Abstract
Bromodomains (BRD) are readers of the epigenetic code that regulate gene transcription through their recognition of acetyl-lysine modified histone tails. Recently, bromodomain-containing proteins such as BRD4 have been demonstrated to be druggable through the discovery of potent inhibitors. These protein–protein interaction inhibitors have the potential to modulate multiple diseases by their profound anti-inflammatory and antiproliferative effects. In order to explore new BRD4 inhibitors as well as lead compounds for the development of new drugs, the secondary metabolites of Alternaria sp. NH-F6, a fungus isolated from deep-sea sediment samples, were analyzed systematically. Five new compounds including two new perylenequinones (1–2), one new alternaric acid (3), 2-(N-vinylacetamide)-4-hydroxymethyl-3-ene-butyrolactone (4), one new cerebroside (5), together with 19 known compounds (6–24) were isolated from the ethyl acetate extracts of this strain. Their structures were elucidated using nuclear magnetic resonance (NMR) and high resolution electrospray ionization mass spectrometry (HR-ESI-MS) analyses. Finally, all these compounds were evaluated for their inhibitory activity against BRD4 protein, and compound 2 exhibited a potent inhibition rate of 88.1% at a concentration of 10 µM. This research provides a new BRD4 inhibitor which may possess potential antitumoral, antiviral, or anti-inflammatory pharmaceutical values.
1. Introduction
Marine-derived fungi have proved to be a major source of natural products due to their complex genetic background and chemodiversity [,]. Marine sediments represent an unexplored resource for the isolation of fungal strains []. Their various secondary metabolites usually have uncommon structures and potent biological activities that might possess substantial pharmaceutical values [].
In the field of drug discovery, epigenetic targets are attracting the attention of more and more researchers. Proteins of this kind of targets are mainly classified into readers, writers, and erasers of marks on histones, DNA, or other nuclear proteins []. These posttranslational marks regulate gene expression after making complex combinations and have been demonstrated as “histone code” []. Bromodomains represent one of the readers of these marks. A bromodomain is an approximately 110 amino acid protein domain that regulates the structure of chromatin, and thereby gene expression, through specially recognizing the acetylated-lysine state of histone tails []. Bromodomain-containing proteins have been associated with a number of diseases, including cancers, human immunodeficiency virus (HIV) infection, and neurological disorders []. Recent studies show that small molecule modulation of the acetyl-lysine binding activity of BRD proteins such as the bromodomain and extra-terminal domain (BET) family protein BRD4 dictates gene transcription outcome in disease models such as lymphoma, ischemia, and HIV-associated kidney disease, indicating BRD4 protein as an attractive drug target for pathologies including cancer and inflammation [].
In order to explore new BRD4 inhibitors and lead compounds for the development of new drugs from marine natural products, the secondary metabolites of Alternaria sp. NH-F6, a fungus isolated from deep-sea sediment samples collected from the South China Sea, were analyzed systematically. As a result, five new compounds including two new perylenequinones (1–2), one new alternaric acid (3), 2-(N-vinylacetamide)-4-hydroxymethyl-3-ene-butyrolactone (4), one new cerebroside (5), together with 19 known compounds (6–24) were isolated from the ethyl acetate extracts of the strain’s fermentation products. All the isolated compounds were evaluated for their inhibitory activity against BRD4 protein. Among them, compound 2 exhibited a potent inhibition rate of 88.1% at a concentration of 10 µM, which suggested that compound 2 might possess potential antitumoral, antiviral, or anti-inflammatory pharmaceutical values. This paper describes the isolation, identification, and inhibitory activity of compounds 1–24 against BRD4 protein and provides a new BRD4 inhibitor which may make a contribution to drug discovery.
2. Results
2.1. Identification and Structure Elucidation
Compounds 1–24 (Figure 1) were obtained by the isolation procedures as described in the materials and methods section.
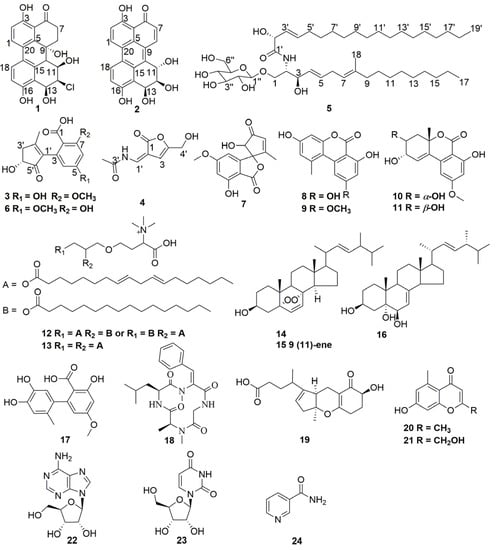
Figure 1.
Structures of compounds 1–24.
Compound 1 was obtained as a red amorphous powder, HR-ESI-MS in negative ion mode showed ion m/z 387.0641 [M − H]−. The 3:1 isotopic peak confirmed that there was a chlorine atom in the structure. From the NMR data (Table 1), the number of carbon and hydrogen atoms were easily observed. ChemDraw software was used to calculate the sum of all atoms’ atomic mass (calculation method see Table S2, Supplementary Data). The molecular formula was assigned as C20H17ClO6, with 12 degrees of unsaturation, calculated for C20H16ClO6 m/z 387.0641 [M − H]−, which was the same as the result of HR-ESI-MS.

