Abstract
Sophora japonica is a native leguminous tree species in China. The high stress tolerance contributes to its long lifespan of thousands of years. The lack of genomic resources greatly limits genetic studies on the stress responses of S. japonica. In this study, RNA-seq was conducted for S. japonica roots grown under short-term 20% polyethylene glycol (PEG) 6000-induced drought stress under normal N and N starvation conditions (1 and 0 mM NH4NO3, respectively). In each of the libraries, we generated more than 25 million clean reads, which were then de novo assembled to 46,852 unigenes with an average length of 1310.49 bp. In the differential expression analyses, more differentially expressed genes (DEGs) were found under drought with N starvation than under single stresses. The number of transcripts identified under N starvation and drought in S. japonica was nearly the same, but more upregulated genes were induced by drought, while more downregulated genes were induced by N starvation. Genes involved in “phenylpropanoid biosynthesis” and “biosynthesis of amino acids” pathways were upregulated according to KEGG enrichment analyses, irrespective of the stress treatments. Additionally, upregulated N metabolism genes were enriched upon drought, and downregulated photosynthesis genes were enriched under N starvation. We found 4,372 and 5,430 drought-responsive DEGs under normal N and N starvation conditions, respectively. N starvation may aggravate drought by downregulating transcripts in the “carbon metabolism”, “ribosome”, “arginine biosynthesis pathway”, “oxidative phosphorylation” and “aminoacyl-tRNA biosynthesis” pathways. We identified 78 genes related to N uptake and assimilation, 38 of which exhibited differential expression under stress. A total of 395 DEGs were categorized as transcription factors, of which AR2/ERF-ERF, WRKY, NAC, MYB, bHLH, C3H and C2C2-Dof families played key roles in drought and N starvation stresses. The transcriptome data obtained, and the genes identified facilitate our understanding of the mechanisms of S. japonica responses to drought and N starvation stresses and provide a molecular foundation for understanding the mechanisms of its long lifespan for breeding resistant varieties for greening.
1. Introduction
Sophora japonica (Chinese scholar tree) is a native deciduous tree species in China belonging to the Fabaceae family. Because of its well-developed root system, large crown and dense foliage, S. japonica is widely used for urban and rural greening in northern China. S. japonica has a long lifespan and many individuals in China have lived for thousands of years, which may be attributed to its strong adaptability and resistance to various abiotic stresses. In recent years, S. japonica, especially the ancient specimens, has been threatened by various environmental stresses as a result of climate change, with drought and nutrient deficiency being the two main factors restricting its growth and development [1]. Drought often occurs in arid and semiarid regions, and is predicted to become more severe because of global warming [2]. Nitrogen (N) is one of the essential macronutrients for plant growth and development. N deficiency often occurs simultaneously with drought because N must be dissolved in water before being taken up by roots. To understand how ancient trees adapt to climate change and to provide a theoretical basis for ancient tree protection, it is urgent to study the physical and molecular mechanisms of stress resistance in S. japonica.
Drought generally leads to physiological, morphological, biochemical and molecular changes in plants [3]. At the physiological level, antioxidative enzymes including superoxide dismutase (SOD), catalase (CAT) and peroxidase (POD), and the osmotic regulator proline play important roles in plant adaption to changes in the environment [4]. The malonaldehyde (MDA) content is considered an indicator of oxidative damage, reflecting the extent of the peroxidation of membrane lipids. Transcription factors (TFs) such as members of the MYB, NAC, WRKY and bZIP families, acting as regulatory proteins, are required in plant responses to various stresses [5]. Two previous physiological studies on S. japonica saplings suggested that the chlorophyll, leaf relative water contents and several physiological indexes were negatively affected by drought [6,7]. However, the lack of whole genome information has greatly limited our understanding of the stress resistance of S. japonica at the molecular level.
Nitrogen is an essential component of various macromolecules such as chlorophylls, nucleic acids, proteins, amino acids and some hormones. The utilization of N by plants mainly includes N uptake, assimilation and metabolism. As two primary inorganic N sources available for higher plants, ammonium (NH4+) and nitrate (NO3−) are first absorbed through roots specifically by ammonium transporters (AMTs) and nitrite transporters (NRTs), respectively. The two ions are then converted into organic N forms that are required for protein formation in plants through a series of metabolic processes. Together with the genes encoding them, enzymes including nitrate reductase (NR), nitrite reductase (NiR), glutamine synthetase (GS), glutamine oxide glutarate aminotransferase (GOGAT) and glutamate dehydrogenase (GDH) play different roles in metabolic processes [8]. Genes related to N usage in plants have been thoroughly studied in many species, especially in model plants such as Arabidopsis, rice and poplar [9,10]. To date, only five nitrogen transporter genes have been homogeneously cloned, but their lengths were all less than 600 bp [11]. S. japonica does not form nodules and cannot fix N [12], so it is more sensitive to the N supply than most legume species that can fix N [13]. Accordingly, identification of these N-related genes is essential for future studies on the mechanisms of N utilization in S. japonica.
Previous studies have shown that the absorption and utilization of water and N are coupled physiological processes [1,14]. Drought significantly affects nitrate uptake, the enzyme activity involved in N metabolism and N-related genes in various species [15,16,17,18]. N form and the levels of N available can also affect root water uptake [19]. Many studies have reported that N supply contributed to enhanced drought tolerance [20,21,22,23,24,25,26]. However, growth inhibition under high nitrate supply was observed in maize [27], and OsNAR2.1 positively regulated drought tolerance and grain yield in rice [28]. In addition, drought tolerance was not influenced by high nutrients in two oak species [29]. These results indicated that the effect of N conditions on drought tolerance varied and may depend on plant species, N form, N concentration and the degree of drought. Up to now, there have been no reports about the effects of N interacting with drought in S. japonica.
At the transcriptional level, the responses of metabolic processes to drought and N starvation have been studied in many plants. In wheat leaves, drought adaption may be related to pathways including transport systems, amino acid metabolism and protein processing pathways [30]. Fatty acid metabolism, starch and sucrose metabolism, and nitrogen metabolism may be involved in the drought response in Caragana korshinskii [31]. Genes in the amino acid metabolism, transport and stress categories were upregulated, whereas genes in hormone metabolism and redox pathways were downregulated in the roots of Arabidopsis thaliana under N starvation [32]. Lu et al. [33] studied the mitigating effect of N supply on drought and found that genes related to the antioxidative system and secondary metabolism were promoted by N addition in poplar. The mechanisms underlying how plants respond to single drought or N deficiency stresses have been widely studied, whereas the effect of N starvation on drought stress has rarely been studied. In recent years, transcriptomics has become an effective strategy to study changes in gene expression or metabolic processes when an organism is under stress, especially in non-modal plants [31,34,35]. There are only two genetic studies on S. japonica, which have only used the aboveground plant parts as sequencing materials [36,37]. Thus, the mechanisms of how S. japonica roots respond to drought and N deficiency at the molecular level remain unknown.
In this study, S. japonica seedlings were exposed to short-term treatments with N starvation and 20% polyethylene glycol(PEG)-6000-induced drought stresses. The leaf chlorophyll content and the MDA and proline contents in both leaves and roots were measured. Simultaneously, twelve fine root samples were sequenced using an Illumina HiSeq 2000 platform. The main objectives of this study were as follows: (1) to understand the molecular regulation of drought and N starvation stresses in S. japonica; (2) to preliminarily explore the effect of N starvation on drought stress; and (3) to identify candidate genes participating in N uptake and metabolism. The S. japonica transcriptome profile and metabolic pathway analysis under N deficiency and drought stress can provide the molecular basis for gene expression, genomics, and functional studies on the abiotic stress response of S. japonica, as well as other leguminous tree species.
