Abstract
Three new cyanobactins, trikoramides B (1)–D (3), have been isolated from the marine cyanobacterium, Symploca hydnoides, following a preliminary bioassay-guided isolation of the two most active polar fractions based on the brine shrimp toxicity assay. These new cyanobactins are new analogues of the previously reported cytotoxic trikoramide A (4) with differences mainly in the C-prenylated cyclotryptophan unit. Their planar structures were elucidated from their 1D and 2D NMR spectral data in combination with the HRMS/MS data. Marfey’s method, 2D NOESY NMR spectroscopic and ECD spectra analyses were used to determine the absolute stereochemistry of trikoramides B (1)–D (3). Trikoramides B (1) and D (3) exhibited cytotoxicity against MOLT-4 acute lymphoblastic leukemia cell line with IC50 values of 5.2 µM and 4.7 µM, respectively. Compounds 1 and 3 were also evaluated for their quorum-sensing inhibitory assay based on the Pseudomonas aeruginosa PAO1 lasB-gfp and rhlA-gfp bioreporter strains. Although trikoramide B (1) exhibited weak quorum-sensing inhibitory activity, the Br-containing trikoramide D (3) exhibited moderate to significant dose-dependent quorum-sensing inhibitory activities against PAO1 lasB-gpf and rhlA-gfp bioreporter strains with IC50 values of 19.6 µM and 7.3 µM, respectively.
1. Introduction
Filamentous cyanobacteria (blue-green microalgae) are oxygen-producing photoautotrophic prokaryotes whose existence dates back to 3.5 billion years ago [1]. These ancient microbes are also known to be prolific producers of novel bioactive natural products. In particular, the order Oscillatoriales accounts to produce most of the isolated bioactive secondary metabolites from marine cyanobacteria, most of which are cyclic or linear peptides [2]. These peptides are assembled by either the nonribosomal or post-translationally modified ribosomal biosynthetic pathways in cyanobacteria. The first ribosomal biosynthetic pathway was described by Schmidt et al. in 2005 for the cyanobactin, patellamide [3]. Cyanobactins are a class of ribosomally synthesized cyclic peptides with post-translational modifications, including formation of azole/azoline rings, D-stereocentres and prenyl group [4]. Several cyanobactins have been found to have anticancer properties (e.g., trunkamide [5]), multidrug resistance-reversing activity (e.g., dendroamides [6]) and antimalarial activity (e.g., venturamides [7]).
Previous efforts to hunt for novel bioactive secondary metabolites in our laboratory has led to the isolation of the cytotoxic prenylated cyanobactin, trikoramide A (4) [8] from Symploca hydnoides. Further investigations into the brine shrimp toxic polar fractions have afforded three new related compounds, trikoramides B (1)–D (3). Both trikoramides B (1) and D (3) exhibited significant cytotoxic activity against MOLT-4 acute lymphoblastic leukemia cell line. Interestingly, the Br-containing trikoramide D (3) exhibited significant quorum-sensing inhibitory (QSI) activities in a dose-dependent manner when tested in the Pseudomonas aeruginosa PAO1 lasB-gpf and rhlA-gfp bioreporter strains. The current study details the purification, structural determination and biological activities of these new trikoramide A related analogues.
2. Results and Discussions
2.1. Isolation and Structural Elucidation
The marine cyanobacterium S. hydnoides was collected by hand at the intertidal shores of Trikora beach, Bintan Island, Indonesia. The sample was extracted exhaustively with CH2Cl2-MeOH (2:1, v/v) and the resulting crude extract was fractionated into nine fractions (1 to 9) using vacuum liquid chromatography (VLC). Fractions 8 and 9, eluted with EtOAc-MeOH (9:1) and EtOAc-MeOH (8:2), respectively, exhibited brine shrimp toxicity with at least 80% lethality when tested at 100 ppm and were further subjected to fractionation by reversed-phase solid-phase extraction (RP-SPE), followed by a series of reversed-phase high performance liquid chromatography (HPLC) purifications to yield white amorphous solids of trikoramide B (1, 1.1 mg), trikoramide C (2, 0.3 mg) and trikoramide D (3, 1.2 mg) (Figure 1).
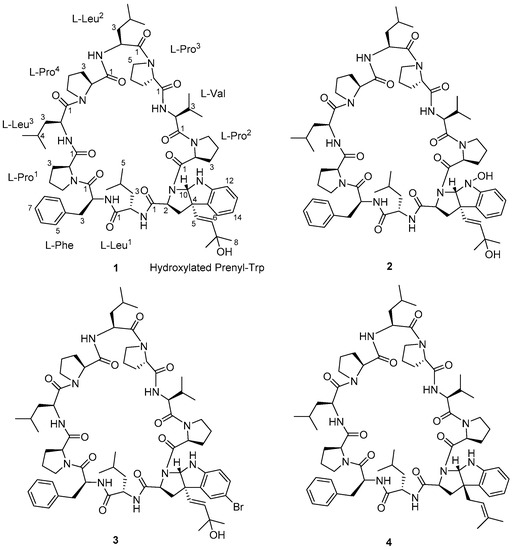
Figure 1.
Structures of trikoramides B (1)–D (3) and A (4).