Table 1.
NMR spectroscopic data for compounds 1 and 2.
The 1H NMR spectrum displayed two sets of ortho-coupled aromatic protons (δH 7.97, H-1, 6.98, H-2, 1H each, d, 8.8 Hz), (δH 6.80, H-17, 7.49, H-18, 1H each, d, 8.5 Hz), five exchangeable hydrogen signals (δH 12.68, 3-OH, 5.01, 9-OH, 5.72, 13-OH, 9.89, 16-OH, 1H each, s), (δH 5.12, 11-OH, 1H, d, 5.0 Hz) along with several sp3 methylene or methine groups. The 13C NMR (Table 1) and heteronuclear singular quantum correlation (HSQC) spectra showed resonances for one carbonyl carbon, four aromatic methine carbons, eight aromatic quaternary carbons, four aliphatic methine carbons, two aliphatic methylene carbons, and one aliphatic quaternary carbon, which characterized 1 as a tetrahydroperylenone []. The NMR data were very similar to those of dihydroalterperylenol in rings A and B, as well as 8β-Chloro-3, 6aα,7β,9β,10-pentahydroxy-9,8,7,6a-tetrahydroperylen-4(6aH)-one in rings D and E compared with the literature []. The p-dihydroxy diphenyl moiety of rings A and C was evidenced by the long range correlations of H-1/C-3, C-5, C-19, H-2/C-4, C-20, 3-OH/C-2, C-4, H-17/C-14, C-16, C-19, H-18/C-15, C-16, C-20 in heteronuclear multiple bond correlation (HMBC) spectrum. HMBC correlations of H-7/C-6, H-7/C-9, H-8/C-5, C-6, C-9, C-10 together with correlations of H-7/H-8 in homonuclear chemical shift correlation spectroscopy (1H-1H COSY) confirmed the structure of ring B. The 11,13-dihydroxy substitution in ring E was assigned by 1H-1H COSY correlations of H-10/H-11, H-11/11-OH, H-11/H-12, H-12/H-13, and H-13/13-OH. The remaining chloride was located only at C-12. The relative configuration of the substitutions in ring E was identified by nuclear overhauser effect spectroscopy (NOESY) and the coupling constants. NOESY spectrum showed correlations of H-11 with H-12, H-12 with H-13, and H-11 with H-13. The coupling constant of J10,11 = 9.3 Hz, suggested that H-10/H-11 was trans-oriented with typical axial-axial interaction []. The small coupling constants of J11,12 = 2.9 Hz and J12,13 = 3.0 Hz were indicative of cis orientation of H-11/H-12/H-13 []. Consequently, the structure of compound 1 was determined to be 12β-Chloro-3,9α,11β,13β,16-pentahydroxy-8,9,10,11,12,13-hexahydro-6(7H)-one (Figure 2).
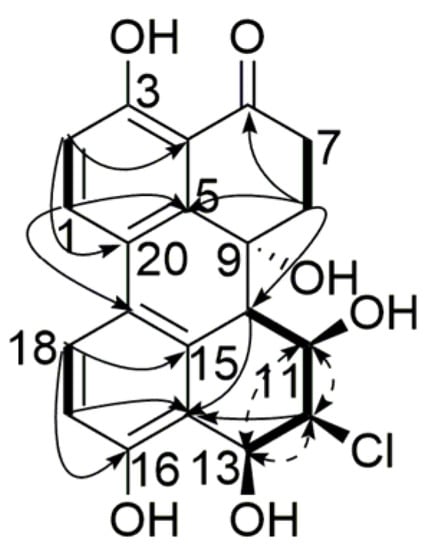
Figure 2.
Key HMBC (  ), 1H-1H COSY (
), 1H-1H COSY (  ) and NOESY (
) and NOESY (  ) correlations of compound 1.
) correlations of compound 1.