2. Materials and Methods
2.1. Plant Materials and Treatments
Seeds of Chinese scholar trees were purchased from Shaanxi Sanyuan Gold Seed Industry Technology Co., Ltd., Yangling Branch. After the seeds germinated, they were then planted in pots (10 L) filled with fine sand. Seedlings were cultivated in a greenhouse (natural light, day/night 28/20 °C, 80% relative humidity), and irrigated every three days with 50 mL of modified Hoagland nutrient solution. After 8 weeks, the sand attached to the seedling roots was washed away with tap water, and uniform seedlings were transferred and acclimated to hydroponic culture for 4 weeks. The nutrient solution used for hydroponics and irrigation was reported in a previous study [18]. The hydroponic solution was renewed every 3 days and aerated continuously by air pumps.
To determine the appropriate time and PEG-6000 concentration for drought treatment, a preliminary experiment was conducted. The plants described above were treated with four PEG concentrations (0%, 10%, 20%, and 30%) for 24 h. Roots were harvested across four time points (0, 6, 12 and 24 h) for POD and SOD activity assays. As the results showed (Figure S1), compared with the control, both activity levels changed rapidly at 0-6 h under all three concentrations of PEG-6000. Three hours was selected as the experimental time for the drought treatment, considering that changes at the transcriptomic level are much more rapid than those at the physiological level, as suggested by a previous study in Arabidopsis [38]. Additionally, the two enzyme activity levels changed relatively slowly under the 10% PEG treatment. The 30% PEG concentration was so high that the cell osmotic balance may have been disrupted at an earlier time point, as SOD activity under 30% PEG decreased continuously at 6–24 h. Therefor, 20% PEG was used to induce moderate drought stress in this study.
In addition to the preliminary experiment, hydroponic-adapted plants were also used for the formal experiment. Briefly, we designed four treatments in this study as described in Figure 1: (a) NN (1 mM NH4NO3 applied for 15 h); (b) NL (1 mM NH4NO3 applied for 12 h, then 20% PEG-6000 applied for 3 h); (c) LN (0 mM NH4NO3 applied for 15 h); (d) LL (0 mM NH4NO3 applied for 12 h, then 20% PEG-6000 applied for 3 h). Leaves and fine roots of the four treatment groups were harvested simultaneously but kept separate. Three independent biological replicates, each containing a pool of three different plants, were sampled. Roots were used for high-throughput transcriptomic profiling and qRT-PCR verification. Both roots and leaves were used for physiological index measurements. The samples were rapidly frozen in liquid nitrogen, and then stored at −80 °C.
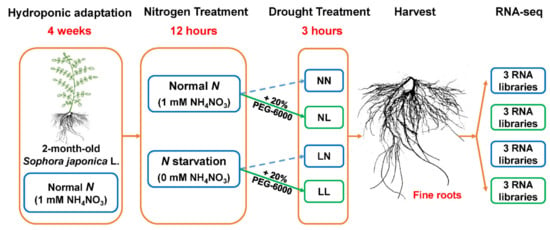
Figure 1.
Experimental design of S. japonica treatments with drought under different nitrogen conditions. The fine roots were used for RNA extraction. NN, normal N supply (1 mM NH4NO3) without drought; NL, normal N supply (1 mM NH4NO3) with drought; LN, N starvation (0 mM NH4NO3) without drought; LL, N starvation (0 mM NH4NO3) with drought.
2.2. Physiological Index Measurements
POD and SOD activities were measured using physiological assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturers’ recommendations. The MDA content was measured using the thiobarbituric acid (TBA) method [39]. The proline content was determined according to the method described by Chołuj et al. [40]. We analyzed the variables using one-way ANOVA in SPSS 23 (SPSS Inc., Chicago, IL, USA) by Tukey’s test at the p < 0.05 probability level.
2.3. Transcriptome Sequencing
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) with RIN > 7.5 in all samples (Table S1). cDNA libraries were generated using the NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) according to the manufacturer’s protocols and were sequenced by Biomarker Technologies Co., Ltd. (Beijing, China) on an Illumina HiSeq 2000 platform. Reads with high quality were necessary for the downstream analyses. Clean data were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from the raw data. The Q30 (the proportion of bases with a quality value greater than 30) and GC-content (the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine or cytosine) of the clean data were calculated at the same time. Then, transcriptome assembly was accomplished using Trinity software [41] with min_kmer_cov set to 2 by default. The assembled sequences were designated as “unigenes” for further annotation.
2.4. Gene Functional Annotation
For functional annotation, all unigenes were searched against public databases including the NR (NCBI nonredundant protein sequences), KOG/COG/eggNOG (Clusters of Orthologous Groups of proteins), Swiss-Prot (a manually annotated and reviewed protein sequence database), KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) databases by using Blast software [42] with an E-value ≤ 10−5. Pfam (Protein family) annotation was performed using HMMER software [43] with an E-value less than 10−10. The plant TF database (http://planttfdb.cbi.pku.edu.cn/ accessed on 25 Mar 2020) was used to search for putative transcription factors in S. japonica.
2.5. Identification of Differentially Expressed Genes (DEGs)
The generated clean data were mapped back to the assembled transcriptome. Gene expression levels were estimated by RSEM [44] and normalized to FPKM (fragments per kilobase per million mapped reads) values. DEGs were identified using the DESeq package [45] with a false discovery rate (FDR) < 0.05 and fold change ≥2 used as thresholds (positive or negative for either over- or underexpression, respectively). In the downstream DEG analyses, GO and KEGG databases were used to classify DEGs, and KEGG enrichment was conducted by KOBAS software [46] to test the statistical enrichment of the DEGs. A heat map showing the expression of selected DEGs was drawn using TBtools [47]. Phylogenetic trees of the identified NRT and AMT proteins with associated proteins in Arabidopsis were constructed via the neighbor-joining (NL, 1000 bootstrap replicates) method with MEGA 7.0 software (Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA). The protein sequences are provided in Table S2.
2.6. qRT-PCR Validation
To validate the reliability of the transcriptome sequencing results, we randomly selected 15 genes and subjected them to quantitative real-time PCR. RNA samples for transcriptome sequencing were also used for RT-qPCR. First-strand cDNA synthesis was performed using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions using an anchored oligo (dT)18 primer and 500 ng of total RNA. Reactions were performed on a LightCycler®96 real-time PCR system (Roche, Mannheim, Germany) using 2 × SYBR real-time PCR premixture (Bioteke, Beijing, China) in a 20 μL reaction under the following conditions: one cycle of 150 s at 95 °C, followed by 45 cycles at 95 °C for 10 s, 58 °C for 20 s and 72 °C for 30 s. The primers used are listed in Table S3. Helicase was used as the reference gene. For each of the selected genes, three biological replicates with three technical replicates were assayed. The relative expression levels were calculated using the 2−ΔΔCT method [48].
