Identification and Functional Analysis of the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family in Liriodendron Hybrids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification, Characteristics and Phylogenetic Analysis of LhPEBPs
2.2. Plant Materials and RNA Extraction
2.3. LhPEBP Gene Cloning and Vector Construction
2.4. Flowering and Germination Analysis in Arabidopsis
2.5. Data Collection and Statistical Analysis
3. Results
3.1. Identification of PEBP Family Members in Liriodendron
3.2. Amino Acid Alignment and Conserved Domain Analysis
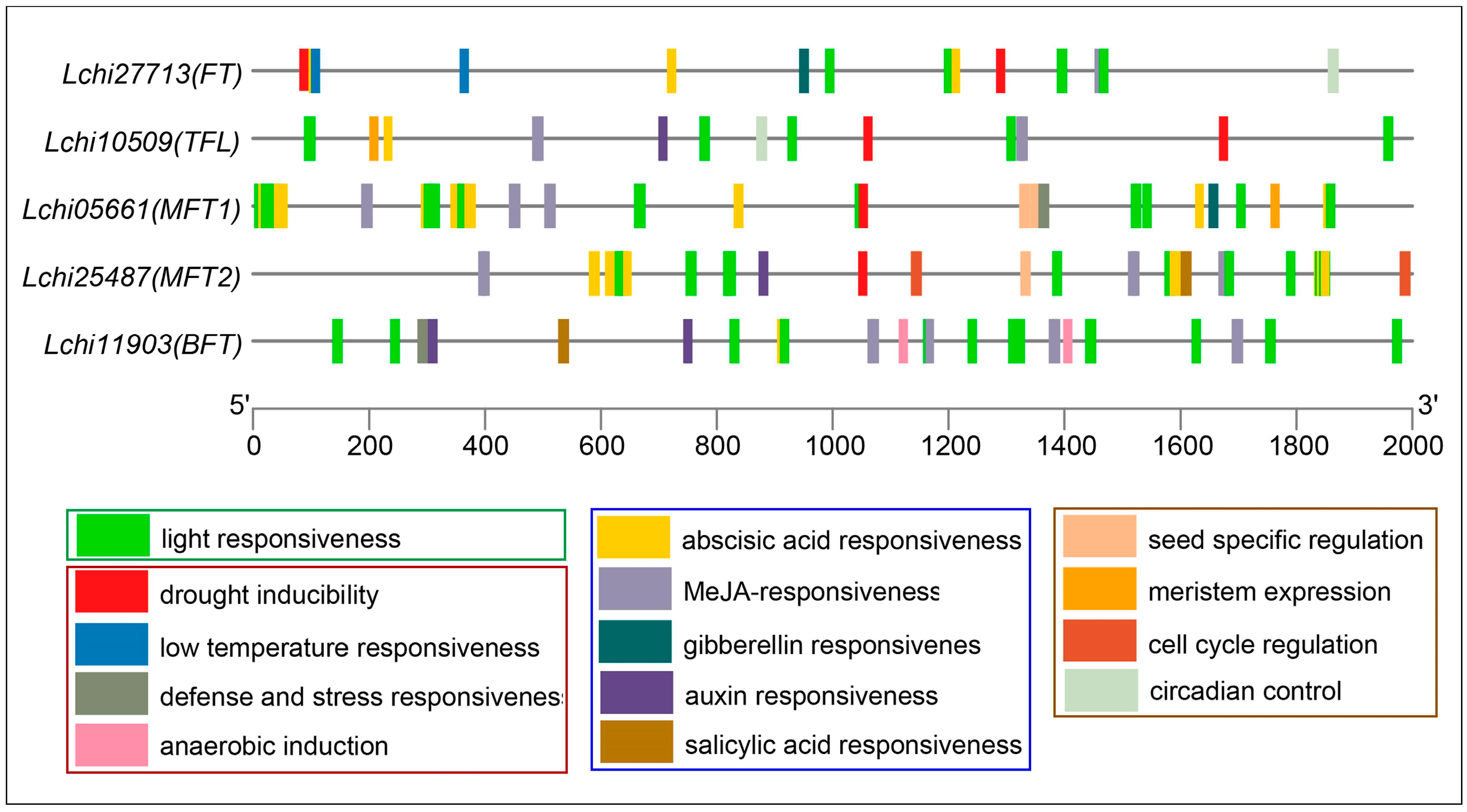
3.3. Cis-Elements Analysis in Promoter of the PEBP Genes
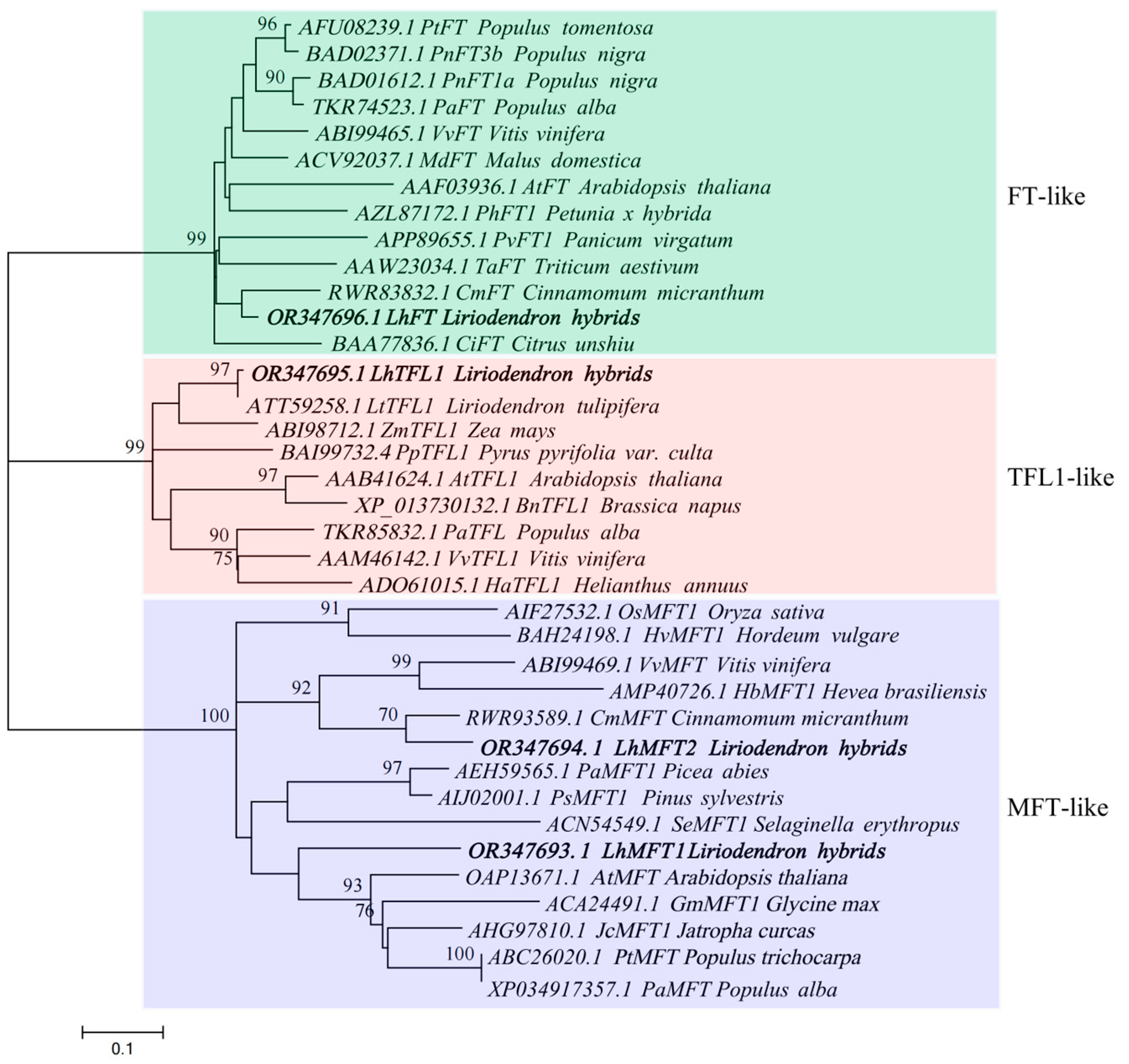
3.4. Phylogenetic Analysis
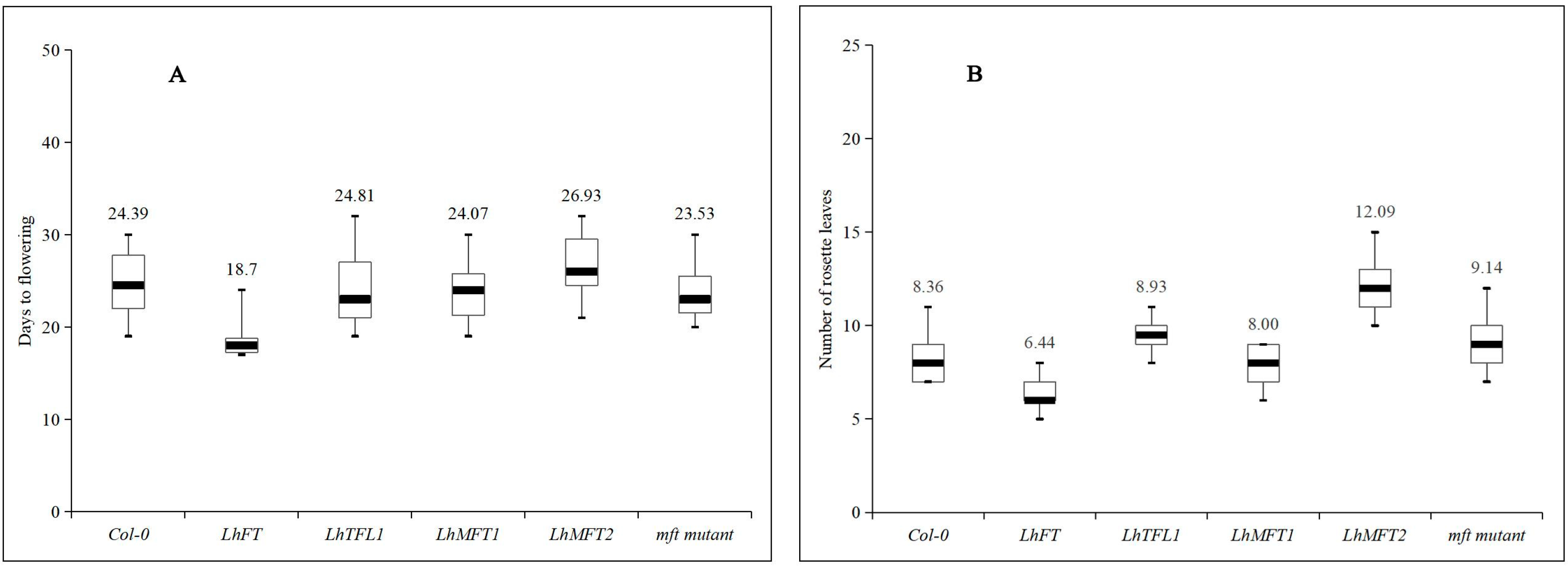
3.5. Flowering Analysis of LhPEBPs in Transgenic Arabidopsis Plants
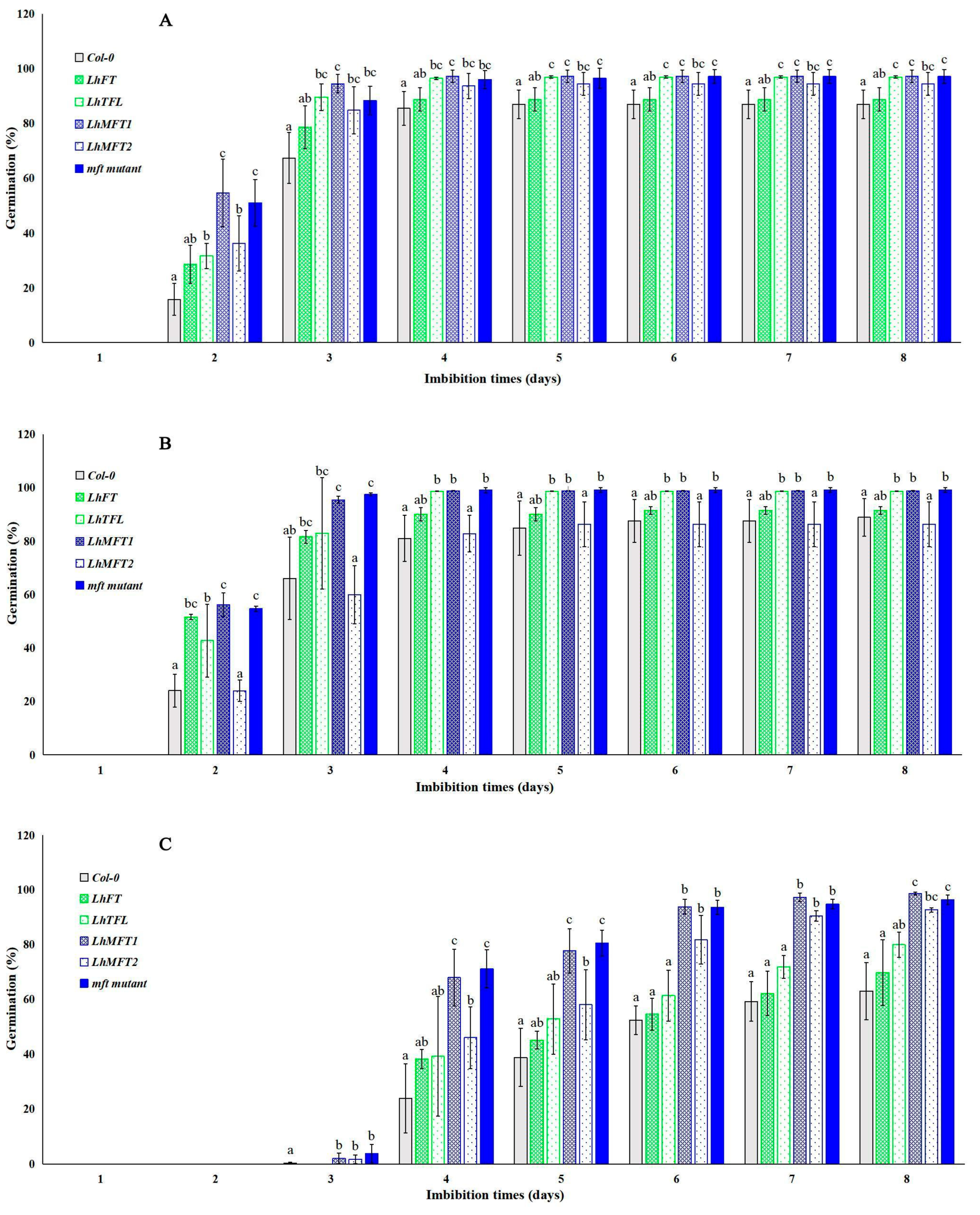
3.6. Germination Analysis of LhPEBPs in Transgenic Arabidopsis Plants
4. Discussion
4.1. The Characteristics and Evolution of PEBP Genes in Liriodendron Hybrids
4.2. The Function of LhFT/LhTFL1 Was Conserved, and LhMFTs Functioned Differently
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Danilevskaya, O.N.; Meng, X.; Hou, Z.L.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef]
- Banfield, M.J.; Brady, R.L. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 2000, 297, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Karlgren, A.; Gyllenstrand, N.; Kallman, T.; Sundstrom, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP Gene Family in Plants: Functional Diversification in Seed Plant Evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef]
- Simon, R.; Igeno, M.I.; Coupland, G. Activation of floral meristem identity genes in Arabidopsis. Nature 1996, 384, 59–62. [Google Scholar] [CrossRef]
