Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108
Abstract
1. Introduction
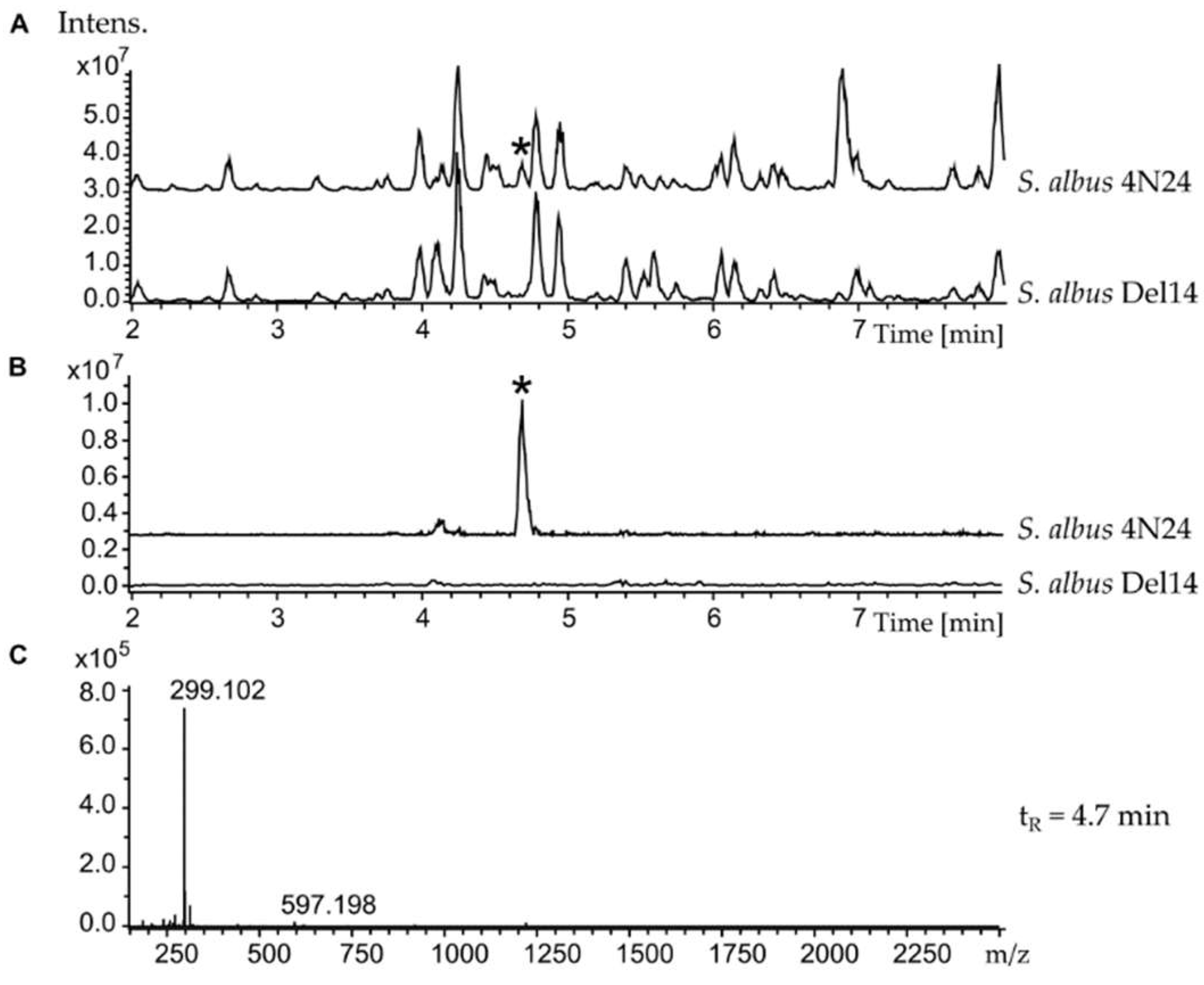
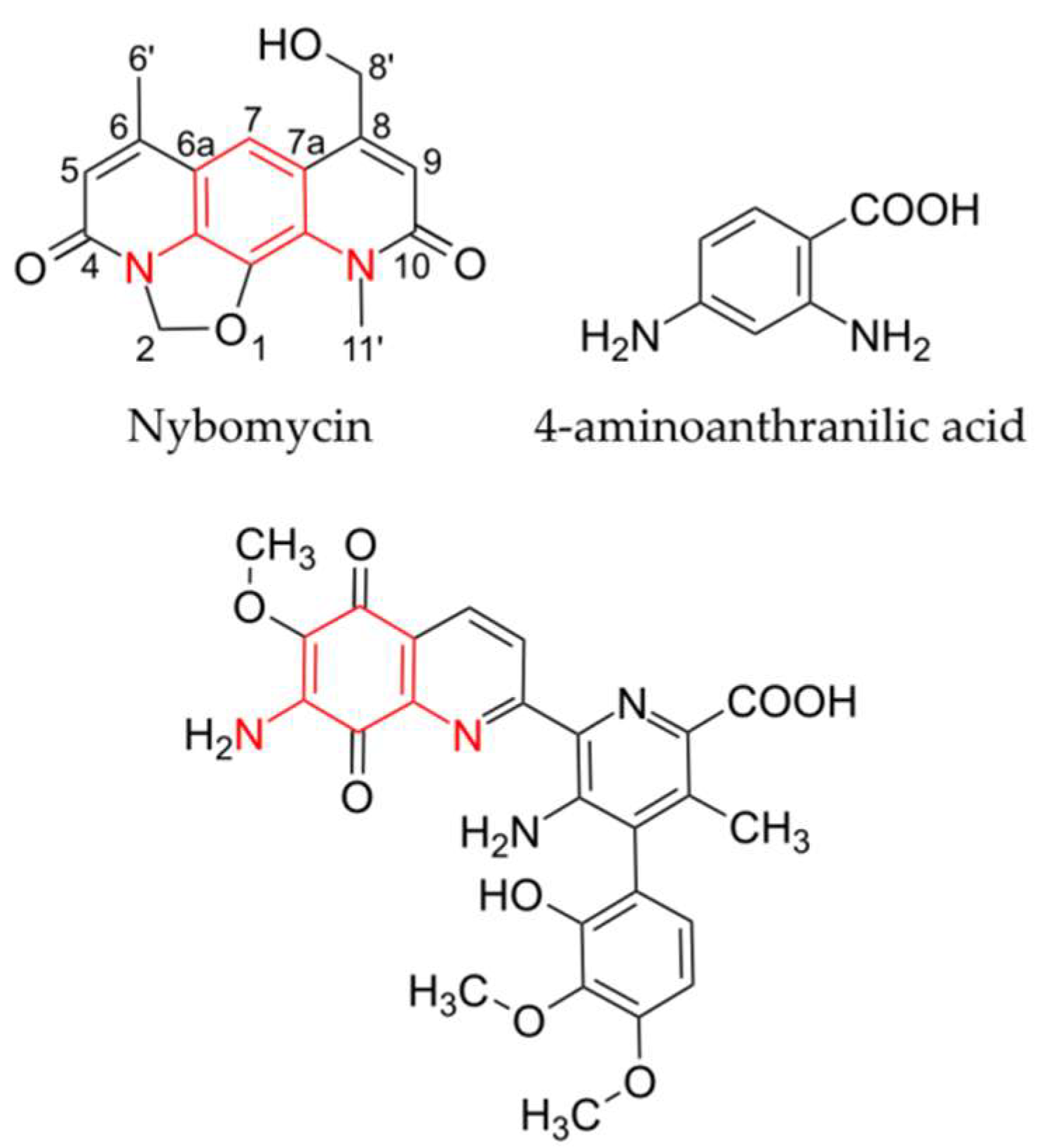
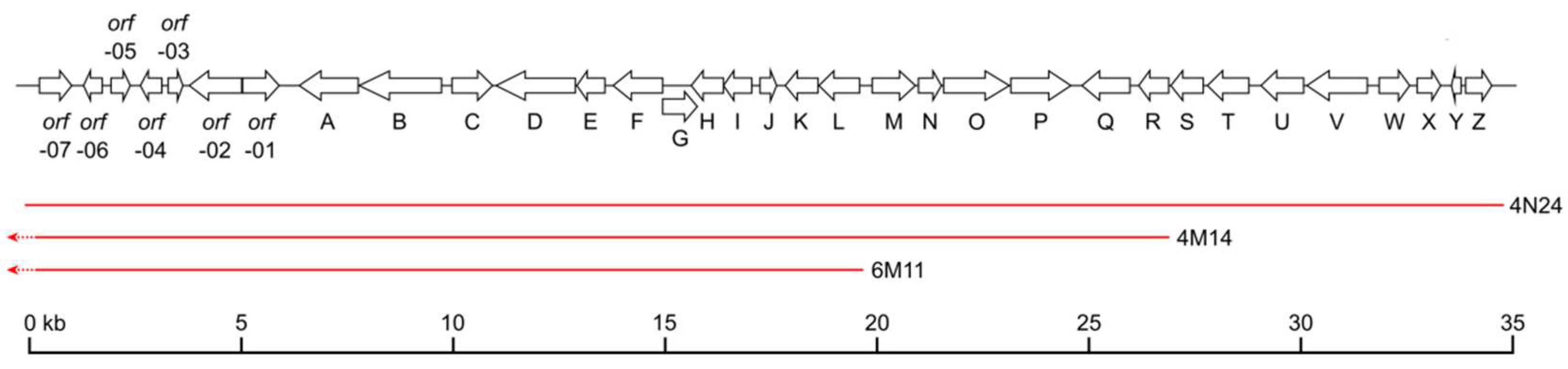
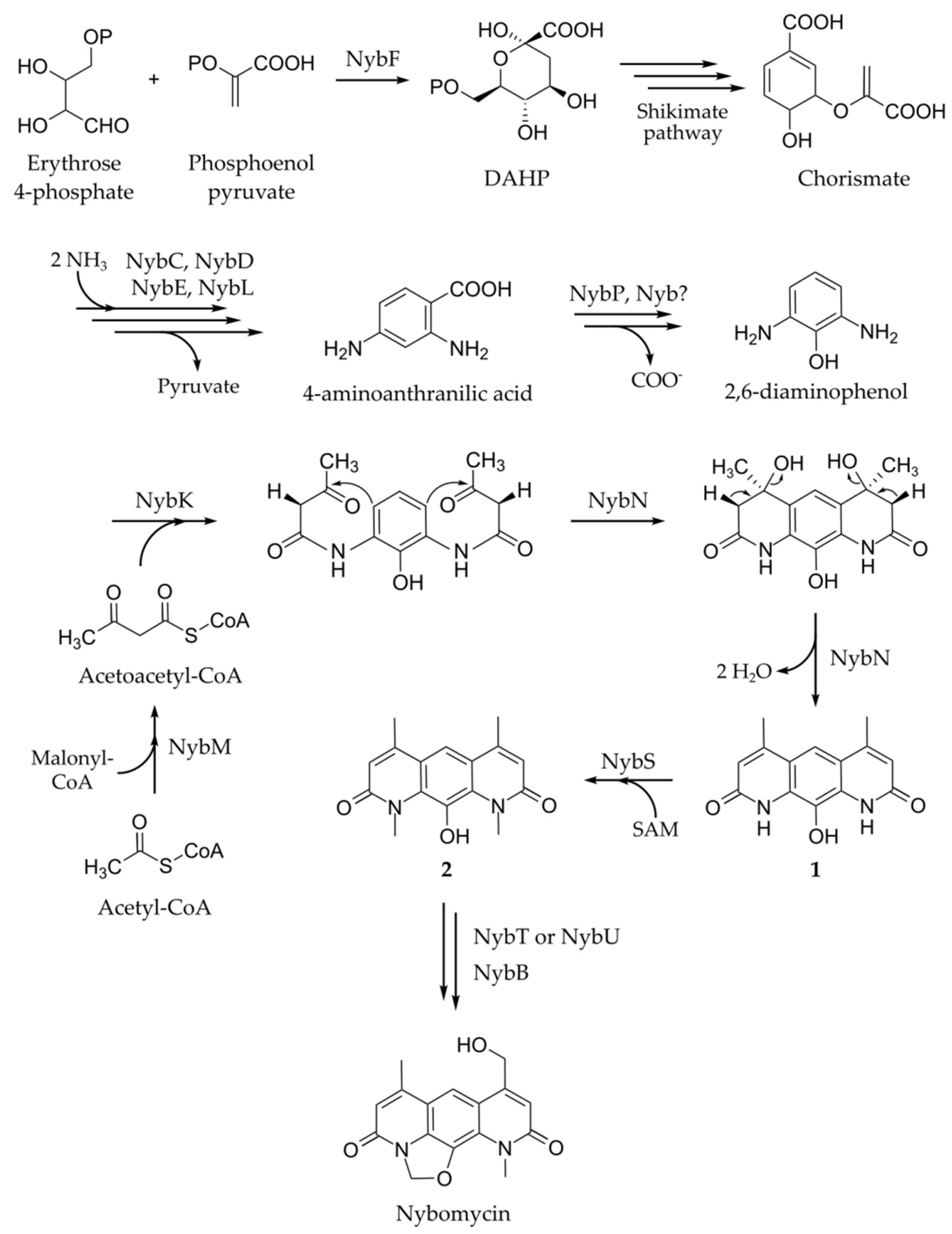
2. Results
Identification of the Nybomycin Gene Cluster through Its Heterologous Expression
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Manipulation of DNA
4.3. Metabolite Extraction and Analysis
4.4. Nybomycin Isolation and 1H-NMR Spectroscopy
4.5. Genome Mining and Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Raju, R.; Gromyko, O.; Andriy, B.; Fedorenko, V.; Luzhetskyy, A.; Muller, R. Oleamycins A and B: New antibacterial cyclic hexadepsipeptides isolated from a terrestrial Streptomyces sp. J. Antibiot. 2014, 67, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Protasov, E.S.; Axenov-Gribanov, D.V.; Rebets, Y.V.; Voytsekhovskaya, I.V.; Tokovenko, B.T.; Shatilina, Z.M.; Luzhetskyy, A.N.; Timofeyev, M.A. The diversity and antibiotic properties of actinobacteria associated with endemic deepwater amphipods of Lake Baikal. Antonie Van Leeuwenhoek 2017, 110, 1593–1611. [Google Scholar] [CrossRef] [PubMed]
- Hahn, D.R.; Graupner, P.R.; Chapin, E.; Gray, J.; Heim, D.; Gilbert, J.R.; Gerwick, B.C. Albucidin: A novel bleaching herbicide from Streptomyces albus subsp. chlorinus NRRL B-24108. J. Antibiot. 2009, 62, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, H.A.; Low-Beinart, L.; Obiajulu, J.U.; Brady, S.F. Natural product discovery through improved functional metagenomics in Streptomyces. J. Am. Chem. Soc. 2016, 138, 9341–9344. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Ruckert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef] [PubMed]