3. Results
3.1. Physiological Characteristics Affected by Drought and N Starvation
After 15 h of exposure to N starvation and 3 h of exposure to 20% PEG, the leaf chlorophyll content and root MDA content did not change under all treatments (Figure 2A,C). The MDA content in leaves increased significantly only under drought with N starvation (LL) when compared with that of the control (NN) (Figure 2B). The proline content in leaves increased significantly only under drought (Figure 2D), and the proline content in roots increased upon drought but decreased under N starvation, whereas it was not affected by drought with N starvation (Figure 2E).
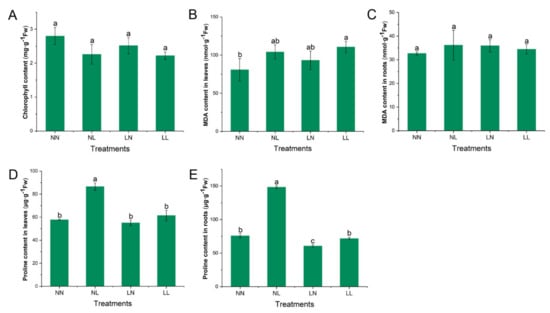
Figure 2.
Physiological analyses of S. japonica under drought and N starvation stresses. (A) Total chlorophyll content; (B) MDA content in leaves; (C) MDA content in roots; (D) Proline content in leaves; (E) Proline content in roots. Data indicate means ± SD. Letters in the same bar indicate significant differences (one-way ANOVA, Tukey’s test; p < 0.05). NN, NL, LN and LL are as defined in Figure 1.
3.2. De Novo Transcriptome Assembly
Transcriptome analysis using the Illumina HiSeq 2000 sequencing platform was performed. We obtained a total of 109.04 Gb clean data. For each cDNA library, 25.72–34.61 million clean (high-quality) reads were generated (Table S4). The Q30 quality scores were over 92.11%, and the GC content was approximately 45% for all samples. The transcriptome was then de novo assembled to 86,672 transcripts (total length of 1.25 × 108 bp), which were grouped into 46,852 trinity ‘unigenes’ with an average length = 1310.49 bp (Table S5). Approximately 48.1% of the unigenes had a length greater than 1000 bp (Figure S2).
Sequence annotation against public databases resulted in the successful annotation of 37,183 (79.36%) unigenes in at least one of the databases (Table S6 and S7). In brief, a total of 35,524 unigenes were annotated against the NR database. Additionally, 24,070 transcripts were assigned to 5,786 GO terms and categorized into 50 functional groups (Figure S3; Table S8), and 18,607 putative proteins were assigned to five main categories that included 131 KEGG pathways (Table S9).
3.3. Differentially Expressed Gene Analyses
Principal component analysis of the gene expression results showed that the main difference between the samples was derived from drought stress, which explained 52.7% of the total detected variation. N status (0 and 1 mM) contributed to another 27.6% of the variability (Figure 3A). The three replicates of each sampling group clustered well together, with the exception of sample LL-1, which clustered with the NL samples. In addition, LL-1 had a low correlation with LL-2 and LL-3 (Table S10). After excluding the outlier LL-1, the correlation among biological replicates was higher than that across samples. A total of 11 samples were used for downstream DEG analyses.
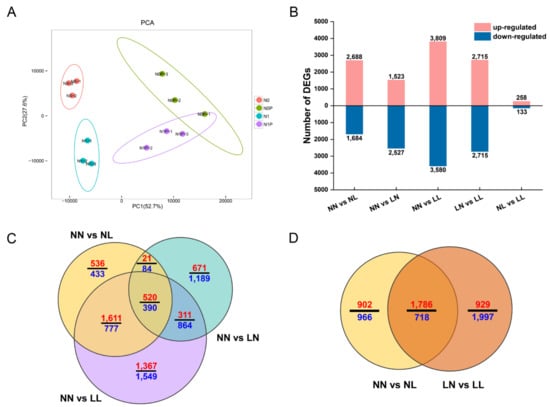
Figure 3.
DEGs of S. japonica roots upon drought stress and N starvation. (A) Principal component analysis (PCA) of all twelve samples used in this study; (B) Numbers of upregulated and downregulated DEGs identified in each comparison of different treatments; (C) Venn diagram of DEGs upon drought treatment and N starvation compared with the control (NN); (D) Venn diagram of DEGs upon drought treatment under two N concentrations. The red and blue numbers in each fraction indicate the numbers of up- and downregulated genes, respectively. NN, NL, LN and LL are as defined in Figure 1.
3.3.1. DEGs Induced by Drought and N Starvation
Under drought and N starvation, we identified 11,808 DEGs from fine roots of S. japonica in five pairwise comparisons. When compared with the control (NN), more DEGs were found under drought with N starvation than under a single stress, with 7,389 genes being differentially expressed. The number of transcripts identified between N starvation and drought in S. japonica varied little, but more upregulated genes were induced by drought, while more downregulated genes were induced by N starvation (Figure 3B). Among all these DEGs, 520 were commonly upregulated by drought, N starvation and drought with N starvation, whereas 390 transcripts were commonly downregulated (Figure 3C). To understand the functions of these genes, the GO database, and KEGG pathway enrichment were used.
When the DEGs were mapped to terms in the GO database and compared to the whole transcriptome background, they were categorized into small functional groups in three main categories (biological process, cellular component, and molecular function) of the GO classification. Under all three stresses, metabolic process, cellular process and single-organism process were dominant within the biological process category. The cell, cell part and membrane were the most frequent groups in the cellular component category. Catalytic activity and binding were major parts of the molecular function category (Table S11).
To further understand the biochemical pathways of DEGs, KEGG analysis was conducted. The numbers of KEGG pathways were individually determined, and the significant pathways were further filtered using a cut-off p-value of 0.05. Genes in the “biosynthesis of amino acids”, “phenylpropanoid biosynthesis” and “plant-pathogen interaction” pathways were upregulated, irrespective of the stress treatments, while “nitrogen metabolism” pathway was enriched under drought and drought with N starvation (Figure 4A,C,E). Remarkably, genes in the “plant hormone signal transduction” and “starch and sucrose metabolism” pathways were downregulated upon drought (Figure 4B), while “photosynthesis” pathway was significantly enriched in downregulated DEGs under N starvation and drought with N starvation (Figure 4D,F). In addition, the number of DEGs in the “ribosome” pathway was much larger under both N starvation and drought with N starvation than under drought (Figure S4).
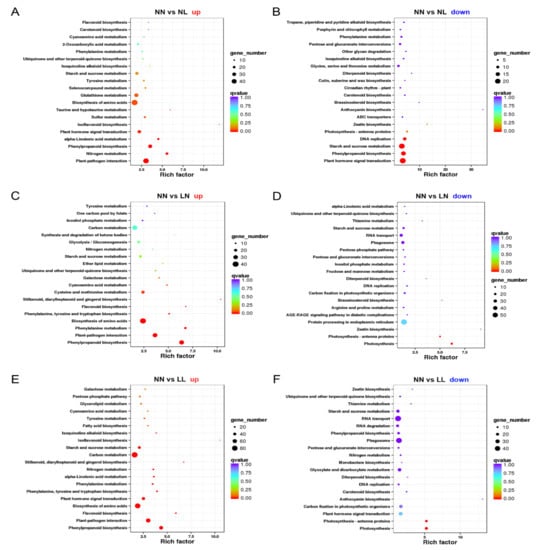
Figure 4.