- Huang, T.; Bohlenius, H.; Eriksson, S.; Parcy, F.; Nilsson, O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering (Retracted Article. See vol 316, pg 367, 2007). Science 2005, 309, 1694–1696. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef]
- Ferrier, T.; Matus, J.T.; Jin, J.; Riechmann, J.L. Arabidopsis paves the way: Genomic and network analyses in crops. Curr. Opin. Biotechnol. 2011, 22, 260–270. [Google Scholar] [CrossRef]
- Shalit, A.; Rozman, A.; Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y.; Lifschitz, E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 2009, 106, 8392–8397. [Google Scholar] [CrossRef] [PubMed]
- Nitcher, R.; Distelfeld, A.; Tan, C.; Yan, L.; Dubcovsky, J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol. Genet. Genom. 2013, 288, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Kawazu, Y.; Fujiyama, R.; Honda, I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 2011, 168, 1602–1607. [Google Scholar] [CrossRef]
- Komiya, R.; Ikegami, A.; Tamaki, S.; Yokoi, S.; Shimamoto, K. Hd3a and RFT1 are essential for flowering in rice. Development 2008, 135, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Pin, P.A.; Nilsson, O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012, 35, 1742–1755. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–132. [Google Scholar] [CrossRef]
- Andre, D.; Marcon, A.; Lee, K.C.; Goretti, D.; Zhang, B.; Delhomme, N.; Schmid, M.; Nilsson, O. FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees. Curr. Biol. 2022, 32, 2988–2996. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1016. [Google Scholar] [CrossRef]
- Nakamura, Y.; Andres, F.; Kanehara, K.; Liu, Y.C.; Doermann, P.; Coupland, G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 2014, 5, 3553. [Google Scholar] [CrossRef]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.I. TFL1-Like Proteins in Rice Antagonize Rice FT-Like Protein in Inflorescence Development by Competition for Complex Formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef]