- English, A.L.; Boufridi, A.; Quinn, R.J.; Kurtboke, D.I. Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species. Synth. Syst. Biotechnol. 2017, 2, 28–38. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, N.L.; Thaker, M.; Koteva, K.; Hughes, D.W.; Wright, G.D.; Nodwell, J.R. Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. J. Antibiot. 2010, 63, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Shima, J.; Hesketh, A.; Okamoto, S.; Kawamoto, S.; Ochi, K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 1996, 178, 7276–7284. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Garcia, I.; Gonzalez, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sanchez-Hidalgo, M.; Brana, A.F.; Mendez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Igarashi, M.; Morimoto, Y.; Baba, T.; Umekita, M.; Akamatsu, Y. Curing bacteria of antibiotic resistance: Reverse antibiotics, a novel class of antibiotics in nature. Int. J. Antimicrob. Agents 2012, 39, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Rückert, C.; Albersmeier, A.; Busche, T.; Jaenicke, S.; Winkler, A.; Friethjonsson, O.H.; Hreggviethsson, G.O.; Lambert, C.; Badcock, D.; Bernaerts, K.; et al. Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 2015, 199, 21–22. [Google Scholar] [CrossRef] [PubMed]
- Strelitz, F.; Flon, H.; Asheshov, I.N. Nybomycin, a new antibiotic with antiphage and antibacterial properties. Proc. Natl. Acad. Sci. USA 1955, 41, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Nadzan, A.M.; Rinehart, K.L., Jr. Letter: Nybomycin. 8. Biosynthetic origin of the central ring carbons studied by 13C-labeled substrates. J. Am. Chem. Soc. 1976, 98, 5012–5014. [Google Scholar] [CrossRef] [PubMed]
- Knöll, W.M.J.; Huxtable, R.J.; Rinehart, K.L., Jr. Nybomycin. VI. Incorporation of acetate-13C, acetate-14C, and methionine-14C. J. Am. Chem. Soc. 1973, 95, 2703–2705. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Kong, D.; He, X.; Zhang, Z.; Han, M.; Xie, X.; Wang, P.; Cheng, H.; Tao, M.; Zhang, L.; et al. Characterization of streptonigrin biosynthesis reveals a cryptic carboxyl methylation and an unusual oxidative cleavage of a N-C bond. J. Am. Chem. Soc. 2013, 135, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Erickson, W.R. Isolation of 4-aminoanthranilic acid: A new shikimate pathway product from Streptomyces flocculus. J. Antibiot. 1988, 41, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Tamir, H.; Srinivasan, P.R. Studies of the mechanism of anthranilate synthase reaction. Proc. Natl. Acad. Sci. USA 1970, 66, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.M.; Raushel, F.M. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry 2005, 44, 6383–6391. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.L.; Clifton, I.J.; Hensgens, C.M.; Shibata, N.; Schofield, C.J.; Hajdu, J.; Baldwin, J.E. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature 1997, 387, 827–830. [Google Scholar] [CrossRef] [PubMed]
- Cremlyn, R.J. An Introduction to Organosulfur Chemistry; John Wiley and Sons: Chichester, UK, 1996. [Google Scholar]