Scatter plot of the top 20 enriched KEGG pathways under drought and N starvation. (A) Upregulated genes under drought; (B) downregulated genes under drought; (C) upregulated genes under N starvation; (D) downregulated genes under N starvation; (E) upregulated genes under drought with N starvation; (F) downregulated genes under drought with N starvation. The rich factor is the ratio of DEG numbers annotated in this pathway term to all gene numbers annotated in this pathway term. Greater rich factor indicates greater intensity. The q-value is the corrected p-value ranging from 0~1. Lower q-value indicates a greater intensity. NN, NL, LN and LL are as defined in Figure 1.
3.3.2. DEG Analyses about the Effect of N Starvation on Drought Stress
To explore the effect of N starvation on drought stress, DEGs between two pairwise comparisons were analyzed, NN vs. NL and LN vs. LL. Upon drought, 2,512 genes were coexpressed (1,786 upregulated and 718 downregulated) under the two N statuses (Figure 3D). With a similar number of upregulated genes, approximately 1,000 more DEGs were downregulated upon drought under N starvation than under normal N condition.
The GO enrichment analysis of the two pairwise comparisons did not reveal differences among treatments (Table S11). Then, the identified DEGs were mapped to pathways of the KEGG database to identify significantly enriched biochemical pathways. Interestingly, the results indicated that genes involved in “starch and sucrose metabolism” were only downregulated under normal N condition (Figure 4B), whereas those related to “carbon metabolism”, “alanine, aspartate and glutamate metabolism” and “RNA transport” were only downregulated under N starvation condition (Figure 5B). Furthermore, the “plant hormone signal transduction” pathway was significantly enriched in both up- and downregulated DEGs under normal N condition but not enriched under N starvation (Figure 4A,B and Figure 5A,B). Additionally, the KEGG map indicated that the pathways of “ribosome” (Figure S5), “arginine biosynthesis pathway” (Figure S6), “oxidative phosphorylation” (Figure S7) and “aminoacyl-tRNA biosynthesis” (Figure S8) were almost downregulated under N deficiency but upregulated under normal N condition.
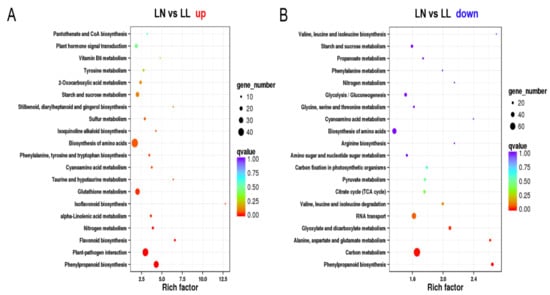
Figure 5.
Scatter plot of the top 20 enriched KEGG pathways upon drought under N starvation. (A) Upregulated genes; (B) Downregulated genes. The rich factor is the ratio of DEG numbers annotated in this pathway term to all gene numbers annotated in this pathway term. Greater rich factor indicates greater intensity. The q-value is the corrected p-value ranging from 0~1. Lower q-value indicates a greater intensity. NN, NL, LN and LL are as defined in Figure 1.
3.4. Candidate Genes Related to N Uptake and Metabolism
According to the NR annotation, 78 unigenes were involved in N uptake and metabolism. Among them, 38 were differentially expressed in at least one comparison (Figure 6; Table S12). In response to drought (NN vs. NL), most predicted NRTs were downregulated (except for c1253196.graph_c0 and c1254472.graph_c0), while two AMTs and all DEGs involved in N assimilation were upregulated. Similarly, most of the DEGs subjected to drought combined with N starvation (NN vs. LL) had consistent expression changes with those under drought, and the differences included a lack of an increase in NRT transcripts and the downregulation of one GS. Conversely, under N starvation (NN vs. LN), all three differentially expressed NRTs had increased transcript abundances, and the expression of only one NR was altered but was downregulated. However, other differentially expressed GOGATs and GDHs were all upregulated, in keeping with under drought treatment and drought with N starvation.
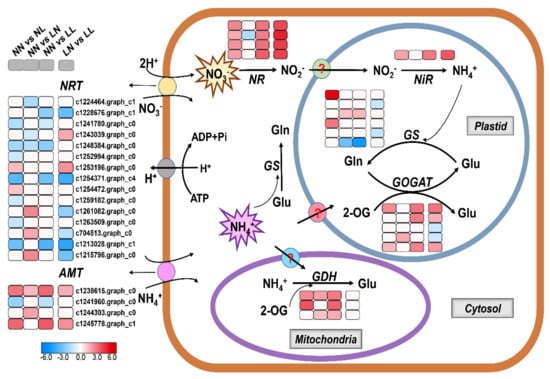
Figure 6.
A conceptual model of nitrogen uptake and metabolism in S. japonica. The four-box strings from left to right indicate different pairwise comparisons (including NN vs. NL, NN vs. LN, NN vs. LL and LN vs. LL). Heat maps were drawn using log2-fold change values. Boxes in white indicate genes expressed normally in the comparisons. NN, NL, LN and LL are as defined in Figure 1.
The expression levels of DEGs predicted to participate in N transport and metabolism varied in response to drought under different N statuses. When N was deficient, the majority of NRTs were also downregulated upon drought, but 11 of 15 NRTs had a different trend compared with those under normal N condition. The expression of all NRs and NiR was enhanced regardless of N concentration, whereas two GS genes and two GOGATs were downregulated and all GDHs were expressed normally under N deficiency condition.
3.5. Transcription Factors
Transcription factors play important roles in upstream regulation when plants suffer from abiotic and biotic stresses. In the present study, we identified 1,051 TFs, which were categorized into 60 TF families. C2H2 was the most abundant family, followed by the C3H and bZIP families. Among them, 395 TFs belonging to 52 families showed different expression in at least one comparison (Figure 7; Table S13). The number of differentially expressed transcription factors (DETs) was higher under drought with N starvation than under either drought or N starvation alone. Most of the AP2/ERF-ERF family DETs were upregulated, whereas bHLH family DETs were downregulated by drought and drought with N starvation. Under N starvation, the enriched upregulated TF groups were members of the WRKY and NAC families, while C2C2-Dof was the largest downregulated family upon N starvation. Furthermore, drought stress increased the expression of genes involved in the AP2/ERF-ERF, WRKY and NAC families but reduced the transcription of genes in the bHLH family, regardless of N status.
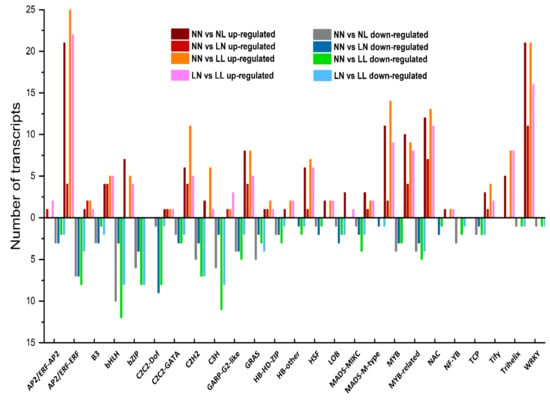
Figure 7.
The top 25 differentially expressed transcription factor families under different pairwise comparisons (including NN vs. NL, NN vs. LN, NN vs. LL and LN vs. LL). NN, NL, LN and LL are as defined in Figure 1.
3.6. Validation of RNA-Seq by qRT-PCR Validation
To verify the RNA-seq data, we validated the expression of 15 genes by qRT-qPCR using the same samples as for RNA-seq. Although the magnitude of the change was not completely equal, the qRT-PCR results of the tested DEGs were generally consistent with the RNA-sequencing data (Figure 8), which means our transcriptome is reliable for further studies.
