Abstract
qRT-PCR is a powerful molecular research tool to study the regulation of gene expression. However, to accurately calculate gene expression levels, an experiment should include proper reference genes that show no changes in their expression level. Pinus massoniana, P. hwangshanensis, and their introgression hybrid in Mountain Lushan, China, are an ideal model for studying introgression and speciation. Although some research on reference gene selection for P. massoniana has been reported before, no studies on this subject have been performed where P. massoniana and its introgression hybrid were evaluated simultaneously. Here, we investigated ten genes (upLOC, SDH, ACT, EF, TOC75, DMWD, FBOX, PGK1, UBQ, and CL2417C7) identified from transcriptome data of these two taxa for reference gene potential. These ten genes were then screened across multiple tissues such as cone, young and mature stems, and young needles according to qRT-PCR thermal cycling and dissociation. Correlation coefficient, amplification efficiency, and cycle threshold value (Ct) range were applied to evaluate the reliability of each gene. The stability of candidate reference gene expression was calculated using three algorithms: geNorm, NormFinder, and BestKeeper. Base on the reliability and stability, we then offered a list of genes of recommended and not recommended for seven different tissue type and species. Our results demonstrated that different sample lines require different genes as reference to evaluate.
1. Introduction
Masson pine (Pinus massoniana) is an economically important conifer species that is naturally distributed in southern China. Its wood is often used for standing timber and furniture manufacturing. Resin, a secretion from its canals, can be used for the maintenance of musical instrument strings, and is an ingredient in several medicines [1,2]. Because of its fast growth and high biomass, it has been planted extensively in south China during the past fifty years for rapid vegetation recovery and additional ecological purposes.
Huangshan pine (P. hwangshanensis) is a species native only to China. Although it grows at a considerably slower rate than P. massoniana, it has ornamental value as a horticultural plant in higher altitude regions.
Around the middle and lower reaches of the Yangtze River, P. massoniana mainly grows at an altitude below 700 m (above sea level, a.s.l.), while P. hwangshanensis mainly grows above 900 m (a.s.l.). On Mountain Lushan, situated in this area, both pines have their own ecological niche according to the distribution mentioned above. An introgression hybrid pine (hereafter referred to as the Z pine) can be found directly adjacent to the habitats of both P. massoniana and P. hwangshanensis (Figure 1). There are several morphological differences between P. massoniana and P. hwangshanensis [3], while the Z pine displays phenotypic characteristics of both parental pines [4,5]. However, it also shows novel phenotypes, such as an ultra-low pinecone ripening and seed germination rate, compared to both P. massoniana and P. hwangshanensis [6]. This distinct characteristic shows that genetic incompatibilities during the process of fertilization and/or even embryonic development might exist between both parents of the Z pine. Our previous research has shown that a group of differentially expressed genes (DEGs) related to reproduction were expressed at a much lower level in the Z pine than in P. massoniana, possibly causing the abnormal reproductive phenotype in the Z pine [7].
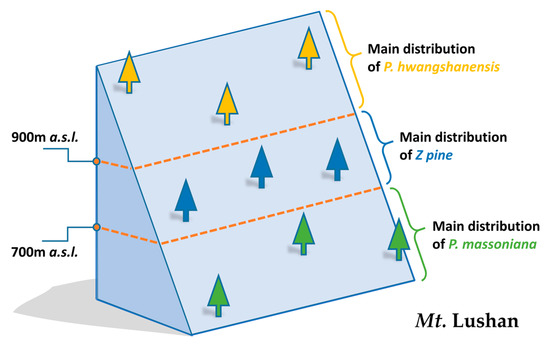
Figure 1.
Schematic indicating the main distribution areas of P. massoniana, P.hwangshanensis, and their introgression hybrid, the Z pine, on Mt. Lushan, Jiangxi Province, China. a.s.l.: above sea level.
Understanding how the expression of certain genes of interest is regulated in space, time, and different tissues is a crucial approach to further understanding about their biological function. Various methods can be applied to study gene expression: With microarrays, northern blots, RNA-seq, and qRT-PCR (quantitative real-time polymerase chain reaction) being considered some of the more common ways, RNA-seq and qRT-PCR being the most popular. After qRT-PCR was introduced in the late twentieth century [8], it has been widely applied to quantify gene expression in life science research and medical diagnosis. qRT-PCR has a series of advantages, including high accuracy, sensitivity, and reproducibility. Nevertheless, due to its high sensitivity, the qRT-PCR assay might be affected by RNA or cDNA quality, primer specificity, amplification efficiency [9], etc. Therefore, optimal normalization methods are required for sample-to-sample comparison. A series of strategies has been introduced for normalizing qRT-PCR data [9,10,11,12,13]. In general, in order to evaluate the expression quantity of a gene, a reference gene is required for various samples and/or experimental conditions (e.g., developmental stages, different organs, treatments, etc.). A reference gene is a gene that shows unchanging expression among all samples and/or experimental treatments [14] and can then be used to normalize and calibrate results [15]. Therefore, obtaining a reliable reference gene is a critical step before being able to perform qRT-PCR in further research.