- Arai, M.; Kamiya, K.; Pruksakorn, P.; Sumii, Y.; Kotoku, N.; Joubert, J.P.; Moodley, P.; Han, C.; Shin, D.; Kobayashi, M. Anti-dormant mycobacterial activity and target analysis of nybomycin produced by a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2015, 23, 3534–3541. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
- Kieser, T.B.M.J.; Buttner, M.J.; Charter, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Rebets, Y.; Kormanec, J.; Lutzhetskyy, A.; Bernaerts, K.; Anne, J. Cloning and expression of metagenomic DNA in Streptomyces lividans and subsequent fermentation for optimized production. In Metagenomics. Methods in Molecular Biology; Streit, W., Daniel, R., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1539. [Google Scholar]
- Buckingham, J. Dictionary of Natural Products; CRC Press/Taylor and Francis Group: London, UK, 1993. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
| Gene | Size (aa) | Proposed Function | GenBank homologue 1 | Identity/Similarity (%) | Streptonigrin Gene Cluster Homologue 2 | Identity/Similarity (%) |
|---|---|---|---|---|---|---|
| orf-07 | 268 | Streptomycin 3’’-adenylyltransferase | WP_037865927.1 | 71/77 | - | - |
| orf-06 | 159 | Hypothetical protein | WP_027736486.1 | 86/93 | - | - |
| orf-05 | 163 | ATP-binding protein | WP_052413949.1 | 76/83 | - | - |
| orf-04 | 332 | Hypothetical protein | WP_055499466.1 | 74/82 | - | - |
| orf-03 | 341 | Hypothetical protein | WP_030379123.1 | 71/80 | - | - |
| orf-02 | 494 | Hypothetical protein | WP_078869279.1 | 70/81 | - | - |