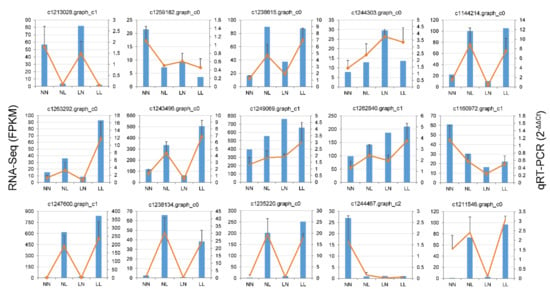
Figure 8.
Comparison of gene expression data obtained from RNA-seq (blue columns) and qRT-PCR (yellow lines) for 15 selected DEGs under four treatments. Error bars represent +SE. NN, NL, LN and LL are as defined in Figure 1.
4. Discussion
Drought and N deficiency negatively affect plant growth, and many physiological and biochemical parameters change to maintain the growth of plants under adverse conditions. In this study, POD and SOD activities rapidly changed in response to drought stress at 0–6 h in S. japonica (Figure S1). Increased POD activity is beneficial to reduce oxidative damage in plant cells. SOD activity decreased first under drought at 6–12 h due to the sudden increase in reactive oxygen species but then increased at 6–12 h, which may be because drought induced SOD resynthesis to enhance drought resistance. The same trend was also observed in Camellia sinensis leaves under cold stress [49]. The chlorophyll content was negatively affected by all the stress treatments, although the change was not significant (Figure 2A), which may have been the result of the short stress duration. Interestingly, the MDA content in leaves noticeably increased under drought combined with N starvation but did not change in roots (Figure 2B,C), which indicated that the cell membrane in leaves may be more sensitive than that in roots of S. japonica. Earlier leaf aging than root aging may be a survival strategy for perennials under adverse conditions [50]. Furthermore, the proline content in roots increased under drought but decreased under N starvation. Notably, the increases were even greater under adequate N condition than under N starvation in both leaves and roots (Figure 2D,E). N is an essential element in proline synthesis. This result may suggest that N deficiency could aggravate drought stress to some extent. However, more efforts are required to explain the underlying mechanisms.
With the rapid development of high-throughput sequencing technology in recent years, transcriptomics provides opportunities for large-scale gene discovery and gene expression studies in nonmodel species without genomic resources. In this regard, we used an RNA-seq approach to explore the molecular mechanisms of the responses to drought and N deficiency stresses in the fine roots of S. japonica. A total of 46,852 unigenes were generated, with a higher N50 value of 1310 bp than two previous sequencing studies of S. japonica [36,37]. Higher quality transcriptome data can enrich the genetic information of S. japonica and provide a good foundation for the study of gene expression in this species.
Plants often activate rapid responses to biotic and abiotic stresses. For example, more than 16% of the genome exhibited altered expression levels in response to drought stress in cotton [51]. Similarly, we observed that 11,809 genes (approximately 25% of all the unigenes) were differentially expressed in fine roots of S. japonica under drought stress and N starvation. In addition, DEGs in different comparisons were categorized into large and varied metabolic pathways (Figure S4). The quick and strong response indicated that S. japonica has a complex regulatory process in response to abiotic stress. Plants are not exposed to only single stresses in nature, as adverse conditions often occur simultaneously. The changes in genes were greater in S. japonica when drought and N deficiency were applied together than when they were applied alone (Figure 3B,C). Whether seedlings were threatened by drought or N starvation, genes related to the “biosynthesis of amino acids” and “phenylpropanoid biosynthesis” were enriched in upregulated pathways in this study. The activation of the phenylpropanoid biosynthesis pathway contributes to the accumulation of various phenolic compounds that have the potential to scavenge harmful reactive oxygen species [52]. Amino acids are not only building blocks of different cellular components, but can also act as compatible osmolytes, regulate pH or act as a nitrogen or carbon reserve when plants are under stress [53]. The two pathways may play key roles in counteracting the adverse effects of stresses in S. japonica. It is worth noting that N starvation decreased the expression of genes in the photosynthesis pathway (Figure 4D), consistent with previous study results, since N is an essential component of chlorophyll synthesis [54]. In addition, downregulated genes in the ribosome pathway accounted for the largest number of DEGs under both N starvation and drought with N starvation (Figure S8B,C), indicating ribosome dysfunction caused by N deficiency, which would further hinder protein synthesis and may result in plant growth being restricted.
Many researchers have focused on the impact of nitrogen concentrations on the effects caused by drought. In S. japonica, more genes were upregulated than downregulated upon drought under normal N condition, which was in line with previous studies in various species [34,51]. However, with little difference in the number of upregulated genes, one thousand more genes were downregulated under N starvation than under normal N condition under drought. In particular, the KEGG pathway of “carbon metabolism” was only enriched in downregulated genes under N starvation, and genes in the KEGG pathways of “ribosome”, “arginine biosynthesis pathway”, “oxidative phosphorylation” and “aminoacyl-tRNA biosynthesis” were markedly downregulated under N starvation. C and N metabolism has been proven to interactively regulate many aspects of the physiology and development of plants [55]. N assimilation requires energy produced by carbon metabolism, and photosynthetic carbon assimilation requires a large amount of nitrogen [56]. Approximately 4-fold more genes involved in carbon metabolism were downregulated under N starvation than under normal N condition, indicating that the balance of carbon and nitrogen metabolism was disturbed, ultimately strengthening drought stress when N was limited. Ribosome and aminoacyl-tRNA underlie protein synthesis and support cell growth [57]. Arginine also plays a key role in stress responses and plant development, as it is a precursor to the synthesis of polyamines (including putrescine, spermidine and spermine) [58,59]. Oxidative phosphorylation in mitochondria affects the oxidative stress response in plants [60]. Decreased activity of COX, which is a terminal oxidase in oxidative phosphorylation pathways, leads to excessive reactive oxygen species. In summary, we speculated that N deficiency may aggravate drought stress by decreasing energy generation and storage, blocking protein synthesis, and reducing the contents of osmotic regulators. This was partly consistent with a previous study on poplars—adequate N could alleviate drought through enhancing expression of genes involved in plant hormone signal transduction and antioxidant defense, and through interactions between carbon metabolism and nitrogen assimilation [33]. However, these details need to be further studied in S. japonica.