However, no single universal reference gene is found acceptable to evaluate all species or samples. Due to the varying PCR amplification efficiency of the target sequence, a different reference gene could be required. Reference genes with poor availability could lead to significant biases and faulty interpretation of the data. Therefore, determining a reference gene is an essential step before starting the experimental process.
Genes such as ACT (actin), β-TUB (β-tubulin), GAPDH (glyceraldehyde 3-phosphate dehydrogenases), EF (elongation factor), and 18S rRNA have commonly been assigned as reference genes in recent research, we call them ‘classical’ reference genes. In addition, several novel reference genes have emerged from recent research. For example, RPL29 (60S ribosomal protein L29) was found to be the most stable reference gene in Bemisia tabaci (Hemiptera: Aleyrodidae) under biotic conditions including host plant, acquisition of a plant virus, developmental stage, tissue from body region of the adult, and whitefly biotype [16]. RPS4 (ribosomal protein S4) and UBQ (ubiquitin), a novel and classical reference gene, were found to be the optimal combination of reference genes used to study turbot (Scophthalmus maximus) gonad development before fertilization [17]. TIP41 (tonoplast intrinsic protein) and NTB (nucleotide tract-binding protein) could serve as reliable reference genes across all tissues and at different developmental stages in bamboo (Phyllostachys edulis) [18].
As the number of transcriptomic studies on various species increased, more and more unigenes were discovered and were assigned as reference genes for non-model species. These novel transcriptomic reference genes usually expressed a better stability than their classical counterparts. Our study is the first attempt to identify reference genes in multiple tissues of P. massoniana and its introgression hybrid. This study could offer a guideline for reference gene identification used for research into hybridization.
2. Materials and Methods
2.1. Preparation of Samples
Cones, mature needles, and young stems of mature P. massoniana and the Z pine were collected. Due to existing individual variation in open-pollinated wild pine, we conducted a mix-up strategy in sample collection to minimize biological variation within each organ sample: Three to five tree individuals were assigned to be collected from, tissues were then harvested and randomized per tissue. After that the mixed materials were subpackaged into RNase-free 50 mL centrifuge tubes or ziplock bags, which were frozen in liquid nitrogen or dry ice immediately before storing in a −80 °C ultra-low temperature freezer.
Young needles were collected from seedlings of P. massoniana and the Z pine. Seeds of each taxa were surface sterilized for 2 h with a 0.4% (w/v) solution of KMnO4 (potassium permanganate) and rinsed 5 times with sterile distilled water. Seeds were then moved into a germination box. A layer of cotton (saturated with sterile distilled water) was placed at the bottom of the germination box for retaining sufficient moisture, seeds were then placed on top. Germination boxes were positioned in a versatile environmental test chamber (MLR-352H, Panasonic Healthcare Co., Ltd., Ehime, Japan) under a 16:8 h day:night photoperiod cycle at 25 °C (day) and 22 °C (night), with a humidity setting of 70% (day) and 75% (night). During germination, seedlings with a root length of more than 1 cm were chosen to transfer to Pindstrup Substrate (Pindstrup Mosebrug A/S, Denmark) inside pots with 16 cm in diameter and grown under conditions identical to those used for germination. Young needles were carefully collected in RNase-free 15 mL centrifuge tubes and were frozen in liquid nitrogen, then transferred into an ultra-low temperature freezer (−80 °C) for storage.

Table 1.
Details of sample collection.

Figure 2.
Representative images of tissue samples collected. (a) P. massoniana cone (MCN); (b) Z pine cone (ZCN); (c) young stem of P. massoniana (MYS); (d) young stem of Z pine (ZYS); (e) mature needle of P. massoniana (MMN); (f) mature needle of Z pine (ZMN); (g) young needle of P. massoniana (MYN); (h) young needle of Z pine (ZYN). Scale = 10 mm.
2.2. Candidate Reference Gene Selection and Primer Design
Candidate reference genes were selected from previous transcriptome data of P. massoniana and Z pine cones [7]. These transcriptome data are available on the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA482692 [7]. Candidate reference genes were screened based on the following criteria: Their FPKM (fragments per kilobase of transcript per million fragments mapped) values were stable at different developmental stages, the coefficient of variation (CV) of the FPKM value can be no larger than 0.5.
According to these criteria, the following ten genes were chosen: upLOC (uncharacterized protein LOC103705956), SDH (succinate dehydrogenase), ACT (actin), EF (elongation factor), TOC75 (protein TOC75-3), DMWD (dystrophia myotonica WD repeat-containing protein), FBOX (F-box family protein), PGK1 (phosphoglycerate kinase 1), UBQ (ubiquitin), and CL2417C7 (unigene with unknown function).