Trikoramide B (1) yielded a [M + H]+ ion at m/z 1244.7446 by HR-ESI-OrbitrapMS and is consistent with the molecular formula of C68H97N11O11 requiring 26 degrees of unsaturation. The 1H NMR spectrum of 1 showed signals typical of a peptidic metabolite with the NH signals observed between δH 6.67–δH 8.02 and α methine protons between δH 3.07–δH 4.75 (Table 1). Initial analysis of the 13C NMR spectrum revealed ten signals characteristic of amide carbonyl groups (δC 170.1, 172.5, 171.7, 172.9, 171.5, 170.5, 170.8, 172.9, 170.3, 170.7) indicating 1 to be a decapeptide as well as an analogue of trikoramide A (4) (Table 1) [8]. Further analysis of the 1D and 2D NMR spectra of 1 established the presence of nine standard amino acids, including 4 × Pro, 1 × Leu, 1 × Val and 1 × Phe, and an atypical amino acid. The identity of the unusual amino acid was determined to be a hydroxylated C-prenylated-cyclotryptophan by a combination of 13C, 1H, COSY, HSQC and HMBC NMR spectra which have been interpreted as follows. From the 13C and DEPT-135 NMR spectra, we observed the presence of four methine carbons (δC 110.5, 119.2, 123.7, 129.3) and two non-protonated aromatic carbons (δC 130.6, 149.2) characteristic of an indoline moiety, which was also evident from the UV absorption at λmax 250 and 295 nm, similar to that of the previously reported trikoramide A (4) [8]. However, the chemical shifts of the olefinic carbons at the prenyl unit in 1 shifted downfield from δ 118.3/5.14 (H-6) and δ 135.8 (C-7) in trikoramide A (4) to δ 126.7/5.81 (H-5) and δ 139.1/5.52 (H-6) in 1. In addition, based on the large coupling constant of 15.0 Hz between H-5 and H-6, these two vicinal protons are trans to each other suggesting a shift in the position of the double bond (H-5/H-6) in the prenyl unit. Furthermore, HMBC correlations between H-5 and H-6 to C-7 (δC 70.4) and presence of a dehydrated protonated molecule [M + H – H2O]+ peak at m/z 1226.7322 confirmed the presence of a tertiary alcohol with two methyl groups attached (Figure 2) at the prenyl residue. The COSY correlation between H-5 and H-6 in combination with several key HMBC correlations, including H-5/C-4, H-5/C-3, H-5/C-15, H-6/CH3-8, and H-6/CH3-9, established that the prenylated unit was attached to the indolic cyclotryptophan unit (Figure 2). This confirms the presence of a C-prenylated cyclotryptophan with a terminal tertiary alcohol group on the prenylated side chain.

Table 1.
NMR data for trikoramide B (1) in CDCl3 (1H 400 MHz, 13C 100 MHz).
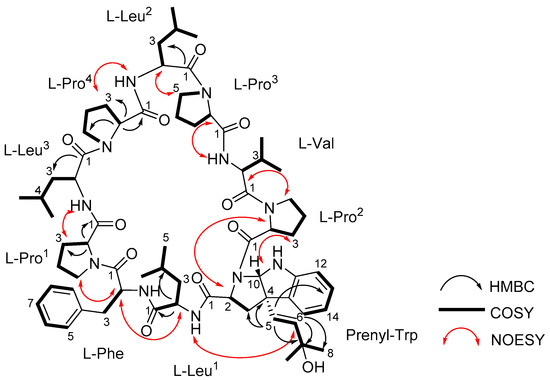
Figure 2.
Structure of trikoramide B (1) with key 2D NMR correlations.
The sequence of the amino acids in 1 was inferred from 2D NMR spectral data as well as MS/MS spectral analysis. Nine key NOESY correlations between H-2 of Phe and H-5a of Pro1, H-2 of Leu1 and H-2 of Phe, N-H of Leu1 and H-6 of Prenyl-Trp, H-2 of Prenyl-Trp and H-2 of Pro2, H-2 of Val and H-5a of Pro2, N-H of Val and H-2 of Pro3, H-2 of Leu2 and H-5a of Pro3, N-H of Leu2 and H-3a of Pro4, H-3a of Pro1 and N-H of Leu3 established the presence of the sequence Leu3-Pro1-Phe-Leu1-Prenyl-Trp-Pro2-Val-Pro3-Leu2-Pro4 (Figure 2). Additional evidence from the fragmentation pattern observed in the MS/MS spectrum of trikoramide B (1) confirms the presence of the aforementioned sequence and the double bond equivalents calculated from its molecular formula suggests the structure to be a cyclic decapeptide, enacting the structure as drawn in 1. The geometries of the amide bonds in the proline residues were determined based on the 13C NMR chemical shift difference at the β and γ positions (Δδβ−γ). Similar to trikoramide A (4), the large difference calculated for Pro1 (Δδβ−γ =8.9 ppm) and Pro2 (Δδβ−γ =8.1 ppm) indicated that their peptide bonds were in cis geometry. For Pro3 (Δδβ−γ =4.5 ppm) and Pro4 (Δδβ−γ = 3.5 ppm) their geometries were determined to be trans which was also further supported by NOESY correlations between H-2 of Leu2 and H-5a/5b of Pro3; H-2 of Leu3 and H-5a/5b of Pro4 (Figure 2).