Plants absorb and utilize inorganic N with the help of genes involved in N transport and metabolism. Our transcriptome analysis first identified major genes involved in N uptake and metabolism in S. japonica roots (Table S12). In the present study, most NRTs were downregulated under drought, while AMTs were upregulated. Similar conclusions were observed in poplar [61]. Absorption of more ammonium than nitrate may result in an increased ammonium to nitrate ratio, which could improve the drought resistance of plants [18,20]. The sharp upregulation of NR and NiR may also contribute to increasing the ammonium to nitrate ratio. Upon drought, GS, GOGAT and GDH were upregulated under normal N but downregulated or unchanged under N starvation, indicating that adequate N may promote the N assimilation process to provide substrates for stress-resistant components. Under N starvation, the expression of three NRTs (c1261082.graph_c0, c704513.graph_c0 and c1215796.graph_c0) increased, but they were expressed normally under drought or drought with N starvation, suggesting that these genes could only be activated by N starvation but not drought. Phylogenetic analysis with NRTs in Arabidopsis showed that c1261082.graph_c0 and c1215796.graph_c0 were separately clustered together with AtNRT1.1 and AtNRT2.5 (Figure S9A). In Arabidopsis, the dual-affinity nitrate transporter NRT1.1 and the high-affinity nitrate transporter NRT2.5 mediate nitrate uptake from soil [62]. AtNRT1.1 also participates in auxin transport to mediate lateral root development [63]. The two identified NRTs in S. japonica may exhibit a similar function when the N concentration was low. The N assimilation process was relatively slow upon N starvation, which may be explained by the fact that plants slowed their growth to require less N when N availability was limited [10]. One AMT gene (c1238615.graph_c0) was significantly induced under all three kinds of stresses and was grouped into the AtAMT1 family (Figure S9B), which consists of high-affinity ammonium transporters, indicating its major role in stress resistance. These N-related genes showed various changes in expression under stress conditions, but their functions need to be further verified. The identification of these N-related genes has greatly promoted our understanding of the N utilization of S. japonica under stress conditions, and provides a molecular basis for further studies.
TFs are important regulators that have various functions in responses to abiotic stresses in plants [5]. In the present study, genes related to the AP2/ERF-ERF, WRKY and NAC families were most abundant among the upregulated TFs under stress. These families have been shown to play an important role in plant-enhanced resistance to stresses [64]. For instance, expression of CsERF1 enhanced cold tolerance in tobacco [65], and overexpression of WRKY11 and ANAC072 increased tolerance to drought and osmotic stresses in Arabidopsis [66,67]. Among the downregulated genes, members of the bHLH and C3H families accounted for the majority upon drought. In contrast to many previous studies indicating that overexpression of several members of the bHLH and C3H families could improve drought resistance [68,69], these TFs may have distinct roles in plants under drought. Furthermore, the C2C2-Dof family had a relatively high proportion of downregulated TFs under N starvation. It was proven that expression of Dof1 in transgenic Arabidopsis improved growth under low-nitrogen conditions [70], highlighting the great roles of C2C2-Dof in N starvation responses. These DEGs in various TF families tightly participate in the molecular regulation of the stress response, which may facilitate the acclimation of S. japonica to adverse environments.
In summary, the physiological and molecular changes play key roles in the resistance to drought and N starvation of S. japonica. At the physiological level, antioxidant enzymes and osmotic regulators help counteract the adverse effects of stress. In present study, S. japonica had high drought tolerance as SOD and POD activities as well as proline contents increased significantly in the roots. At the molecular level, the activation of “phenylpropanoid biosynthesis” and “biosynthesis of amino acids” pathways, the enhanced ability of N uptake and metabolism and the upregulated expressions of transcription factors in AP2/ERF-ERF, WRKY and NAC families may be beneficial to plant-enhanced resistance to either drought or N starvation. On the other hand, N deficiency could aggravate drought stress, as N starvation slowed down the processes of “photosynthesis”,“ribosome”, “arginine biosynthesis”, “oxidative phosphorylation” and “aminoacyl-tRNA biosynthesis”, indicating that adequate N may be necessary for improving drought tolerance. These results suggest that Sophora japonica can activate stress pathways rapidly via modulating gene expressions in response to drought and N starvation. Thus, studying these genes could improve our knowledge of resistance mechanisms in S. japonica roots to short-term environmental stresses.
5. Conclusions
In conclusion, we performed a combination of transcriptome sequencing and differential expression analyses to identify pathways and genes altered under drought, N starvation and drought with N starvation treatments. Several physiological traits showed various trends, and transcripts in various metabolic processes reacted rapidly and strongly in response to drought and N starvations stresses. Genes in pathways of phenylpropanoid biosynthesis and biosynthesis of amino acids were greatly activated, and they may play important roles in stress resistance in roots of S. japonica. N deficiency may aggravate drought stress by decreasing energy generation and storage, blocking protein synthesis, and reducing the contents of osmotic regulators. Additionally, many N-related genes and TFs were identified differentially expressed and played key roles in drought and N starvation stresses in this study, but the functions of these genes need to be further studied in the future. The rapid and strong transcriptomic changes of Sophora japonica roots to short-term drought and N starvation reflect an initial response to abiotic stresses, which is a basis of the adaptation to long-term stresses of ancient trees. Our research is the first to explore the molecular mechanism of -and provide valuable information on differentially expressed metabolic pathways and genes of S. japonica under abiotic stresses. This research also reveals the molecular basis for protecting ancient tree resources and breeding resistant varieties for greening.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/f12050650/s1, Figure S1: POD and SOD activity assays of S. japonica roots under four PEG-6000 concentrations (0%, 10%, 20%, 30%) across four time points (0, 6, 12 and 24 h). (A) POD activity (U·g−1min−1); (B) SOD activity (U·g−1Fw). CK denotes without PEG-6000, Figure S2: Length distribution of assembled unigenes, Figure S3: Functional annotation of assembled sequences based on Gene Ontology (GO) categorization, Figure S4: DEGs annotations in various KEGG pathways under four pairwise comparisons, Figure S5: DEGs involved in ribosome pathway upon drought. (A) Under normal N condition, NN vs. NL; (B) Under N starvation condition, LN vs. LL. Transcripts significantly up-regulated are highlighted in red, significantly down-regulated are highlighted in green. Blue indicates both significantly up- and down-regulated genes exist, Figure S6: DEGs involved in arginine biosynthesis pathway upon drought. (A) Under normal N condition, NN vs. NL; (B) Under N starvation condition, LN vs. LL. Transcripts significantly up-regulated are highlighted in red, significantly down-regulated are highlighted in green. Blue indicates both significantly up- and down-regulated genes exist, Figure S7: DEGs involved in oxidative phosphorylation pathway upon drought. (A) Under normal N condition, NN vs. NL; (B) Under N starvation condition, LN vs. LL. Transcripts significantly up-regulated are highlighted in red, significantly down-regulated are highlighted in green. Blue indicates both significantly up- and down-regulated genes exist, Figure S8: DEGs involved in aminoacyl-tRNA biosynthesis pathway upon drought. (A) Under normal N condition, NN vs. NL; (B) Under N starvation condition, LN vs. LL. Transcripts significantly up-regulated are highlighted in red, significantly down-regulated are highlighted in green. Blue indicates both significantly up- and down-regulated genes exist, Figure S9: Neighbor-joining phylogenetic analysis of (A) NTRs and (B) AMTs with related genes in Arabidopsis. The trees were constructed by neighbor-joining phylogeny test, and 1000bootstrao replicates. The protein sequences are provided in Table S3. The green triangles represent the genes proved to have functions to transports nitrate in Arabidopsis. The red stars represent the identified genes involved in N uptake and metabolism at present study, Table S1: RNA sample details including quality and concentration measurements, Table S2: Gene accession numbers of Arabidopsis thaliana used in phylogenetic analysis, Table S3: The primers used for qRT-PCR, Table S4: Summary of sequences analysis, Table S5: Length distribution of assembled unigenes, Table S6: Summary of the annotation results, Table S7: Information of unigene annotations to public databases, Table S8: GO annotation of unigenes, Table S9: KEGG pathway annotations of unigenes, Table S10: The correlation of three biological repeats for transcriptome data, Table S11: GO classifications of DEGs in paired comparisons of different treatments, Table S12: Candidate differentially expressed genes related to nitrogen uptake and metabolism, Table S13: Predicted differentially expressed transcription factors in S. japonica roots transcriptome under drought and N starvation treatments.