Primer-BLAST [19] and Beacon Designer (v7.90, PREMIER Biosoft International, Palo Alto, CA, USA) were used for primer design. The main parameter settings of Primer-BLAST were as follows: PCR product size (10–250 bp), Primer melting temperature (Tm) (Min 59–Opt 60–Max 61 °C), Primer size (Min 18–Opt 20–Max 25 bp), Primer GC content (45%–55%), other parameters were set as default. In Beacon Designer, the main parameter setting were as follows: Tm (60 ± 1 °C), Primer length (18–24 bp), Amplicon length (80–150 bp), GC% (30%–80%), other parameters were set as default.
Details of candidate reference genes and their corresponding primers are listed in Table 2.

Table 2.
Reference genes and their primers in this study.
2.3. RNA Extraction, cDNA Template Preparation, and Quantitative Real-Time PCR (qRT-PCR)
Total RNA extraction of all samples was conducted using a Bioteke Plant RNA Extraction Kit (RP3301, Bioteke Corporation, Beijing, China). The 260 nm/280 nm UV absorption value of each RNA sample was measured using a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) to confirm its purity. The RIN (RNA integrity number) was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
cDNA templates were synthesized using the Vazyme HiScript II Q RT SuperMix for qPCR (R223-01, Vazyme Biotech Co.,Ltd., Nanjing, China). A total of 1 μg RNA was chosen for cDNA synthesis of each sample. After that, each reaction (20 μL) was diluted to 200 μL (10-fold diluted) and stored at −20 °C.
An Applied Biosystems 7500 PCR cycler (Thermo Fisher Scientific Corporation, CA, USA) was used to run the qRT-PCR. The Vazyme ChamQ SYBR qPCR Master Mix (Q331-02, Vazyme Biotech Co.,Ltd., Nanjing, China) was chosen as reaction reagent kit. For each qRT-PCR sample, 4 μL cDNA was added to a 20 μL reaction volume, containing 10 μL of 2× ChamQ SYBR qPCR Master Mix (Low ROX Premixed) and 0.4 μL of each primer. Thermal cycling was performed with an auto increment step of 30 s at 95 °C, and followed by 40 cycles of 10 s at 95 °C, 34 s at 60 °C. Dissociation analysis was performed with a default step of 15 s at 95 °C, and followed by 60 s at 60 °C, then 15 s at 95 °C. After running the thermal cycling and dissociation, the amplification and melting curves were obtained for evaluating the quality of the candidate reference genes amplification product. Each sample was tested with three technical replicates.
To assess the qRT-PCR amplification efficiency, the original cDNA solution was diluted 10-, 50-, 250-, 1250-, 6250-fold, respectively. All these solutions were used as templates for qRT-PCR amplification. A standard curve for each reference gene was drawn by using the average value of the three Ct (cycle threshold) technical replicates and its corresponding log solution concentration. The amplification efficiency was calculated using a standard Formula (1), where AE is the amplification efficiency and slope is the slope of the standard curve. The slope and correlation coefficient were automatically computed by Microsoft Office Excel when drawing the standard curve.
2.4. Stability Evaluation of Candidate Reference Genes
Three different algorithms, including geNorm [20], NormFinder [21], and BestKeeper [22] were assigned as tools to evaluate the stability of each candidate reference gene. All three algorithms are operated in Microsoft Office Excel, where geNorm and NormFinder need to enable ‘Macros’ in Excel to launch, while BestKeeper does not need this operation. The Ct relative expression value was the only correct data format to put into geNorm and NormFinder before evaluating, while in BestKeeper the Ct value can be put in directly.
The Excel Add-in geNorm searches for the one or two most stable reference genes by computing the gene expression normalization factor, an M value, according to the geometric mean of a series of candidate reference genes. The M value indicates the stability level of every single gene, with the least stable gene having the maximum M value and the most stable gene the minimum M value. Usually, genes with an M value higher than 1.5 would be identified as unstable. Furthermore, a pairwise variation value offered by geNorm determines how many genes should be chosen as additional reference gene. The pairwise variation has a vn/vn+1 form value: If this value is bigger than 0.15 (threshold of default), then the ideal number of combinations would be ‘n+1′. Otherwise it would be ‘n’ as recommended.
The Excel-based VBA (Visual Basic for Applications) applet NormFinder provides an algorithm evaluating stability of a group of genes. It can assess the expression variation of entire groups of genes with intra- and inter-group comparisons simultaneously. Like geNorm, it uses an M value to indicate the stability of a gene, with a lower number equating to better stability.