The absolute stereochemical configurations of the usual amino acids in trikoramide B (1) were determined by acid hydrolysis followed by derivatizing with Marfey’s reagent 1-fluoro-2,4-dinitro-phenyl-5-L-alanine amide (L-FDAA), revealing the L-configuration of Pro, Val, Leu and Phe residues [9]. For the hydroxylated Prenyl-Trp residue, acid hydrolysis of 1 with 1% phenol yielded tryptophan residue from the Prenyl-Trp residue and subsequent derivatization with Marfey’s reagent afforded L-Trp, indicating the absolute configuration of C-2 on Prenyl-Trp residue to be of S configuration. Analysis of the NOESY spectrum of 1 showed absence of cross peak between H-5 and H-10 of the Prenyl-Trp residue, indicating that they were on the different face of the tricyclic indole ring. This suggested that there were only two possible absolute configurations of the Prenyl-Trp unit at (2S, 4S, 10S) or (2S, 4R, 10R). The electronic circular dichroism (ECD) spectrum of trikoramide B (1) was investigated to determine the absolute stereochemistry at C-4 and C-10 of the Prenyl-Trp residue. The ECD spectrum of 1 was compared to the previously reported ECD spectrum of motobamide, a cyclic peptide containing similar Prenyl-Trp unit, which showed a negative Cotton effect at 295 nm [10]. Takahashi et al. designed two model compounds with the absolute stereochemistries of 2S, 4S, 10S and 2S, 4R, 10R of the Prenyl-Trp and calculated the theoretical ECD spectra of the model compounds and compared with the experimental ECD spectrum of motobamide [10]. The resulting ECD spectrum for the (2S, 4S, 10S) model compound exhibited negative Cotton curves similar to that of motobamide and its absolute stereochemistry was directly correlated [10]. Similarly, the ECD spectrum of trikoramide B (1) portrayed negative Cotton effect at 295 nm, contributed solely by the Prenyl-Trp unit, and is similar to the ECD spectra of the simplified model compound with 2S, 4S, 10S as well as motobamide (Figure 3). Furthermore, the ECD spectrum of the previously isolated trikoramide A (4) was obtained and it showed negative Cotton curves similar to that of 1, suggesting the absolute stereochemistry of the hydroxylated Prenyl-Trp residue to be 2S, 4S, 10S (Figure 3).
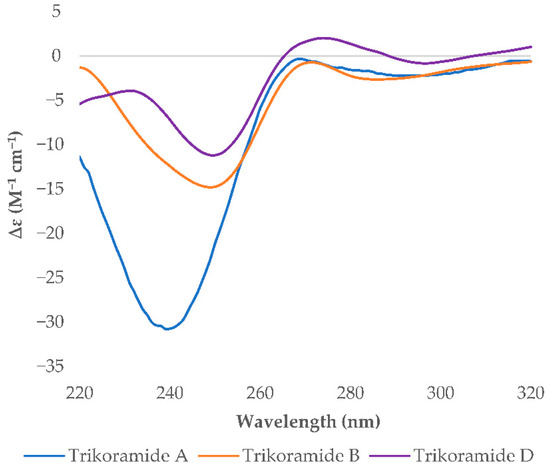
Figure 3.
Experimental ECD spectra of trikoramides A (4), B (1) and D (3).
Trikoramide C (2) gave a [M + H]+ protonated molecule at m/z 1260.7386 by HR-ESI-OrbitrapMS, consistent with the molecular formula of C68H97N11O12 with an additional 16 mass units equating to an additional oxygen atom compared to trikoramide B (1). Further analysis of the 1H NMR spectrum of 2 in conjunction with the HR-ESI-OrbitrapMS spectrum established the presence of an unusual hydroxylamine group on the Prenyl-Trp residue. The 1H NMR spectrum of trikoramide C (2) showed similar chemical shifts to that of 1. However, the chemical shifts of the H-2α proton and the H-10 singlet proton signals of the Prenyl-Trp group shifted downfield from δH 3.86/ δH 5.42 in 1 to δH 3.94/ δH 5.63 in 2, indicating the presence of a nearby electronegative atom. Further evidence from the [M − H2O − O]+ peak from the HR-ESI-OrbitrapMS and the absence of NH proton signal on the Prenyl-Trp residue from 1H NMR spectrum confirms the substitution of a hydroxyl group on the indole nitrogen where the loss of 16 mass units was accounted for by the loss of oxide from the hydroxylamine group. Moreover, the chemical shifts, coupling constant values and MS/MS fragmentation pattern due to the hydroxylated prenyl side chain in 2 is similar to that in 1, suggesting that the chemical nature of this prenyl side chain is retained in trikoramide C (Figure 4). As the quantity of the isolated trikoramide C (2) was obtained in the sub milligram range (about 0.3 mg), 13C signals of the quaternary carbons in the compound are mostly indiscernible due to low signal to noise ratio and hence not reported (Table 2).
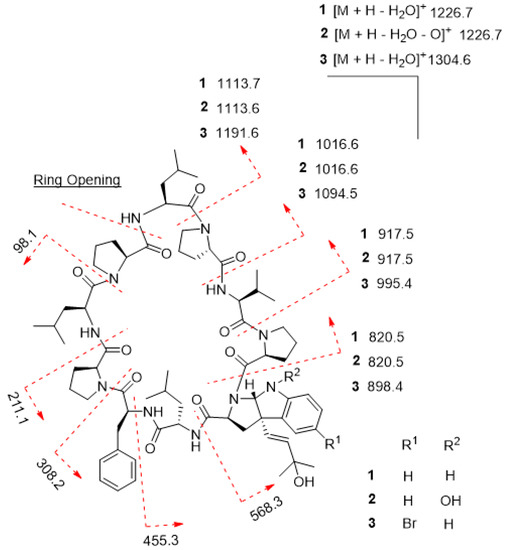
Figure 4.
MS/MS fragmentation of trikoramides B (1)–D (3).

Table 2.
NMR data of trikoramides C (2) and D (3) in CDCl3 (1H 400 MHz, 13C 100 MHz).