| orf-01 | 242 | Hypothetical protein | - | - | - | - |
| nybA | 475 | 3-carboxy-cis,cis-muconate cycloisomerase | WP_066029238.1 | 66/85 | stnL (AFW04563.1) | 71/76 |
| nybB | 669 | FAD-binding protein | WP_066029239.1 | 77/84 | stnK1 (AFW04562.1) | 66/75 |
| nybC | 325 | NADPH:quinone reductase | WP_066029240.1 | 81/87 | stnH1 (AFW04558.1) | 63/71 |
| nybD | 638 | Anthranilate synthase | WP_079145437.1 | 81/87 | stnM1 (AFW04564.1) | 65/75 |
| nybE | 227 | Isochorismatase | WP_066029243.1 | 84/89 | stnM2 (AFW04565.1) | 66/76 |
| nybF | 402 | DAHP synthase | WP_066029245.1 | 81/87 | stnM3 (AFW04567.1) | 62/69 |
| nybG | 279 | Hypothetical protein | - | - | - | - |
| nybH | 257 | Vicinal oxygen chelate protein | WP_066029246.1 | 64/72 | - | - |
| nybI | 222 | NAD(P)H:dehydrogenase | WP_066029248.1 | 90/95 | - | - |
| nybJ | 135 | Hypothetical protein | WP_066029250.1 | 76/86 | - | - |
| nybK | 266 | N-acetyltransferase | WP_066029251.1 | 80/88 | - | - |
| nybL | 333 | Amidohydrolase | WP_066029254.1 | 84/90 | - | - |
| nybM | 354 | Acetoacetyl-CoA synthase | WP_066029256.1 | 82/87 | - | - |
| nybN | 182 | Aromatase/cyclase | WP_066029258.1 | 78/85 | stnI (AFW04559.1) | 54/67 |
| nybO | 550 | Long-chain acyl-CoA synthetase | WP_066029260.1 | 85/90 | stnJ (AFW04560.1) | 68/81 |
| nybP | 476 | Salicylate hydroxylase | WP_079145438.0 | 74/79 | stnH2 (AFW04561.1) | 62/72 |
| nybQ | 376 | Hypothetical protein | WP_030685222.1 | 57/69 | - | - |
| nybR | 238 | NAD-dependent epimerase | WP_066029265.1 | 82/89 | - | - |