Finally, the Excel-based tool BestKeeper, computes a series of indicators such as standard deviation, correlation coefficient, coefficient of variable, aiming to find which genes are more stable. Here, we adopted the correlation coefficient as a single parameter to decide the most stable gene, the closer to 1 the better.
3. Results
3.1. Reference Gene Evaluation
Ten candidate reference genes were tested in different P. massoniana and Z pine tissues (cone, young stem, mature needle, and young needle). The PCR product melting curves showed that all genes and primers had their unique amplification specificity (Figures S1–S8). Correlation coefficient (R), slope and amplification efficiency (AE) of each gene under every tissue were calculated (Figures S1–S8), also. The correlation coefficient ranged from 0.9610 to 0.9999, while the amplification efficiency range is 1.22 to 2.86. Concerning the correlation coefficient, in both species and all tissues, EF was greater than 0.99, while ACT, PGK1, and CL2417C7 were greater than 0.985. One sample, amplification of the FBOX gene in MYN tissue, had the lowest correlation coefficient at 0.9610. The SDH and FBOX genes showed the highest amplification efficiency across all samples, while upLOC, ACT, TOC75, PGK1, and CL2417C7 had a lower value (Figure 3).
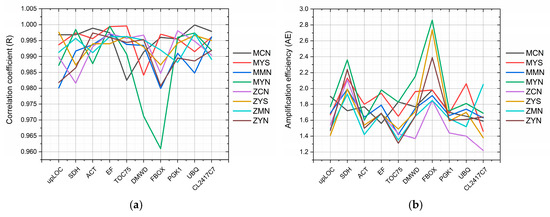
Figure 3.
Correlation coefficient and amplification efficiency of ten candidate reference genes across all samples. (a) Correlation coefficient; (b) amplification efficiency. The legend on the right displays colored sample code information, see Table 1.
3.2. Z pine Has a Higher Expression Abundance than P. massoniana Across Ten Candidate Genes
The expression level of ten candidate reference genes in all assayed tissues (cone, young stem, mature needle, young needle) of P. massoniana and Z pine has a wide range according to their Ct (~16 to ~28 for both species) (Figure 4). EF has the lowest Ct value in both species, indicating that it shows the highest expression abundance. The FBOX Ct value is the highest, indicating that it has the lowest expression abundance. In addition, Ct fluctuations in PGK1 are smallest in P. massoniana, while TOC75 and UBQ expression varies the least in Z pine, indicating that their expression abundance is very similar across different organs for their respective species. SDH shows the highest variation in expression level among all genes tested, which was not really to serve as a reference gene anymore. Overall, Ct values from P. massoniana are significantly higher than those of the Z pine, possibly reflecting that the Z pine has a higher basic expression abundance. All genes show a Ct distribution range between 15 and 30, which is an acceptable range for a potential reference gene.
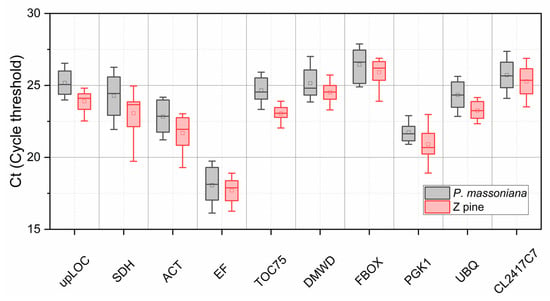
Figure 4.
Range of Ct values for ten reference gene candidates across different P. massoniana and Z pine tissues. Ct distribution is presented as a box-plot, indicating the interquartile range (box), median (horizon line in box), average (small square), maximum and minimum value (upper and lower whisker cap).
3.3. Candidate Reference Genes Show Variable Stability Across Sample Lines
The stability of these ten genes was assessed using the M and r (correlation coefficient) values derived from either geNorm, NormFinder, and BestKeeper. Ranking the M and r value of each gene offers information about their relative performance; therefore, we created a parameter called ‘ranking score’ which is the sum of the M and r values, with smaller values being better (Table S1). Both the exact values (of M and r) and ranking score were assigned to decide which gene would perform best as a reference gene.
Within both cone samples (MCN + ZCN), UBQ, DMWD, and ACT were expressed at the most stable level in geNorm, NormFinder, and BestKeeper, respectively (Figure 5a), with ACT having the lowest ranking score (Table S1). This makes ACT the most stably expressed reference gene in cone tissue, while EF and TOC75 showed the most unstable expression. Within young stem samples (MYS + ZYS), PGK1 was the most stably expressed according to all three algorithms, followed by ACT and EF; TOC75 was the most unstably expressed in young stem tissue (Figure 5b). Within mature needle tissue (MMN + ZMN), FBOX, DMWD, and upLOC showed the most stable expression according to geNorm, NormFinder, and BestKeeper, respectively (Figure 5c). According to their ranking score, FBOX was the most stably expressed (Table S1), CL2417C7 the most unstable. Considering that FBOX has a relatively high amplification efficiency (Figure 3b), which may cause uncertainty in normalization, it was excluded from the recommended list. Within young needle tissue (MYN + ZYN), almost all genes, excluding CL2417C7 which was more unstable, performed similarly, with upLOC, UBQ, DMWD, and ACT showing the highest stability (Figure 5d).