A high-resolution mass spectrum of trikoramide D (3) yielded a [M + NH4 ion peak at 1339.6805 by HR-ESI-OrbitrapMS for a molecular formula of C68H9679BrN11O11 with an isotopic pattern indicative of a mono brominated decapeptide (Figure S3 in Supplementary Material). The 1H and 13C NMR chemical shifts of trikoramide D (3) are similar to that of trikoramide B (1) but only differs in the aromatic chemical shifts of the indole ring of the hydroxylated Prenyl-Trp residue. A substituted aromatic residue of the tricyclic moiety was immediately evident from the absence of an aromatic methine carbon C-14 (δC 119.0) in 1 which was replaced by a more relatively shielded quaternary carbon (δC 110.2). The substituent was assigned as Br and was located at meta position on the basis of HMBC (H-13/C-14, H-13/C-15) and COSY (H-12/H-13) NMR spectra. Since the 1H NMR spectra of trikoramides B (1)–D (3) as well as A (4) showed similar chemical shifts and have similar negative cotton curves in the ECD spectra, trikoramides C and D were assumed to have the same absolute configuration as 1.
2.2. Biosynthetic Pathway of the Trikoramides
Cyanobactins are ribosomally synthesized and post-translationally modified peptides (RiPPs) encompassing post-translational modifications [11], such as heterocyclization (e.g., patellamide A) [12], oxidation, methylation (e.g., microcyclamide) [13] and prenylation (e.g., trunkamide) [5]. Cyanobactin biosynthesis begins with the precursor E-peptide, comprising of the N-terminal sequence recognized by the cleaving/modifying enzymes. Subsequently, the precursor peptide or recognition sequence is cleaved, leaving a free amine for macrocyclization and other transformation, such as cyclization and prenylation of tryptophan residue as in the case of trikoramides [14]. Prenylation happens in tandem with the formation of the new ring to form the tricyclic cyclotryptophan unit as evident from the studies done by Parajuli et al. on the biosynthetic pathway of kawaguchipeptins [15].
Three new cyanobactins, trikoramides B (1)–D (3) have been isolated from the marine cyanobacterium S. hydnoides. They are structurally related to the previously isolated trikoramide A (4) [8] with differences only in the C-prenylated cyclotryptophan residue. One could envision that trikoramides B (1)–D (3) underwent further post-translational enzymatic modifications by hydroxylase and tryptophan brominase giving rise to the unusual oxygenated isoprene, hydroxylamine and brominated indoline ring systems. Incidentally, a recent paper by Nguyen and co-workers reported the presence of broad-spectrum regiospecific peptidyl tryptophan-6-brominase on RiPP substrate. This brominase enzyme was detected through the mining for halogenating enzymes in sponge metagenomes [16]. To our knowledge, trikoramides C (2) and D (3) are the first hydroxylamine- and brominated cyclotryptophan-containing cyanobactins, respectively, from S. hydnoides.
2.3. Biological Activity of the Trikoramides
P. aeruginosa is a Gram-negative bacterial pathogen which causes acute and severe chronic infections, such as cystic fibrosis (CF) and diffuse panbronchiolitis (DPB) in patients [17]. There has been an increasing trend to further understand the mechanism of bacterial pathogenesis and intercellular microbial communication which has led to potential strategies to treat the widespread multidrug resistant bacterial diseases [18]. One such strategy is the uncovering of quorum-sensing (QS) system used by the bacteria to contemporise physiological activities of bacteria based on cell density through genetic manipulation [19]. The QS system involves synthesis of autoinducers (AI), such as N-acyl homoserine lactone, intracellularly by Gram-negative bacteria, which are actively or passively exchanged with the surrounding environment [20]. Gene expression related to the physiological activities is then triggered when the cell density is sufficient for the concentration of autoinducers to surpass the threshold [20]. Therefore, recent years has seen intensive research focusing on the interference of the QS system in pathogenic bacteria for development of novel therapeutics.
Although most of the QS inhibitory (QSI) molecules discovered to date are from marine invertebrates, such as corals and sponges, there has been reports of QS inhibitory compounds from marine cyanobacteria. These cyanobacterial QSI compounds include honaucins A–C [21], 8-epi malyngamide C [22] and lyngbyastatin 3 [23], which has motivated the investigation of QSI compounds from these microbes in our research. Several these compounds have been reported to possess significant QSI activity. For instance, extracts of a well-studied cyanobacterium Leptolyngbya crossbyana produced small molecular weight compounds, honaucins A–C, which exhibited dose-dependent QSI activity with IC50 values of 5.6, 17.6 and 14.6 µM, respectively against Vibrio harveyi BB120 [21]. 8-epi malyngamide C, isolated from the extracts of Lyngbya majuscula collected at various sites in Florida, was able to reduce 3-oxo-C12-HSL induced signaling in a LasR-based QS reporter (pSB1075) at a concentration of 10 µM [22]. QS inhibitory activities were not only limited to small molecular weight compounds but also present in larger compounds, such as peptidic lyngbyastatin 3, also isolated from L. majuscula, which inhibited the QS of bacterial reporter C. violaceum CV017 with an MIC of 12 µM [23].
The biological activity of trikoramide C (2) was not evaluated due to its minute quantity. Trikoramide B (1) exhibited moderate quorum-sensing inhibitory activity in assays based on P. aeruginosa PAO1 lasB-gfp and rhlA-gfp bioreporter strains, showing approximately 52% reduction in fluorescence when tested at an initial concentration of 100 µM. However, dose-dependent response was not observed when 1 was tested at concentrations ranging from 100 µM to 1.563 µM. Conversely, the brominated trikoramide D (3) exhibited significant QS inhibition in both PAO1 lasB-gfp and rhlA-gfp bioreporter strains in a dose-dependent manner (Figure 5) with low micromolar IC50 values of 19.6 µM and 7.3 µM, respectively (Table 3 and Figure 6). The higher potency of 3 could potentially be contributed by the brominated indole ring at the hydroxylated Prenyl-Trp residue.