- Shannon, S.; Meekswagner, D.R. A Mutation in the Arabidopsis Tfl1 Gene Affects Inflorescence Meristem Development. Plant Cell 1991, 3, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Sohn, E.J.; Rojas-Pierce, M.; Pan, S.; Carter, C.; Serrano-Mislata, A.; Madueno, F.; Rojo, E.; Surpin, M.; Raikhel, N.V. The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc. Natl. Acad. Sci. USA 2007, 104, 18801–18806. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, H.; Szankowski, I.; Waidmann, S.; Peil, A.; Trankner, C.; Hanke, M.V. The MdTFL1 gene of apple (Malus x domestica Borkh.) reduces vegetative growth and generation time. Tree Physiol. 2012, 32, 1288–1301. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Gaston, A.; Remay, A.; Thouroude, T.; Jeauffre, J.; Kawamura, K.; Oyant, L.H.S.; Araki, T.; Denoyes, B.; Foucher, F. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012, 69, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; He, X.H.; Yu, H.X.; Mo, X.; Fan, Y.; Fan, Z.Y.; Xie, X.J.; Liu, Y.; Luo, C. Overexpression of four MiTFL1 genes from mango delays the flowering time in transgenic Arabidopsis. BMC Plant Biol. 2021, 21, 407. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Z.K.; Liu, Y.F.; Liu, T.F.; Li, Q.; Ji, Y.Y.; Li, C.C.; Fang, C.; Wang, M.; Wu, M.; et al. Functional Evolution of Phosphatidylethanolamine Binding Proteins in Soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef]
- Yoo, S.Y.; Kardailsky, I.; Lee, J.S.; Weigel, D.; Ahn, J.H. Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol. Cells 2004, 17, 95–101. [Google Scholar]