Author Contributions
Conceptualization, J.T. and Z.Z.; methodology, J.T. and Y.P.; software, J.T.; validation, J.T. and Y.P.; formal analysis, J.T.; investigation, J.T. and Y.P.; data curation, J.T; writing—original draft preparation, J.T.; writing—review and editing, J.T., Y.P. and Z.Z.; visualization, J.T.; supervision, J.T. and Z.Z.; project administration, Z.Z.; funding acquisition, Z.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Forestry Industry Research Special Funds for Public Welfare Projects (China), grant number 201404302.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw transcriptome datasets generated for this study was deposited in the NCBI Sequence Reads Archive (SRA) with the accession number PRJNA665329. Data is contained within the article or supplementary figures and supplementary tables.
Acknowledgments
We sincerely thank Sen Meng for his great help in the experimental design and Yang Gao for her valuable comments on the manuscript. We also thank Hailan Zhu for her assistance in laboratory.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Lei, D.; Lu, Z.; Gao, L.; Guo, S.; Shen, Q. Is nitrogen a key determinant of water transport and photosynthesis in higher plants upon drought stress? Front. Plant Sci. 2018, 9, 1143. [Google Scholar] [CrossRef]
- Mackay, A. Climate change 2007: Impacts, adaptation and vulnerability. Contribution of working group II to the fourth assessment report of the intergovernmental panel on climate change. J. Environ. Qual. 2008, 37, 2407. [Google Scholar] [CrossRef]
- Salehi-Lisar, S.Y.; Bakhshayeshan-Agdam, H. Drought Stress in Plants: Causes, Consequences, and Tolerance. In Drought Stress Tolerance in Plants; Hossain, M.A., Wani, S.H., Bhattacharjee, S., Burritt, D.J., Tran, L.-S.P., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–16. [Google Scholar]
- Morgan, J.M. Osmoregulation and water stress in higher plants. Ann. Rev. Plant Physiol. 1984, 35, 299–319. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xue, X.; Zhou, X. Response of growth and other physiological characteristics of Sophora Japonica L. saplings to drought stress. IOP Conf. Ser. Earth Environ. Sci. 2018, 170, 052029. [Google Scholar] [CrossRef]
- Quy, N.V.; Yi, Z.Q.; Zhong, Z. Cytokinin ameliorates the abiotic stress severity in Chinese Scholartree (Sophora Japonica L.) through regulation of chlorophyll fluorescence, antioxidative response and proline metabolism. Res. J. Biotechnol. 2017, 12, 11–18. [Google Scholar]
- Rennenberg, H.; Wildhagen, H.; Ehlting, B. Nitrogen nutrition of poplar trees. Plant Biol. 2010, 12, 275–291. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Cheng, Y.-H.; Chen, K.-E.; Tsay, Y.-F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.-B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef]
- Su, L.; Meng, S.; Zhang, S.; Li, Y.; Zhao, Z. Mechanism of molecular responses of nitrogen transport to drought stress in Sophora japonica. J. Northwest For. Univ. 2017, 32, 1–11. (In Chinese) [Google Scholar] [CrossRef]
- Yin, A.; Jin, H.; Han, Z.; Han, S. Study on the relationship between chromosome numbers and nodulation of 18 species of leguminous trees. Sci. Silvae Scnicae 2006, 42, 26–28. (In Chinese) [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Yu, Y.; Cui, H.; Wang, R.; Guo, W. Increased nitrogen supply promoted the growth of non-N-fixing woody legume species but not the growth of N-fixing Robinia pseudoacacia. Sci. Rep. 2018, 8, 17896. [Google Scholar] [CrossRef]
- Shangguan, Z.; Shao, M.; Dyckmans, J. Effects of nitrogen nutrition and water deficit on net photosynthetic rate and chlorophyll fluorescence in winter wheat. J. Plant Physiol. 2000, 156, 46–51. [Google Scholar] [CrossRef]
- Buljovcic, Z.; Engels, C. Nitrate uptake ability by maize roots during and after drought stress. Plant Soil 2001, 229, 125–135. [Google Scholar] [CrossRef]
- Fotelli, M.N.; Rennenberg, H.; Geβler, A. Effects of drought on the competitive interference of an early successional species (Rubus fruticosus) on Fagus sylvatica L. seedlings: 15N uptake and partitioning, responses of amino acids and other N compounds. Plant Soil 2002, 4, 311–320. [Google Scholar] [CrossRef]
- Geßler, A.; Jung, K.; Gasche, R.; Papen, H.; Heidenfelder, A.; Börner, E.; Metzler, B.; Augustin, S.; Hildebrand, E.; Rennenberg, H. Climate and forest management influence nitrogen balance of European beech forests: Microbial N transformations and inorganic N net uptake capacity of mycorrhizal roots. Eur. J. For. Res. 2005, 124, 95–111. [Google Scholar] [CrossRef]
- Meng, S.; Zhang, C.; Su, L.; Li, Y.; Zhao, Z. Nitrogen uptake and metabolism of Populus simonii in response to PEG-induced drought stress. Environ. Exp. Bot. 2016, 123, 8–87. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Wignes, J.A.; Kaiser, B.N. Root hydraulic and aquaporin responses to N availability. In Plant Aquaporins: From Transport to Signaling; Chaumont, F., Tyerman, S.D., Eds.; Springer: Cham, Switzerland, 2017; pp. 207–236. [Google Scholar]
- Ding, L.; Gao, C.; Li, Y.; Li, Y.; Zhu, Y.; Xu, G.; Shen, Q.; Kaldenhoff, R.; Kai, L.; Guo, S. The enhanced drought tolerance of rice plants under ammonium is related to aquaporin (AQP). Plant Sci. 2015, 234, 14–21. [Google Scholar] [CrossRef]
- Guo, J.; Yang, Y.; Wang, G.; Yang, L.; Sun, X. Ecophysiological responses of Abies fabri seedlings to drought stress and nitrogen supply. Physiol. Plant. 2010, 139, 335–347. [Google Scholar] [CrossRef]
- Villar-Salvador, P.; Peñuelas, J.L.; Jacobs, D.F. Nitrogen nutrition and drought hardening exert opposite effects on the stress tolerance of Pinus pinea L. seedlings. Tree Physiol. 2013, 33, 221–232. [Google Scholar] [CrossRef]
- Bascuñán-Godoy, L.; Sanhueza, C.; Hernández, C.E.; Cifuentes, L.; Pinto, K.; Álvarez, R.; González-Teuber, M.; Bravo, L.A. Nitrogen supply affects photosynthesis and photoprotective attributes during drought-induced senescence in quinoa. Front. Plant Sci. 2018, 9, 994. [Google Scholar] [CrossRef] [PubMed]
- Gorska, A.; Ye, Q.; Holbrook, N.M.; Zwieniecki, M.A. Nitrate control of root hydraulic properties in plants: Translating local information to whole plant response. Plant Physiol. 2008, 148, 1159. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Li, Y.; Wang, Y.; Gao, L.; Wang, M.; Chaumont, F.; Shen, Q.; Guo, S. Root ABA accumulation enhances rice seedling drought tolerance under ammonium supply: Interaction with aquaporins. Front. Plant Sci. 2016, 7, 1206. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Chen, D.; Min, D.; Li, W.; Xu, Z.; Zhou, Y.; Li, L.; Chen, M.; Ma, Y. AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 457, 433–439. [Google Scholar] [CrossRef]