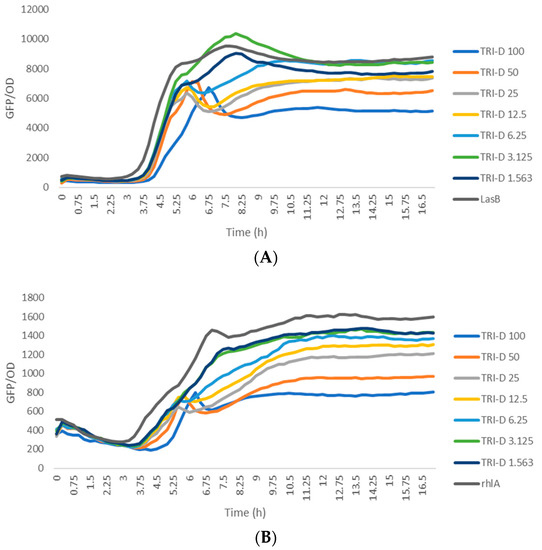
Figure 5.
Dose response curves of trikoramide D (3) when incubated with P. aeruginosa PAO1 lasB-gfp (A) and rhlA-gfp (B) bioreporter strains.

Table 3.
Biological activities of trikoramides A (4), B (1) and D (3).
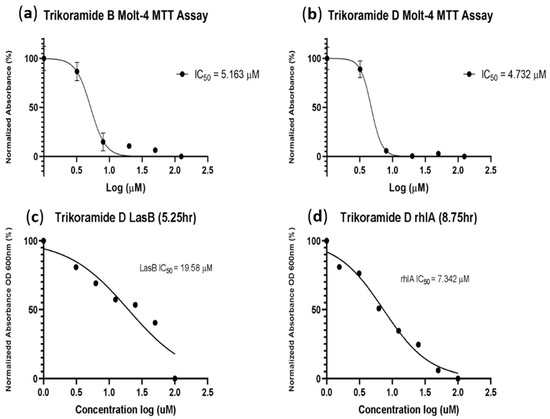
Figure 6.
(a,b) showing log concentrations (µM) of trikoramides B (1) and D (3) against normalized absorbance (%); (c,d) for the MTT assay performed on MOLT-4 human leukemia cell line and quorum-sensing inhibitory assay based on P. aeruginosa PAO1 lasB-gfp and rhlA-gfp bioreporter strains.
Trikoramide B (1) and D (3) were also investigated for their cytotoxic activity based on the MTT assay using MOLT-4 acute lymphoblastic leukemia cell line (Table 3 and Figure 6). Both compounds exhibited similar significant cytotoxic activities, with IC50 values of trikoramides B and D at 5.2 µM and 4.7 µM, respectively (Table 3 and Figure 6). In addition to the trikoramides, a recently reported C-prenylated cyclotryptophan-containing cyanobactin, motobamide, was reported from Leptolyngbya sp. with antitrypanosomal property but with weak cytotoxicity activity [10].
3. Experimental
3.1. General Experimental Procedures
Optical rotations were measured on Anton Paar Polarimeter while UV and IR spectral readings were measured on a PerkinElmer UV-Visible spectrophotometer and a PerkinElmer spectrum 100 FT-IR spectrophotometer, respectively. ECD spectra were recorded on a ChirascanTM circular dichroism spectrometer. All NMR spectra were recorded in CDCl3 on a 400 MHz Bruker NMR Spectrometer (400.13 MHz 1H, 100.61 MHz 13C) using residual solvent signals as internal references (referenced to residual CDCl3 observed at δH 7.24 or δC 77.0) with chemical shifts given in ppm downfield from TMS. Isolation and purification of the trikoramides were conducted on Shimadzu LC-8A preparative LC coupled to a Shimadzu SPD-M10A VP diode array detector HPLC. High-resolution MS data and MS/MS data were acquired on Q ExactiveTM Plus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a heated electrospray ionization (H-ESI) probe.
3.2. Sample Collection
Marine cyanobacterial samples, with cell morphology resembling that of Symploca hydnoides Kützing ex Gomont 1892 (Microcoleaceae), was collected in April 2018 by hand from the intertidal shores of Trikora beach, Bintan Island (GPS 1.1546879, 104.5783424). Samples were subsequently stored in 70% EtOH at −20 °C before workup at NIE. The cyanobacterial sample was subsequently confirmed as S. hydnoides based on phylogenetic analysis in a previous study [10]. The voucher specimen, TLT/Tri/22Apr2018/001, is deposited at Natural Sciences and Science Education, National Institute of Education, Singapore.