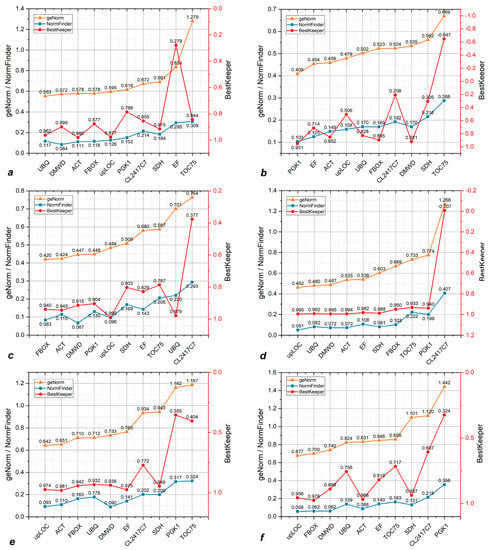
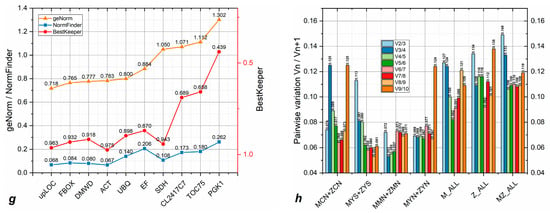
Figure 5.
Stability values per gene as calculated using three separate algorithms (geNorm, NormFinder, and BestKeeper). Figures display the stability value data per gene of: (a) MCN+ZCN; (b) MYS+ZYS; (c) MMN+ZMN; (d) MYN+ZYN; (e) all P. massoniana samples; (f) all Z pine samples; (g) all P. massoniana and Z pine samples; and (h) pairwise variation calculated by geNorm.
Analyzing the combined data from all P. massoniana tissues (Figure 5e), we found that upLOC and ACT showed the most stable expression using the geNorm and BestKeeper algorithms. Even though DMWD was deemed the most stable using NormFinder, it had a relatively low ranking using the remaining two algorithms. Therefore, we conclude that ACT and upLOC are the most stably expressed potential reference genes in P. massoniana. According to Figure 5e and Table S1, TOC75 was the most unstably expressed gene we analyzed across all P. massoniana samples. We then found upLOC and FBOX to be the most stably expressed reference genes across all Z pine tissues (Figure 5f). However, FBOX was not included in our recommended list due to its higher amplification efficiency (Figure 3b). In addition, we found that the PGK1 gene is not suitable for gene expression normalization in the Z pine due to its low ranking across all three algorithms. Finally, combining data from both P. massoniana and the Z pine within all tissue types, we conclude that upLOC, ACT, FBOX, and DMWD in that order are the most suitable for functioning as reference gene, with only PGK1 being unsuitable for this purpose (Figure 5g). We listed all (non-) recommended reference genes for each tissue type and species in Table 3.

Table 3.
The recommended and NOT recommended reference genes for each tissue type/species.
We next analyzed the optimal number of reference genes that should be used for relative expression normalization using geNorm (Figure 5h). According to the criteria mentioned in Section 2.4, all pairwise variations of all tissue types expressed below 0.15 (threshold of default), therefore a combination of two genes would be the optimal number for every normalization.
4. Discussion
Acquisition of compatible reference genes is a prerequisite for obtaining reliable qRT-PCR results. The utility of a reference gene must be experimentally validated for particular tissues or cell types and specific experimental designs [14]. Several traditional reference genes such as the actin, elongation factor, and GAPDH gene families have been frequently used in a number of different species, especially in model organisms like Danio rerio [23], Drosophila melanogaster [24], Schmidtea mediterranea, and Macrostomum lignano [25]. However, they are not always suitable as reference gene under every possible experimental condition. As the scope of research continues to expand, other unconventional novel reference genes like uroporphyrin III C-methyltransferase, HcaT MFS transporter, L-idonate/5-ketogluconate/gluconate transporter in Escherichia coli [26], Type II metacaspases and GYF domain-containing protein in Brassica napus [27], have emerged. Evaluation of reference genes via transcriptome research in conifers has been reported in recent years [28]. However, reference gene selection for more than one conifer taxa simultaneously has rarely been described [29].