| nybS | 253 | SAM-dependent methyltransferase | WP_079145439.1 | 89/94 | - | - |
| nybT | 333 | Isopenicillin N synthase family oxygenase | WP_078974705.1 | 80/89 | - | - |
| nybU | 342 | Isopenicillin N synthase family oxygenase | WP_078974705.1 | 72/83 | - | - |
| nybV | 495 | MFS transporter | WP_079145411.1 | 81/87 | - | - |
| nybW | 243 | Transcriptional regulator | WP_079145410.1 | 82/93 | - | - |
| nybX | 197 | Transcriptional regulator | WP_025356654.1 | 89/93 | - | - |
| nybY | 87 | Hypothetical protein | WP_086560781.1 | 68/81 | - | - |
| nybZ | 219 | Transcriptional regulator | WP_057613815.1 | 79/86 | - | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez Estévez, M.; Myronovskyi, M.; Gummerlich, N.; Nadmid, S.; Luzhetskyy, A. Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108. Mar. Drugs 2018, 16, 435. https://doi.org/10.3390/md16110435
Rodríguez Estévez M, Myronovskyi M, Gummerlich N, Nadmid S, Luzhetskyy A. Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108. Marine Drugs. 2018; 16(11):435. https://doi.org/10.3390/md16110435
Chicago/Turabian StyleRodríguez Estévez, Marta, Maksym Myronovskyi, Nils Gummerlich, Suvd Nadmid, and Andriy Luzhetskyy. 2018. "Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108" Marine Drugs 16, no. 11: 435. https://doi.org/10.3390/md16110435
APA StyleRodríguez Estévez, M., Myronovskyi, M., Gummerlich, N., Nadmid, S., & Luzhetskyy, A. (2018). Heterologous Expression of the Nybomycin Gene Cluster from the Marine Strain Streptomyces albus subsp. chlorinus NRRL B-24108. Marine Drugs, 16(11), 435. https://doi.org/10.3390/md16110435