3.3. Extraction and Isolation of Compounds
Marine cyanobacterial samples (ca. 2.0 L, wet weight) were thawed and extracted exhaustively with 2:1 CH2Cl2/MeOH. After the solvent was evaporated in vacuo, 2.19 g of a crude organic extract was obtained. The extract was then extracted using normal phase Si gel column chromatography using stepwise gradient with increasing polarity of 100% hexanes, 9:1 hexanes/EtOAc, 4:1 hexanes/EtOAc, 3:2 hexanes/EtOAc, 2:3 hexanes/EtOAc, 1:4 hexanes/EtOAc, 100% EtOAc, 9:1 EtOAc/MeOH, 8:2 EtOAc/MeOH. Fractions 8 and 9, eluted with 9:1 EtOAc/MeOH and 8:2 EtOAc/MeOH, respectively, was subjected to solid-phase fractionation on a Sep-Pak C18 cartridge (Phenomenex, Torrance, CA, USA) using 100% MeOH to remove pigments. The resulting filtrate was further subjected to semi-preparative RP-HPLC separation (Phenomenex Luna 5μm Phenyl-Hexyl, 250 × 10 mm, 85% MeOH/H2O in 60 min at 3.0 mL/min, detected at 210 nm, 230 nm and 290 nm) to yield semi-pure trikoramides B (1)–D (3). A final purification was achieved using semi-preparative RP-HPLC (Phenomenex Kinetex 5 μm C18, 250 × 4.6 mm, 70% MeOH/H2O) to yield pure trikoramides B (1)−D (3) (1, 1.1 mg, tR = 11.4 min; 2, 0.3 mg, tR = 25.5 min; 3, 1.2 mg, tR = 45.3 min).
3.4. Compound Characterization Data
Trikoramide B (1): white amorphous solid; –108 (c 0.15, MeOH); IR (Nujol) vmax 3436, 2953, 1651, 1458, 722 cm−1; UV (MeOH) λmax (log ε) 215 (2.91), 295 (2.84), 425 (2.76) nm; 1H and 13C NMR data (CDCl3, 400.13 and 100.61 MHz, respectively), see Table 1 and Supplementary Data; HR-ESI-OrbitrapMS m/z 1244.7446 [M + H]+ (calcd for C68H98N11O11, 1244.7441, mass error, 0.401689 ppm).
Trikoramide C (2): white amorphous solid; –105 (c 0.05, MeOH); IR (Nujol) vmax 3436, 2953, 1651, 1458, 722 cm−1; UV (MeOH) λmax (log ε) 215 (2.91), 295 (2.84), 425 (2.76) nm; 1H and 13C NMR data (CDCl3, 400.13 and 100.61 MHz, respectively), see Table 2 and Supplementary Data; HR-ESI-OrbitrapMS m/z 1260.7386 [M + H]+ (calcd for C68H98N11O12, 1260.7402, mass error, −1.261957 ppm).
Trikoramide D (3): white amorphous solid; –103 (c 0.07, MeOH); IR (Nujol) vmax 3436, 2953, 1651, 1458, 722 cm−1; UV (MeOH) λmax (log ε) 215 (2.91), 295 (2.84), 425 (2.76) nm; 1H and 13C NMR data (CDCl3, 400.13 and 100.61 MHz, respectively), see Table 2 and Supplementary Data; HR-ESI-OrbitrapMS m/z 1339.6805 [M + NH4]+ (calcd for C68H96 79BrN11O11, 1339.6812, mass error, −0.522512 ppm).
3.5. Marfey’s Analysis of Amino Acid Residues in 1
Acid hydrolysis of trikoramide B (1, 200 μg) was achieved in 1 mL of 1% phenol in 6N HCl placed in a sealed reaction vial purged with N2 gas at 110 °C for 5 h. Trace HCl was then removed in vacuo and the resulting hydrolysate was redissolved in 0.1 mL of H2O. A 1% solution of L-FDAA (1-fluoro-2,4-dinitrophenyl-5-L-alaninamide) (200 μL) in acetone and 1N NaHCO3 (100 μL) was added to the aqueous hydrolysate and the mixture subsequently was heated at 80 °C for 10 min. Once the resulting mixture was cooled to rt, it was sequentially quenched with 2N HCl (100 μL), then dried under vacuum and resuspended in 1:1 H2O/CH3CN for RP-HPLC analysis. Each HPLC analysis was carried out using a Phenomenex Kinetex C18 column (250 × 4.6 mm, 2.6 μm) and an isocratic elution at 40% CH3CN–60% 0.05 M trifluoroacetic acid with 0.5 mL/min flow rate over 60 min. The retention times tRL/tRD in min of the L-DAA monoderivatized standards were Pro (10.01/10.67), Val (14.27/20.31), Leu (22.06/34.08), Phe (21.28/30.01), Trp (18.72/22.65). The derivatized hydrolysate peaks of 1 gave retention times at 9.91 min, 14.27 min, 18.85 min, 21.31 min and 22.03 min which corresponded to L-Pro, L-Val, L-Trp, L-Phe and L-Leu, respectively.
3.6. MOLT-4 Acute Lymphoblastic Leukemia Cell Line Assay
Assessment of the cytotoxicity of compounds 1 and 3 were carried out using the MTT bioassay based on the MOLT-4 (T lymphoblast; acute lymphoblastic leukemia), cancer cell line over a 3-day procedure. On the first day, 1 and 3 were prepared in a 96-well microtiter plate at 10 mM stock concentration dissolved in 100% DMSO, conducted in triplicate. The mixtures were then added with RPMI media, supplemented with fetal calf serum; and serial diluted to give concentrations of 125, 50, 20, 8 and 3.2 µM. To each of the concentration, 10 µL of the diluted compound 1 was combined with 70 µL of the cancer cells. The plate was incubated for 24 h in a 37 °C, 5% CO2 incubator. On day 2, 20 µL of MTT solution were added to each of the wells and incubated for 3 h. Another 100 µL of lysing buffer was added to each well thereafter and incubated overnight. On day 3, the microtiter plate was measured at OD570 nm, and the results were tabulated.