- Lu, K.X.; Guo, Z.Y.; Di, S.Y.; Lu, Y.Y.; Muhammad, I.A.R.; Rong, C.Y.; Ding, Y.F.; Li, W.Q.; Ding, C.Q. OsMFT1 Inhibits Seed Germination by Modulating Abscisic Acid Signaling and Gibberellin Biosynthesis under Salt Stress in Rice. Plant Cell Physiol. 2023, 64, 674–685. [Google Scholar] [CrossRef]
- Xi, W.Y.; Liu, C.; Hou, X.L.; Yu, H. MOTHER OF FT AND TFL1 Regulates Seed Germination through a Negative Feedback Loop Modulating ABA Signaling in Arabidopsis. Plant Cell 2010, 22, 1733–1748. [Google Scholar] [CrossRef]
- Li, Q.; Fan, C.M.; Zhang, X.M.; Wang, X.; Wu, F.Q.; Hu, R.B.; Fu, Y.F. Identification of a Soybean MOTHER OF FT AND TFL1 Homolog Involved in Regulation of Seed Germination. PLoS ONE 2014, 9, e99642. [Google Scholar] [CrossRef]
- Yu, X.L.; Liu, H.; Sang, N.; Li, Y.F.; Zhang, T.T.; Sun, J.; Huang, X.Z. Identification of cotton MOTHER OF FT AND TFL1 homologs, GhMFT1 and GhMFT2, involved in seed germination. PLoS ONE 2019, 14, e0215771. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, G.F.; Wu, H.; Fan, X.W.; Liang, L.W.; Zhao, H.; Li, S.L.; Hu, Y.; Liu, H.Y.; Ayaad, M.; et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 2020, 103, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, G.F.; Hu, Y.; Liu, H.Y.; Bai, X.F.; Qin, R.; Xing, Y.Z. OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J. Exp. Bot. 2018, 69, 4283–4293. [Google Scholar] [CrossRef]
- Cai, Z.D.; Xian, P.Q.; Cheng, Y.B.; Zhong, Y.W.; Yang, Y.; Zhou, Q.H.; Lian, T.X.; Ma, Q.B.; Nian, H.; Ge, L.F. MOTHER-OF-FT-AND-TFL1 regulates the seed oil and protein content in soybean. New Phytol. 2023, 239, 905–919. [Google Scholar] [CrossRef]
- Parks, C.R.; Wendel, J.F. Molecular Divergence between Asian and North-American Species of Liriodendron (Magnoliaceae) with Implications for Interpretation of Fossil Floras. Am. J. Bot. 1990, 77, 1243–1256. [Google Scholar] [CrossRef]