In this paper, we aimed to determine optimal reference genes from P. massoniana and its introgression hybrid, the Z pine, in each of several chosen tissues: cone, young stem, mature and young needle. P. massoniana, and Z pine display significant phenotypic differences not only in appearance, but also at the transcriptome level [7]. Finding appropriate reference genes for both taxa is an essential step for downstream experiments. After conducting sequence alignment and annotation using our previously generated transcriptomic data, we selected ten candidate reference genes (upLOC, SDH, ACT, EF, TOC75, DMWD, FBOX, PGK1, UBQ, and CL2417C7).
A series of assessments were conducted before calculating their stability value. All candidate genes and their primers should generate a single PCR product according to their melting curve (Figures S1–S8), which is a basic requirement for any reference gene. The correlation coefficient would be good if it closer to 1 and the amplification efficiency would be suitable when it is not too high. We found that most of our selected 10 genes indeed adhered to these criteria. Their amplification efficiency ranged from 1.22 to 2.86, which is higher than in some species like Davidia involucrata [30], Euphorbia esula [31], Phyllostachys edulis [18], but close to more related species such as Pinus pinaster and Picea abies [29]. Furthermore, recent research has reported both low [32] and high [33] amplification efficiencies for P. massoniana genes. What causes this difference is still poorly understood? FBOX has a higher amplification efficiency compared to the other genes (Figure 3b); considering the accuracy of normalization, it was disqualified from becoming a reference gene (Table 3), although it was higher-ranked in the MMN+ZMN (all mature needle) and Z_ALL (all Z pine) samples.
During a qRT-PCR reaction, fluorescence values are recorded during every cycle and represent the amount of product amplified up until that point in the amplification reaction [9]. If the sample contains more DNA template, it would take less cycles to reach the point at which the fluorescence signal could be detected [34]. This point is called the cycle threshold [9]. The cycle threshold, or Ct, is the main parameter obtained from a qRT-PCR reaction. Lower Ct values equate to a higher gene abundance. We found that the Z pine shows overall lower Ct values for all tested genes, in other words, it shows higher expression of these genes compared to P. massoniana. It is currently not clear why this is the case, however, since significant expression level differences exist between these two taxa, it is advised to perform stability expression analysis separately for each species if possible.
We used three different algorithms to evaluate the expression stability of candidate reference genes: geNorm [20], NormFidner [21], and BestKeeper [22]. From each algorithm, we used both the stability value and the ranking score of each gene to determine their suitability as reference gene across multiple tissue types. The three algorithms did not always give the same ranking of gene expression stability (Figure 5a–g). In particular, geNorm and NormFinder showed a similar ranking across most samples, while BestKeeper gave a significantly different outcome. The reason for this phenomenon could be that geNorm and NormFinder requires a ΔCt input while BestKeeper uses the raw Ct data. This difference in reference gene evaluation could also be seen in Undaria pinnatifida [35]. However, studies have also been reported in which NormFinder and BestKeeper provided similar outcomes [36]. Multiple control reference genes are essential if possible [10]. Therefore, the parameter of pairwise variation in geNorm was calculated (Figure 5h) as a reference.
For each tissue type and species, we found different recommended and not recommended reference genes (Table 3). The upLOC gene is the most widely suitable reference gene, as it can be used for young and mature needle tissues, as well as combined tissue samples. In addition, ACT is a suitable reference gene for cone and mature needle tissue and combined tissue samples. Several candidate reference genes, such as TOC75 and CL2417C7 showed too much expression variability in several tissues to be usable as a reference gene. Interestingly, the PGK1 gene, which performs well as a reference gene for young stem tissue, showed too much expression variation in combined tissue samples. This analysis shows that suitability as a reference gene depends on tissue type, which could indicate that such genes show clear tissue specific expression. These findings suggest that prior to analyzing gene expression in a novel tissue type, reference gene suitability should be examined.
The actin gene family originates from a common ancestor in various species [37]. Some species have three types of actin genes: α-, β- and γ-actin. In plants, more than ten types of actin exist. β-actin has been widely used as reference gene across countless studies. Yet, even though actin is a celebrity in the ‘reference gene club’, it could not always be successfully applied as a reference gene; for example, in Medicago sativa [38], Symbiodinium [39], and Ganoderma lucidum [40]. However, we tested ACT in normalization of a series target genes of cones from P. massoniana and Z pine in a recent study (as introduced in Table 3) and obtained an ideal result [7], which indicated that despite ACT not always being reliable, it might be in this case anyway. Phosphoglycerate kinase 1 (PGK1) is an ATP-generating glycolytic enzyme that forms part of the glycolytic pathway. Reports of this enzyme mainly concern human health and disease [41,42], where it also acts as reference gene in human pathology examinations [43]. Little research has been published on dystrophia myotonica WD repeat-containing protein (DMWD), which plays a role in myotonic dystrophy, a complex neuromuscular disorder in mammals [44,45]. The uncharacterized protein LOC103705956 (upLOC) is a novel gene that was initially found in transcriptome data of P. massoniana and Z pine cones [7]. It showed high expression stability across multiple tissues in this study (Table 3), implying that it could be a potential valuable reference gene used for expression normalization, perhaps even for P. hwangshanensis and other species in the Pinus genus.
