northeast China. Iforest–Biogeosciences For. 2015, 8, 642. [Google Scholar] [CrossRef]
- Sharma, R.P.; Breidenbach, J. Modeling Height-Diameter Relationship for Norway spruce, Scots pine, and downy birch using Norwegian national forest inventory data. For. Sci. Technol. 2015, 11, 44–53. [Google Scholar] [CrossRef]
- Wang, Y.; Lemay, V.; Baker, T.G. Modelling and prediction of dominant height and site index of Eucalyptus globulus plantations using a nonlinear mixed-effects model approach. Can. J. For. Res. 2007, 37, 1390–1403. [Google Scholar] [CrossRef]
- Wang, T.; Wang, G.; Innes, J.L.; Seely, B.; Chen, B. ClimateAP: An Application for Dynamic Local Downscaling of Historical and Future Climate Data in Asia Pacific. Front. Agr. Sci. Eng. 2017, 4, 448–458. [Google Scholar] [CrossRef]
- Wang, T.; Hamann, A.; Spittlehouse, D.L.; Murdock, T.Q. Murdock: ClimateWNA—High-Resolution Spatial Climate Data for Western North America. J. Appl. Meteor. Clim. 2012, 51, 16–29. [Google Scholar] [CrossRef]
- Wang, J.-F.; Li, X.-H.; Christakos, G.; Liao, Y.-L.; Zhang, T.; Gu, X.; Zheng, X.-Y. Geographical Detectors-Based Health Risk Assessment and Its Application in the Neural Tube Defects Study of the Heshun Region, China. Int. J. Geogr. Inf. Sci. 2010, 24, 107–127. [Google Scholar] [CrossRef]
- Dai, S.; Zheng, X.; Gao, L.; Xu, C.; Zuo, S.; Chen, Q.; Wei, X.; Ren, Y. Improving Plot-Level Model of Forest Biomass: A Combined Approach Using Machine Learning with Spatial Statistics. Forests 2021, 12, 1663. [Google Scholar] [CrossRef]
- Fang, J.Y.; Chen, A.P.; Peng, C.H.; Zhao, S.Q.; Ci, L. Changes in Forest Biomass Carbon Storage in China between 1949 and 1998. Science 2001, 292, 2320–2322. [Google Scholar] [CrossRef] [PubMed]
- Jagodzinski, A.M.; Dyderski, M.K.; Gesikiewicz, K.; Horodecki, P. Effects of Stand Features on Aboveground Biomass and Biomass Conversion and Expansion Factors Based on a Pinus Sylvestris L. Chronosequence in Western Poland. Eur. J. For. Res. 2019, 138, 673–683. [Google Scholar] [CrossRef]
- Jagodzinski, A.M.; Dyderski, M.K.; Gesikiewicz, K.; Horodecki, P. Tree and Stand Level Estimations of Abies Alba Mill. Aboveground Biomass. Ann. For. Sci. 2019, 76, 56. [Google Scholar] [CrossRef]
- Wang, G.; Guan, D.; Xiao, L.; Peart, M.R. Forest Biomass-Carbon Variation Affected by the Climatic and Topographic Factors in Pearl River Delta, South China. J. Environ. Manag. 2019, 232, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ji, X.; Deane, D.C.; Wu, L.; Chen, S. Spatiotemporal Distribution and Driving Factors of Forest Biomass Carbon Storage in China: 1977–2013. Forests 2017, 8, 263. [Google Scholar] [CrossRef]
- Wang, L.; Silván-Cárdenas, J.L.; Sousa, W.P. Neural Network Classification of Mangrove Species from Multi-seasonal Ikonos Imagery. Photogramm. Eng. Remote Sens. 2008, 74, 921–927. [Google Scholar] [CrossRef]
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random Forests for Classification in Ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef] [PubMed]
- Muukkonen, P.; Heiskanen, J. Estimating Biomass for Boreal Forests Using ASTER Satellite Data Combined with Standwise Forest Inventory Data. Remote Sens. Environ. 2005, 99, 434–447. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Funahashi, K.-I. On the Approximate Realization of Continuous Mappings by Neural Networks. Neural Netw. 1989, 2, 183–192. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Zong, H. Support Vector Machine Classification over Encrypted Data. Appl. Intell. 2022, 52, 5938–5948. [Google Scholar] [CrossRef]
- Dhanda, P.; Nandy, S.; Kushwaha, S.P.S.; Ghosh, S.; Murthy, Y.V.N.K.; Dadhwal, V.K. Optimizing Spaceborne LiDAR and Very High Resolution Optical Sensor Parameters for Biomass Estimation at ICESat/GLAS Footprint Level Using Regression Algorithms. Prog. Phys. Geogr. 2017, 41, 247–267. [Google Scholar] [CrossRef]
- Kim, K.I.; Jung, K.; Park, S.H.; Kim, H.J. Support Vector Machines for Texture Classification. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 1542–1550. [Google Scholar] [CrossRef]
- Zeng, W.; Duo, H.; Lei, X.; Chen, X.; Wang, X.; Pu, Y.; Zou, W. Individual Tree Biomass Equations and Growth Models Sensitive to Climate Variables for Larix Spp. in China. Eur. J. For. Res. 2017, 136, 233–249. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, K.; Liu, L.; Wang, S.; Liu, H. Retrieval of Mangrove Aboveground Biomass at the Individual Species Level with WorldView-2 Images. Remote Sens. 2015, 7, 12192–12214. [Google Scholar] [CrossRef]