- Chen, T.T.; Sheng, Y.; Hao, Z.D.; Long, X.F.; Fu, F.F.; Liu, Y.; Tang, Z.H.; Ali, A.; Peng, Y.; Lu, L.; et al. Transcriptome and proteome analysis suggest enhanced photosynthesis in tetraploid Liriodendron sino-americanum. Tree Physiol. 2021, 41, 1953–1971. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Igasaki, T.; Watanabe, Y.; Nishiguchi, M.; Kotoda, N. The FLOWERING LOCUS T/TERMINAL FLOWER 1 family in Lombardy poplar. Plant Cell Physiol. 2008, 49, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.X.; Guo, X.; Hao, Y.J.; Lu, G.; Li, D.; Lu, J.X.; Zhang, T. Genome-wide characterization of PEBP gene family in Perilla frutescens and PfFT1 promotes flowering time in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1026696. [Google Scholar] [CrossRef]
- Venail, J.; Santos, P.H.D.; Manechini, J.R.; Alves, L.C.; Scarpari, M.; Falcao, T.; Romanel, E.; Brito, M.; Vicentini, R.; Pinto, L.; et al. Analysis of the PEBP gene family and identification of a novel FLOWERING LOCUS T orthologue in sugarcane. J. Exp. Bot. 2022, 73, 2035–2049. [Google Scholar] [CrossRef]
- Wang, L.S.; Li, H.Y.; He, M.; Dong, L.D.; Huang, Z.R.; Chen, L.Y.; Nan, H.Y.; Kong, F.J.; Liu, B.H.; Zhao, X.H. GIGANTEA orthologs, E2 members, redundantly determine photoperiodic flowering and yield in soybean. J. Integr. Plant Biol. 2023, 65, 188–202. [Google Scholar] [CrossRef]
- Yang, Z.H.; Chen, L.; Kohnen, M.V.; Xiong, B.; Zhen, X.; Liao, J.K.; Oka, Y.; Zhu, Q.; Gu, L.F.; Lin, C.T.; et al. Identification and Characterization of the PEBP Family Genes in Moso Bamboo (Phyllostachys heterocycla). Sci. Rep. 2019, 9, 14998. [Google Scholar] [CrossRef]
- Zhao, S.L.; Wei, Y.R.; Pang, H.G.; Xu, J.F.; Li, Y.L.; Zhang, H.X.; Zhang, J.G.; Zhang, Y.X. Genome-wide identification of the PEBP genes in pears and the putative role of PbFT in flower bud differentiation. PeerJ 2020, 8, e8928. [Google Scholar] [CrossRef]