5. Conclusions
In this study we tested ten candidate reference genes, both classic and novel, from the transcriptome data of P. massoniana and Z pine cones. Our results show that ACT and upLOC are the most consistently well behaving reference genes taken across all tissue types and species, while PGK1 is the most stably expressed specifically in young stem samples. Our study has further shown that reference genes should be chosen carefully, for no single gene can fit every tissue even in the same or closely related species.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4907/10/9/787/s1, specificity of primers and standard curves tested in Figure S1: cone of P. massoniana (MCN), Figure S2: young stem of P. massoniana (MYS), Figure S3: mature needle of P. massoniana (MMN), Figure S4: young needle of P. massoniana (MYN), Figure S5: cone of Z pine (ZCN), Figure S6: young stem of Z pine (ZYS), Figure S7: mature needle of Z pine (ZMN), Figure S8: young needle of Z pine (ZYN); Table S1: Ranking of three algorithms in sample lines.
Author Contributions
Conceptualization, J.X., T.Y. and J.S.; Formal analysis, J.M., W.J. and L.Y.; Funding acquisition, J.X. and J.S.; Investigation, J.M. and W.J.; Project administration, J.X.; Writing-original draft, J.M.; Writing-review & editing, J.M.
Funding
This research was funded by National Natural Science Foundation of China (31270661) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Acknowledgments
The authors are grateful to Youxin Du, Qiang Huang and Benzhong Zhou of the Lushan Botanical Garden, Jiangxi Province and Chinese Academy of Sciences, for assistance with sample collection on Mountain Lushan. The authors are also indebted to two reviewers for their insightful comments on this article. Special thanks go to editors for their help in formulating the revisions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mills, J.S.; White, R. Natural resins of art and archaeology their sources, chemistry, and identification. Stud. Conserv. 1977, 22, 12–31. [Google Scholar]
- Yang, N.; Liu, L.; Tao, W.; Duan, J.; Tian, L. Diterpenoids from Pinus massoniana resin and their cytotoxicity against A431 and A549 cells. Phytochemistry 2010, 71, 1528–1533. [Google Scholar] [CrossRef] [PubMed]
- Editorial Board of Flora of China, Chinese Academy of Science. Flora of China, 1st ed.; Science Press: Beijing, China, 1978; Volume 7, pp. 263, 266. (In Chinese) [Google Scholar]
- Luo, S.; Zou, H.; Liang, S. Study on the introgressive hybridization between Pinus hwangshanensis and P. massoniana. Sci. Silvae Sin. 2001, 37, 118–122. (In Chinese) [Google Scholar]
- Zhai, D.; He, Z.; Feng, J.; Zheng, Y. Study on introgression between Pinus hwangshanensis and Pinus massoniana by using inter-simple sequence repeat marker (ISSR). For. Sci. Technol. 2012, 37, 4–6. (In Chinese) [Google Scholar]
- Li, S.; Chen, Y.; Gao, H.; Yin, T. Potential chromosomal introgression barriers revealed by linkage analysis in a hybrid of Pinus massoniana and P. hwangshanensis. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef]
- Mo, J.; Xu, J.; Cao, Y.; Yang, L.; Yin, T.; Hua, H.; Zhao, H.; Guo, Z.; Yang, J.; Shi, J. Pinus massoniana introgression hybrids display differential expression of reproductive genes. Forests 2019, 10, 230. [Google Scholar] [CrossRef]
- Weis, J.H.; Tan, S.S.; Martin, B.K.; Wittwer, C.T. Detection of rare mRNAs via quantitative RT-PCR. Trends Genet. 1992, 8, 263–264. [Google Scholar] [CrossRef]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. BioTechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Karge, W.H.; Schaefer, E.J.; Ordovas, J.M. Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol. Biol. 1998, 110, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xie, W.; Wang, S.; Wu, Q.; Yang, N.; Yang, X.; Pan, H.; Zhou, X.; Bai, L.; Xu, B. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS ONE 2013, 8, e53006. [Google Scholar] [CrossRef] [PubMed]