- Han, M.; Xing, Y.; Li, G.; Huang, J.; Cai, L. Comparison of the accuracy of the maximum canopy height and biomass inversion of the data of different GEDI algorithm groups. J. Cent. South Univ. For. Technol. 2022, 42, 72–82. [Google Scholar] [CrossRef]
- Konopka, B.; Pajtik, J.; Moravcik, M.; Lukac, M. Biomass Partitioning and Growth Efficiency in Four Naturally Regenerated Forest Tree Species. Basic Appl. Ecol. 2010, 11, 234–243. [Google Scholar] [CrossRef]
- Santoro, M.; Beaudoin, A.; Beer, C.; Cartus, O.; Fransson, J.B.S.; Hall, R.J.; Pathe, C.; Schmullius, C.; Schepaschenko, D.; Shvidenko, A.; et al. Forest Growing Stock Volume of the Northern Hemisphere: Spatially Explicit Estimates for 2010 Derived from Envisat ASAR. Remote Sens. Environ. 2015, 168, 316–334. [Google Scholar] [CrossRef]
- Che, J. Optimal Sub-Models Selection Algorithm for Combination Forecasting Model. Neurocomputing 2015, 151, 364–375. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, W.; Xiao, J.; Xie, L. Estimating the grassland aboveground biomass based on remote sensing data and machine learning algorithm. J. Glaciol. Geocryol. 2023, 45, 1–10. [Google Scholar]
- Li, X.; Wu, B.; Su, X.; Chen, Y.; Peng, Y.; Yu, Y.; Fan, X. Study on Estimation Model of Eucalyptus Accumulation in Guangxi Based on Decision Tree Integrated Learning. J. Agric. Sci. Technol. 2020, 22, 81–90. [Google Scholar] [CrossRef]
- Saint-Andre, L.; M’bou, A.T.; Mabiala, A.; Mouvondy, W.; Jourdan, C.; Roupsard, O.; Deleporte, P.; Hamel, O.; Nouvellon, Y. Age-Related Equations for above- and below-Ground Biomass of a Eucalyptus Hybrid in Congo. For. Ecol. Manag. 2005, 205, 199–214. [Google Scholar] [CrossRef]
- Niklas, K.; Tiffney, B. The Quantification of Plant Biodiversity Through Time. Philos. Trans. R. Soc. B-Biol. Sci. 1994, 345, 35–44. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Beier, C.M.; Johnson, L.; Phoenix, D.B. A COMPARISON OF DECISION TREE-BASED MODELS FOR FOREST ABOVE-GROUND BIOMASS ESTIMATION USING A COMBINATION OF AIRBORNE LIDAR AND LANDSAT DATA. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2021, V-3-2021, 235–241. [Google Scholar] [CrossRef]
- Laurance, W.F.; Andrade, A.S.; Magrach, A.; Camargo, J.L.C.; Campbell, M.; Fearnside, P.M.; Edwards, W.; Valsko, J.J.; Lovejoy, T.E.; Laurance, S.G. Apparent Environmental Synergism Drives the Dynamics of Amazonian Forest Fragments. Ecology 2014, 95, 3018–3026. [Google Scholar] [CrossRef]
- Main-Knorn, M.; Cohen, W.B.; Kennedy, R.E.; Grodzki, W.; Pflugmacher, D.; Griffiths, P.; Hostert, P. Monitoring Coniferous Forest Biomass Change Using a Landsat Trajectory-Based Approach. Remote Sens. Environ. 2013, 139, 277–290. [Google Scholar] [CrossRef]
- Oliveira, C.P.d.; Ferreira, R.L.C.; da Silva, J.A.A.; Lima, R.B.d.; Silva, E.A.; Silva, A.F.d.; Lucena, J.D.S.d.; dos Santos, N.A.T.; Lopes, I.J.C.; Pessoa, M.M.d.L.; et al. Modeling and Spatialization of Biomass and Carbon Stock Using LiDAR Metrics in Tropical Dry Forest, Brazil. Forests. 2021, 12, 473. [Google Scholar] [CrossRef]
- Wu, Z.; Dai, E.; Ge, Q.; Xi, W.; Wang, X. Modelling the integrated effects of land use and climate change scenarios on forest aboveground biomass: A case study in Taihe County of China. J. Geogr. Sci. 2017, 27, 205–222. [Google Scholar] [CrossRef]
- Macave, O.A.; Ribeiro, N.S.; Ribeiro, A.I.; Chaúque, A.; Bandeira, R.; Branquinho, C.; Washington-Allen, R. Modelling Aboveground Biomass of Miombo Woodlands in Niassa Special Reserve, Northern Mozambique. Forests 2022, 13, 311. [Google Scholar] [CrossRef]
- Cornejo, S.; Becker, N.; Hemp, A.; Hertel, D. Effects of land-use change and disturbance on the fine root biomass, dynamics, morphology, and related C and N fluxes to the soil of forest ecosystems at different elevations at Mt. Kilimanjaro (Tanzania). Oecologia 2023, 201, 1089–1107. [Google Scholar] [CrossRef]
- Ryan, C.M.; Williams, M.; Grace, J. Above- and Belowground Carbon Stocks in a Miombo Woodland Landscape of Mozambique. Biotropica 2011, 43, 423–432. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).