3.7. Quorum-Sensing Inhibitory Assay
The anti-quorum-sensing bioassay was carried out using Pseudomonas aeruginosa PAO1 lasB-gfp and rhlA-gfp reporter strains. Compounds 1 and 3 were prepared in a 96-well microtiter plate at 10 mM stock concentration dissolved in 100% DMSO, conducted in triplicate. Compounds 1 and 3 were then mixed with ABTGC medium; and serial diluted to give a concentration of 20 µM in the first dilution factor (with 0.2% of DMSO). A total of seven dilution factors, down to 0.3125 µM were done. An overnight culture of PAO1 lasB-gfp strain [24], grown in lysogeny broth at 37 °C, 200 rpm, was diluted in ABTGC medium to an optical density of 0.02 at OD600 which correspond to 2.5 × 107 CFU/mL. An equal amount of the bacterial suspension was added to reach a final test concentration of 10, 5, 2.5, 1.25, 0.625, 0.3125, and 0.1563 µM. A DMSO control, media control, and culture control were used, and the microtiter plates were incubated at 37 °C in a Tecan Infinite 200 Pro plate reader to measure the cell density (OD600) and green fluorescence protein fluorescence (excitation at 483 nm, emission at 535 nm) with 15 min intervals for up to 16 h. Similar procedure was carried out using the PAO1 rhlA-gfp biosensor strain.
4. Conclusions
Further chemical investigation on the brine shrimp toxic polar VLC fractions obtained from the CH2Cl2-MeOH (2:1) extracts of the marine cyanobacterium, S. hydnoides, from Trikora beach, Bintan, yielded three new cyanobactins trikoramides B (1)–D (3). Compounds 1–3 contained either a hydroxylamine or bromine atom at the Prenyl-Trp residue, which are considered rare post-modifications in the cyanobactin RiPPs pathway. The C-prenylated Trp unit has been observed in compounds, such as kawaguchipeptins A–B [25,26], trikoramide A [10] and motobamide A [12] which have all been isolated from cyanobacteria. Trikoramides B and D possessed significant cytotoxicity against MOLT-4 acute lymphoblastic leukemia cell line with an IC50 value of 5.2 µM and 4.7 µM, respectively. In addition, the Br-containing trikoramide D exhibited significant dose-dependent response in QS inhibitory assay based on P. aeruginosa lasB-gfp and rhlA-gfp bioreporter strains with IC50 values of 19.6 and 7.3 µM, respectively. Bromination on the prenyl-cyclotryptophan moiety of trikoramide D (3) could play a significant role in the QS inhibition in a dose-dependent manner. In fact, previous studies have suggested that the bromination of biologically active compounds increases their effectiveness by enhancing membrane permeability and decreasing metabolic degradation [27]. Due to the relatively large molecular weight of this class of cyanobactins, the addition of bromine may indeed have increased the compound permeability through the membrane and caused significant QS inhibitory activity. A recent study of mono brominated indoles by Kemp et al. sheds light on the effect of bromination on the QS inhibitory activity where the bromination led to the reduction of the IC50 by 2 to 13 folds when tested on E. coli. [28]. It would therefore be valuable to further study the effect of additional halogenation on this class of cyanobactins on enhancing the QS inhibitory activity against P. aeruginosa. In summary, this study reveals the rich bioactivities of secondary metabolites from the marine cyanobacterium, S. hydnoides.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/md19100548/s1, Figures S1–S3: HR mass spectra and MS/MS spectra of trikoramides B–D, Figures S4–S11, S12–S14, S15–S21: 1D and 2D NMR spectra of trikoramides B–D.
Author Contributions
L.T.T. supervised and visualized the experiments as well as elaborated and reviewed the manuscript. M.Y.P. isolated, purified and elucidated the structure from planar to their absolute stereochemistries and contributed to the writing of the manuscript. T.M.B.G. conducted and interpreted the results of the biological assays. J.X.G. provided HRMS data. All authors have read and agreed to the published version of the manuscript.
Funding
The facilities and equipment support were provided partially by the National Institute of Education, Singapore. This research is funded by the National Research Foundation, Prime Minister’s Office, Singapore under its Marine Science Research and Development Program (Award Nos. MSRDP-P15 and MSRDP-P34), NIE AcRF grant (RI 2/20/TLT) as well as the Nanyang Technological University Research Scholarship (Reg No. 200604393R).