- Bennett, T.; Dixon, L.E. Asymmetric expansions of FT and TFL1 lineages characterize differential evolution of the EuPEBP family in the major angiosperm lineages. BMC Biol. 2021, 19, 181. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.H.; Tan, B.Y.; Robinson, K.M.; Michelson, I.H.; Johansson, A.; Nystedt, B.; Scofield, D.G.; Nilsson, O.; Jansson, S.; et al. A major locus controls local adaptation and adaptive life history variation in a perennial plant. Genome Biol. 2018, 19, 72. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Adams, J.P.; Kim, H.J.; No, K.; Ma, C.P.; Strauss, S.H.; Drnevich, J.; Vandervelde, L.; Ellis, J.D.; Rice, B.M.; et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 2011, 108, 10756–10761. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Hao, Z.D.; Peng, Y.; Liu, S.Q.; Hu, L.F.; Shen, Y.B.; Shi, J.S.; Chen, J.H. Morphological, phenological, and transcriptional analyses provide insight into the diverse flowering traits of a mutant of the relic woody plant Liriodendron chinense. Hortic. Res. 2021, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1(BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.J.; Calonje, M.; Martinez-Zapater, J.M. The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 2007, 63, 637–650. [Google Scholar] [CrossRef]
- Herath, D.; Voogd, C.; Mayo-Smith, M.; Yang, B.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype. Plant Biotechnol. J. 2022, 20, 2064–2076. [Google Scholar] [CrossRef]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef]
- Wang, L.L.; Yan, J.P.; Zhou, X.; Cheng, S.Y.; Chen, Z.X.; Song, Q.L.; Liu, X.M.; Ye, J.B.; Zhang, W.W.; Wu, G.X.; et al. GbFT, a FLOWERING LOCUS T homolog from Ginkgo biloba, promotes flowering in transgenic Arabidopsis. Sci. Hortic. 2019, 247, 205–215. [Google Scholar] [CrossRef]