- Wilkinson, S.; Bacon, M.A.; Davies, W.J. Nitrate signalling to stomata and growing leaves: Interactions with soil drying, ABA, and xylem sap pH in maize. J. Exp. Bot. 2007, 58, 1705–1716. [Google Scholar] [CrossRef]
- Chen, J.; Qi, T.; Hu, Z.; Fan, X.; Zhu, L.; Iqbal, M.F.; Yin, X.; Xu, G.; Fan, X. OsNAR2.1 positively regulates drought tolerance and grain yield under drought stress conditions in rice. Front. Plant Sci. 2019, 10, 197. [Google Scholar] [CrossRef]
- Kleiner, K.W.; Abrams, M.D.; Schultz, J.C. The impact of water and nutrient deficiencies on the growth, gas exchange and water relations of red oak and chestnut oak. Tree Physiol. 1992, 11, 271–287. [Google Scholar] [CrossRef]
- Li, H.; Li, M.; Wei, X.; Zhang, X.; Xue, R.; Zhao, Y.; Zhao, H. Transcriptome analysis of drought-responsive genes regulated by hydrogen sulfide in wheat (Triticum aestivum L.) leaves. Mol. Genet. Genom. 2017, 292, 1091–1110. [Google Scholar] [CrossRef]
- Long, Y.; Liang, F.; Zhang, J.; Xue, M.; Zhang, T.; Pei, X. Identification of drought response genes by digital gene expression (DGE) analysis in Caragana korshinskii Kom. Gene 2020, 725, 144170. [Google Scholar] [CrossRef]
- Krapp, A.; Berthomé, R.; Orsel, M.; Mercey-Boutet, S.; Yu, A.; Castaings, L.; Elftieh, S.; Major, H.; Renou, J.P.; Daniel-Vedele, F. Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol. 2011, 157, 1255–1282. [Google Scholar] [CrossRef]
- Lu, M.; Chen, M.; Song, J.; Wang, Y.; Pan, Y.; Wang, C.; Pang, J.; Fan, J.; Zhang, Y. Anatomy and transcriptome analysis in leaves revealed how nitrogen (N) availability influence drought acclimation of Populus. Trees 2019, 33, 1003–1014. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, L.; Zhao, Z.; Li, Y.; Zhou, K.; Su, L.; Zhou, Q. Root transcriptome sequencing and differentially expressed drought-responsive genes in the Platycladus orientalis (L.). Tree Genet. Genomes 2016, 12, 79. [Google Scholar] [CrossRef]
- Dharshini, S.; Hoang, N.V.; Mahadevaiah, C.; Sarath Padmanabhan, T.S.; Alagarasan, G.; Suresha, G.S.; Kumar, R.; Meena, M.R.; Ram, B.; Appunu, C. Root transcriptome analysis of Saccharum spontaneum uncovers key genes and pathways in response to low-temperature stress. Environ. Exp. Bot. 2020, 171, 103935. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, Y.; Guo, W.; Xu, X.J.; Wang, Q. De novo assembly and characterization of Sophora japonica transcriptome using RNA-seq. Biomed. Res. Int. 2014, 2014, 750961. [Google Scholar] [CrossRef]
- Zhang, F.S.; Wang, Q.Y.; Pu, Y.J.; Chen, T.Y.; Qin, X.M.; Gao, J. Identification of genes involved in flavonoid biosynthesis in Sophora japonica through transcriptome sequencing. Chem. Biodivers. 2017, 14, e1700369. [Google Scholar] [CrossRef]
- Kreps, J.A.; Wu, Y.; Chang, H.S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Fu, L.; Ding, Z.; Han, B.; Hu, W.; Li, Y.; Zhang, J. Physiological investigation and transcriptome analysis of polyethylene glycol (PEG)-induced dehydration stress in cassava. Int. J. Mol. Sci. 2016, 17, 283. [Google Scholar] [CrossRef]
- Chołuj, D.; Karwowska, R.; Ciszewska, A.; Jasińska, M. Influence of long-term drought stress on osmolyte accumulation in sugar beet (Beta vulgaris L.) plants. Acta Physiol. Plant. 2008, 30, 679. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.; Zhang, J.; Zhang, Z.; Miller, W.E.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein databases search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Prec. 2010, 11, R106. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, J.; Jiang, C.; Li, W. Effect of low temperature stress on SOD activity, soluble protein content and soluble sugar content in Camellia sinensis leaves. J. Anhui Agric. Univ. 2011, 38, 24–26. (In Chinese) [Google Scholar]
- Wu, Q.; Xia, R.; Zhang, Q. A review of progress in response to fruit trees under water stress. Chin. Agric. Sci. Bull. 2003, 32, 72–76. (In Chinese) [Google Scholar] [CrossRef]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Attal, N.B.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genom. 2014, 14, 11–22. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Ali, Q.; Athar, H.-U.-R.; Haider, M.Z.; Shahid, S.; Hussain, S.M. Role of amino acids in improving abiotic stress tolerance to plants. In Plant Tolerance to Environmental Stress: Role of Phytoprotectants; Mirza, H., Masayuki, F., Hirosuke, O., Tofazzal, I.M., Eds.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Luo, J.; Zhou, J.; Li, H.; Shi, W.; Polle, A.; Lu, M.; Sun, X.; Luo, Z.B. Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 2015, 35, 1283–1302. [Google Scholar] [CrossRef]
- Cui, G.; Zhang, Y.; Zhang, W.; Lang, D.; Zhang, X.; Li, Z.; Zhang, X. Response of carbon and nitrogen metabolism and secondary metabolites to drought stress and salt stress in plants. J. Plant Biol. 2019, 62, 387–399. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Lempiäinen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Pathak, M.R.; Teixeira da Silva, J.A.; Wani, S.H. Polyamines in response to abiotic stress tolerance through transgenic approaches. GM Crop. Food 2014, 5, 87–96. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Li, L.; Huang, S.; Pong Lee, C.; Millar, A.H.; Taylor, N.L. Mitochondrial composition, function and stress response in plants. J. Integr. Plant Biol. 2012, 54, 887–906. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, S.; Li, M.; Zhao, Z. Transcriptomic insight into nitrogen uptake and metabolism of Populus simonii in response to drought and low nitrogen stresses. Tree Physiol. 2018, 38, 1672–1684. [Google Scholar] [CrossRef]
- Tsay, Y.F.; Chiu, C.C.; Tsai, C.B.; Ho, C.; Hsu, P.K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Zhang, J.; Chen, J.; Wu, T.; Zhu, S.; Yan, S.; Zhao, X.; Zhong, G. Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci. Hortic. 2014, 174, 65–76. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef]
- Liu, H.; Yang, W.; Liu, D.; Han, Y.; Zhang, A.; Li, S. Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 2011, 38, 417–427. [Google Scholar] [CrossRef]
- Jiang, A.L.; Xu, Z.S.; Zhao, G.Y.; Cui, X.Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Genome-wide analysis of the C3H zinc finger transcription factor family and drought responses of members in Aegilops tauschii. Plant Mol. Biol. Rep. 2014, 32, 1241–1256. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Akiyama, A.; Kisaka, H.; Uchimiya, H.; Miwa, T. Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. USA 2004, 101, 7833–7838. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).