Acknowledgments
The authors acknowledge Marshall Ong Ji Fa, Nursheena Parveen and Jasmine Tong Jielin for sample collection. We also acknowledge Guang Lei Ma for assistance with the ChirascanTM circular dichroism spectrometer at the School of Biological Sciences, NTU.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schopf, J.; Packer, B. Early Archean (3.3-billion to 3.5-billion-year-old) microfossils from Warrawoona Group, Australia. Science 1987, 237, 70–73. [Google Scholar] [CrossRef]
- Tidgewell, K.; Clark, B.R.; Gerwick, W.H. 2.06–The Natural Products Chemistry of Cyanobacteria. In Comprehensive Natural Products II; Liu, H.-W., Mander, L., Eds.; Elsevier: Oxford, UK, 2010; pp. 141–188. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 2005, 102, 7315–7320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaspars, M. The origins of cyanobactin chemistry and biology. Chem. Commun. 2014, 50, 10174–10176. [Google Scholar] [CrossRef]
- Carroll, A.; Coll, J.; Bourne, D.; Macleod, J.; Zabriskie, T.; Ireland, C.; Bowden, B. Patellins 1–6 and trunkamide A: Novel cyclic hexa-, hepta- and octa-peptides from colonial ascidians, Lissoclinum sp. Aust. J. Chem. 1996, 49, 659–667. [Google Scholar] [CrossRef]
- Ogino, J.; Moore, R.E.; Patterson, G.M.L.; Smith, C.D. Dendroamides, new cyclic hexapeptides from a blue-green alga. Multidrug-Resistance reversing activity of dendroamide A. J. Nat. Prod. 1996, 59, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Linington, R.G.; González, J.; Ureña, L.-D.; Romero, L.I.; Ortega-Barría, E.; Gerwick, W.H. Venturamides A and B: antimalarial constituents of the Panamanian marine cyanobacterium Oscillatoria sp. J. Nat. Prod. 2007, 70, 397–401. [Google Scholar] [CrossRef]
- Phyo, M.Y.; Ding, C.Y.G.; Goh, H.C.; Goh, J.X.; Ong, J.F.M.; Chan, S.H.; Yung, P.Y.M.; Candra, H.; Tan, L.T. Trikoramide A, a prenylated cyanobactin from the marine cyanobacterium Symploca hydnoides. J. Nat. Prod. 2019, 82, 3482–3488. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Iwasaki, A.; Kurisawa, N.; Suzuki, R.; Jeelani, G.; Matsubara, T.; Sato, T.; Nozaki, T.; Suenaga, K. Motobamide, an antitrypanosomal cyclic peptide from a Leptolyngbya sp. marine cyanobacterium. J. Nat. Prod. 2021, 84, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652. [Google Scholar] [CrossRef]
- In, Y.; Doi, M.; Inoue, M.; Ishida, T.; Hamada, Y.; Shioiri, T. Patellamide A, a cytotoxic cyclic peptide from the ascidian Lissoclinum patella. Acta Cryst. C 1994, 50, 432–434. [Google Scholar] [CrossRef]
- Ishida, K.; Nakagawa, H.; Murakami, M. Microcyclamide, a cytotoxic cyclic hexapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 2000, 63, 1315–1317. [Google Scholar] [CrossRef]
- Sivonen, K.; Leikoski, N.; Fewer, D.P.; Jokela, J. Cyanobactins—Ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 2010, 86, 1213–1225. [Google Scholar] [CrossRef] [Green Version]
- Parajuli, A.; Kwak, D.H.; Dalponte, L.; Leikoski, N.; Galica, T.; Umeobika, U.; Trembleau, L.; Bent, A.; Sivonen, K.; Wahlsten, M.; et al. A unique tryptophan C-prenyltransferase from the kawaguchipeptin biosynthetic pathway. Angew. Chem. Int. Ed. Engl. 2016, 55, 3596–3599. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.A.; Lin, Z.; Mohanty, I.; Garg, N.; Schmidt, E.W.; Agarwal, V. An obligate peptidyl brominase underlies the discovery of highly distributed biosynthetic gene clusters in marine sponge microbiomes. J. Am. Chem. Soc. 2021, 143, 10221–10231. [Google Scholar] [CrossRef]
- Paul, D.; Gopal, J.; Kumar, M.; Manikandan, M. Nature to the natural rescue: Silencing microbial chats. Chem.-Biol. Interact. 2018, 280, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Nijnik, A.; Philpott, D.J. Modulating immunity as a therapy for bacterial infections. Nat. Rev. Microbiol. 2012, 10, 243–254. [Google Scholar] [CrossRef]
- Winzer, K.; Hardie, K.R.; Williams, P. Bacterial cell-to-cell communication: Sorry, can’t talk now–gone to lunch! Curr. Opin. Microbiol. 2002, 5, 216–222. [Google Scholar] [CrossRef]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Mascuch, S.J.; Villa, F.A.; Byrum, T.; Teasdale, M.E.; Smith, J.E.; Preskitt, L.B.; Rowley, D.C.; Gerwick, L.; Gerwick, W.H. Honaucins A−C, potent inhibitors of inflammation and bacterial quorum sensing: Synthetic derivatives and structure-activity relationships. Chem. Biol. 2012, 19, 589–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, J.C.; Teplitski, M.; Gunasekera, S.P.; Paul, V.J.; Luesch, H. Isolation and biological evaluation of 8-epi-malyngamide C from the Floridian marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2010, 73, 463–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobretsov, S.; Teplitski, M.; Bayer, M.; Gunasekera, S.; Proksch, P.; Paul, V.J. Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 2011, 27, 893–905. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Ishida, K.; Matsuda, H.; Murakami, M.; Yamaguchi, K. Kawaguchipeptin A, a novel cyclic undecapeptide from cyanobacterium Microcystis aeruginosa (NIES-88). Tetrahedron 1996, 52, 9025–9030. [Google Scholar] [CrossRef]
- Ishida, K.; Matsuda, H.; Murakami, M.; Yamaguchi, K. Kawaguchipeptin B, an antibacterial cyclic undecapeptide from the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 1997, 60, 724–726. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; El-Alfy, A.T.; Ezel, K.; Radwan, M.O.; Shilabin, A.G.; Kochanowska-Karamyan, A.J.; Abd-Alla, H.I.; Otsuka, M.; Hamann, M.T. Marine inspired 2-(5-Halo-1H-indol-3-yl)-N,N-dimethylethanamines as modulators of serotonin receptors: An example illustrating the power of bromine as part of the uniquely marine chemical space. Mar. Drugs 2017, 15, 248. [Google Scholar] [CrossRef] [Green Version]
- Kemp, C.A.; McCullough, D.K.; Bialonska, D.; Johnson, P.J.T. Effect of bromination on the quorum sensing-inhibiting properties of indole-3-carboxaldehydes in Chromobacterium violaceum AHL system. Microbiol. Res. 2021, 12, 25. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).