- Qin, Z.R.; Wu, J.J.; Geng, S.F.; Feng, N.; Chen, F.J.; Kong, X.C.; Song, G.Y.; Chen, K.; Li, A.L.; Mao, L.; et al. Regulation of FT splicing by an endogenous cue in temperate grasses. Nat. Commun. 2017, 8, 14320. [Google Scholar] [CrossRef]
- Coelho, C.P.; Minow, M.A.; Chalfun, A.; Colasanti, J. Putative sugarcane FT/TFL1 genes delay flowering time and alter reproductive architecture in Arabidopsis. Front. Plant Sci. 2014, 5, 221. [Google Scholar] [CrossRef]
- Samarth; Lee, R.; Kelly, D.; Turnbull, M.H.; Macknight, R.; Poole, A.M.; Jameson, P.E. A novel TFL1 gene induces flowering in the mast seeding alpine snow tussock, Chionochloa pallens (Poaceae). Mol. Ecol. 2022, 31, 822–838. [Google Scholar] [CrossRef]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.; Wang, A.K.; Sun, S.L.; Liang, S.; Wang, X.J.; Ye, Q.S.; Li, H.Q. Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tissue Organ 2012, 111, 143–151. [Google Scholar] [CrossRef]
- Bi, Z.H.; Li, X.; Huang, H.S.; Hua, Y.W. Identification, Functional Study, and Promoter Analysis of HbMFT1, a Homolog of MFT from Rubber Tree (Hevea brasiliensis). Int. J. Mol. Sci. 2016, 17, 247. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Abe, F.; Kawahigashi, H.; Nakazono, K.; Tagiri, A.; Matsumoto, T.; Utsugi, S.; Ogawa, T.; Handa, H.; Ishida, H.; et al. A Wheat Homolog of MOTHER OF FT AND TFL1 Acts in the Regulation of Germination. Plant Cell 2011, 23, 3215–3229. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Gao, Y.R.; Wei, W.; Zhang, K.; Feng, J.Y. Strawberry MOTHER OF FT AND TFL1 regulates seed germination and post-germination growth through integrating GA and ABA signaling in Arabidopsis. Plant Cell Tissue Organ 2016, 126, 343–352. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Yano, K.; Wang, F.M.; Mori, M.; Kawamura, M.; Koketsu, E.; Hattori, M.; Ordonio, R.L.; Huang, P.; et al. Genome-wide association study identifies a gene responsible for temperature-dependent rice germination. Nat. Commun. 2022, 13, 5665. [Google Scholar] [CrossRef]