- Robledo, D.; Hernández-Urcera, J.; Cal, R.M.; Pardo, B.G.; Sánchez, L.; Martínez, P.; Viñas, A. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genom. 2014, 15, 648. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–11. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Dodd, A.; Lai, D.; Mcnabb, W.C.; Love, D.R. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim. Biophys. Sin. 2007, 39, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Ponton, F.; Chapuis, M.; Pernice, M.; Sword, G.A.; Simpson, S.J. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 2011, 57, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Plusquin, M.; DeGheselle, O.; Cuypers, A.; Geerdens, E.; Van Roten, A.; Artois, T.; Smeets, K. Reference genes for qPCR assays in toxic metal and salinity stress in two flatworm model organisms. Ecotoxicology 2012, 21, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zhou, L.; Lim, Q.; Zou, R.; Stephanopoulos, G.; Too, H. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Behringer, D.; Zimmermann, H.; Ziegenhagen, B.; Liepelt, S. Differential gene expression reveals candidate genes for drought stress response in Abies alba (Pinaceae). PLoS ONE 2015, 10, e124564. [Google Scholar] [CrossRef]
- de Vega-Bartol, J.J.; Santos, R.R.; Simões, M.; Miguel, C.M. Normalizing gene expression by quantitative PCR during somatic embryogenesis in two representative conifer species: Pinus pinaster and Picea abies. Plant Cell Rep. 2013, 32, 715–729. [Google Scholar] [CrossRef]
- Ren, R.; Huang, F.; Gao, R.; Dong, X.; Peng, J.; Cao, F.; Li, M. Selection and validation of suitable reference genes for RT-qPCR analysis in dove tree (Davidia involucrata Baill.). Trees 2019, 1–13. [Google Scholar] [CrossRef]
- Chao, W.S.; Doğramaci, M.; Foley, M.E.; Horvath, D.P.; Anderson, J.V. Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS ONE 2012, 7, e42839. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.; Hu, Y.; Tan, J.; Jia, J.; Xu, H.; Chen, X. Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees 2016, 30, 685–696. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, Q.; Dong, H.; Zhou, Z.; Hao, Y.; Chen, X.; Xu, L. Selection of reference genes for real-time quantitative PCR in Pinus massoniana post nematode inoculation. PLoS ONE 2016, 11, e147224. [Google Scholar] [CrossRef] [PubMed]
- Gibson, U.E.; Heid, C.A.; Williams, P.M. A novel method for real time quantitative RT-PCR. Genome Res. 1996, 6, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, H.; Shan, T.; Pang, S. Selection of reference genes for real-time RT-PCR normalization in brown alga Undaria pinnatifida. J. Appl. Phycol. 2019, 31, 787–793. [Google Scholar] [CrossRef]
- Wang, B.; Du, H.; Yao, Z.; Ren, C.; Ma, L.; Wang, J.; Zhang, H.; Ma, H. Validation of reference genes for accurate normalization of gene expression with quantitative real-time PCR in Haloxylon ammodendron under different abiotic stresses. Physiol. Mol. Biol. Plants 2018, 24, 455–463. [Google Scholar] [CrossRef]
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fu, Y.; Ban, L.; Wang, Z.; Feng, G.; Li, J.; Gao, H. Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet. Syst. 2015, 90, 175–180. [Google Scholar] [CrossRef]
- Chong, G.; Kuo, F.; Tsai, S.; Lin, C. Validation of reference genes for cryopreservation studies with the gorgonian coral endosymbiont Symbiodinium. Sci. Rep. 2017, 7, 39396. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, J.; Ji, A.; Zhu, Y.; Zhang, X.; Hu, Y.; Song, J.; Chen, S. Genome-wide selection of superior reference genes for expression studies in Ganoderma lucidum. Gene 2015, 574, 352–358. [Google Scholar] [CrossRef]
- Duan, Z.; Lamendola, D.E.; Yusuf, R.Z.; Penson, R.T.; Preffer, F.I.; Seiden, M.V. Overexpression of human phosphoglycerate kinase 1 (PGK1) induces a multidrug resistance phenotype. Anticancer Res. 2002, 22, 1933–1941. [Google Scholar]
- Wang, J.; Ying, G.; Wang, J.; Jung, Y.; Lu, J.; Zhu, J.; Pienta, K.J.; Taichman, R.S. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 2010, 70, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, V.R.; Whistler, T.; Janna’R, M.; Unger, E.R.; Rajeevan, M.S. Identification of phosphoglycerate kinase 1 (PGK1) as a reference gene for quantitative gene expression measurements in human blood RNA. BMC Res. Notes 2011, 4, 324. [Google Scholar] [CrossRef] [PubMed]
- Jansen, G.; Bächner, D.; Coerwinkel, M.; Wormskamp, N.; Hameister, H.; Wieringa, B. Structural organization and developmental expression pattern of the mouse WD-repeat gene DMR-N9 immediately upstream of the myotonic dystrophy locus. Hum. Mol. Genet. 1995, 4, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.J.; McCurrach, M.; Rundle, S.A.; Harley, H.G.; Crow, S.R.; Sohn, R.; Thirion, J.; Hamshere, M.G.; Buckler, A.J.; Harper, P.S. Genomic organization and transcriptional units at the myotonic dystrophy locus. Genomics 1993, 18, 673–679. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).