| Gene Name | Accession No. | CDS Length (bp) | Putative Protein Length (aa) | aa Identities to Arabidopsis thaliana (%) | aa Identities to Populus alba (%) |
|---|---|---|---|---|---|
| LhFT | OR347696 | 525 | 174 | 78.74 | 87.93 |
| LhTFL | OR347695 | 522 | 173 | 75.72 | 79.19 |
| LhMFT1 | OR347693 | 522 | 173 | 70.52 | 69.94 |
| LhMFT2 | OR347694 | 525 | 174 | 63.79 | 64.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, M.; Liu, L.; Hu, P.; Yu, X.; Zhou, H.; Liu, S.; Liu, T.; Yu, F.; Yang, A. Identification and Functional Analysis of the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family in Liriodendron Hybrids. Forests 2023, 14, 2103. https://doi.org/10.3390/f14102103
Hu M, Liu L, Hu P, Yu X, Zhou H, Liu S, Liu T, Yu F, Yang A. Identification and Functional Analysis of the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family in Liriodendron Hybrids. Forests. 2023; 14(10):2103. https://doi.org/10.3390/f14102103
Chicago/Turabian StyleHu, Miao, Lipan Liu, Ping Hu, Xiaoling Yu, Hua Zhou, Shujuan Liu, Tengyun Liu, Faxin Yu, and Aihong Yang. 2023. "Identification and Functional Analysis of the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family in Liriodendron Hybrids" Forests 14, no. 10: 2103. https://doi.org/10.3390/f14102103
APA StyleHu, M., Liu, L., Hu, P., Yu, X., Zhou, H., Liu, S., Liu, T., Yu, F., & Yang, A. (2023). Identification and Functional Analysis of the Phosphatidylethanolamine-Binding Protein (PEBP) Gene Family in Liriodendron Hybrids. Forests, 14(10), 2103. https://doi.org/10.3390/f14102103