Compound 2 was obtained as a red amorphous powder, and its molecular formula was assigned as C20H14O6, with 14 degrees of unsaturation, from its HR-ESI-MS (m/z 351.0862 [M + H]+, calcd. for C20H15O6 351.0863) and NMR data (Table 1). The 1H NMR spectrum displayed three sets of ortho-coupled aromatic protons (δH 9.21, H-1, 7.47, H-2, 1H each, d, 9.2 Hz), (δH 7.04, H-7, 8.79, H-8, 1H each, d, 10.0 Hz), (δH 7.43, H-17, 8.81, H-18, 1H each, d, 9.2 Hz), four exchangeable hydrogen signals (δH 15.23, 3-OH, 10.54, 16-OH, 1H each, s), (δH 5.51, 12-OH, 1H, d, 5.4 Hz), (δH 5.72, 13-OH, 1H, d, 5.3 Hz) along with three sp3 methine groups (δH 5.33, H-11, 1H, d, 8.5 Hz), (δH 3.92, H-12, 1H, ddd, 8.5, 5.4, 3.3 Hz), (δH 5.65, H-13, 1H, dd, 5.3, 3.3 Hz). The 13C NMR (Table 1) and HSQC spectra showed resonances for 1 carbonyl carbon, 6 aromatic methine carbons, 10 aromatic quaternary carbons, and 3 aliphatic methine carbons. These signals together indicated that compound 2 had similar structure to compound 1. HMBC correlations of H-7/C-4, C-9 and H-8/C-5, C-6, C-10 suggested that the additional aromatic methine and quaternary carbons were located in ring B and ring C, respectively. 1H-1H COSY correlations of H-11/H-12, H-12/12-OH, and H-12/H-13 confirmed the 11,12,13-trihydroxy substitution in ring E. The relative configuration of the substitutions in ring E was identified by the same method described above. The absent correlation signal of H-11/H-13 in the NOESY spectrum, together with the coupling constants of J11,12 = 8.5 Hz and J12,13 = 3.3 Hz suggested that H-11/H-12 was trans-oriented while H-12/H-13 was cis-oriented. Consequently, the structure of compound 2 was determined to be 3,11α,12β,13β,16-pentahydroxy-11,12-dihydroperylen-6(13H)-one (Figure 3).
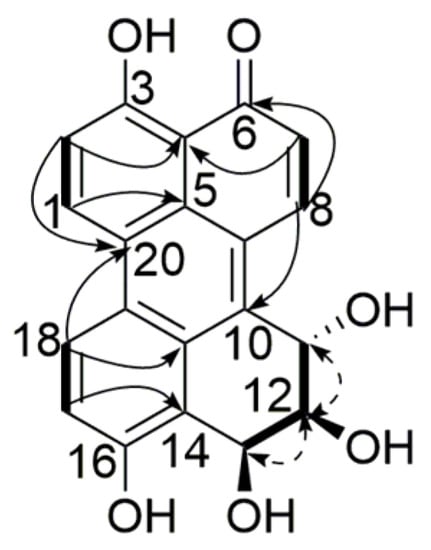
Figure 3.
Key HMBC (  ), 1H-1H COSY (
), 1H-1H COSY (  ) and NOESY (
) and NOESY (  ) correlations of compound 2.
) correlations of compound 2.
Compound 3 was obtained as a viscous, yellow oil, and its molecular formula was assigned as C14H14O6, with eight degrees of unsaturation, from its HR-ESI-MS (m/z 277.0724 [M − H]−, calcd. for C14H13O6 277.0718) and NMR data (Table 2). The 1H NMR spectrum displayed two aromatic protons (δH 6.35, H-6, 5.95, H-4, 1H each, s), one methoxyl group (δH 3.74, 7-OCH3, 3H, s), one methyl group (δH 1.90, 2′-CH3, 3H, s), one methine group (δH 4.21, H-4′, 1H, d, 6.5 Hz), and one methylene group (δH 2.90, H-3′a, 1H, dd, 17.1, 6.5 Hz), (δH 2.41, H-3′b, 1H, d, 17.1 Hz). The 13C NMR spectrum exhibited a total of 14 carbon resonances including 10 sp2 carbon atoms and 4 sp3 carbon atoms (Table 2). The data displayed similar signals to alternarienonic acid (6) reported in the literature [] except the 13C chemical shifts at C-1, C-5, and C-7. This indicated that the substituent groups at C-1, C-5, and C-7 were different from those of compound 6. 1H-1H COSY correlation of H-4/H-6 along with HMBC correlations of H-4/C-1′, C-6, C-7, H-6/C-2, C-4, C-7, 7-OCH3/C-1, C-6, C-7, 2′-CH3/C-2, C-4 indicated that the carboxyl, methoxyl, and hydroxyl groups were located at C-1, C-7, and C-5, respectively. The NMR signals at C-4′ (δH 4.21, δC 71.4) were exactly similar to those of compound 6, so the configuration of C-4′ was determined to be R. Consequently, the structure of compound 3 was assigned as shown, and this compound has been named alternarienonic acid B (Figure 4).

Table 2.
NMR spectroscopic data for compound 3.
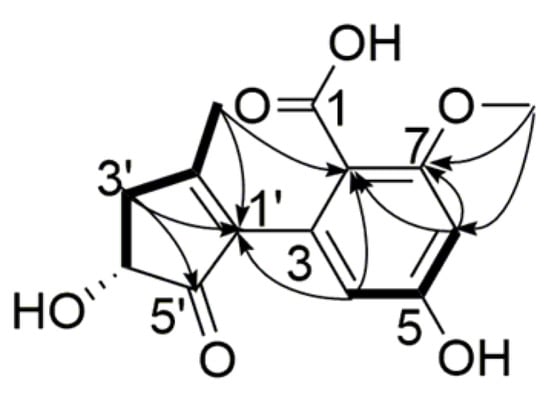
Figure 4.
Key HMBC (  ) and 1H-1H COSY (
) and 1H-1H COSY (  ) correlations of compound 3.
) correlations of compound 3.
Compound 4 was obtained as a white powder, and its molecular formula was assigned as C8H9NO4, with five degrees of unsaturation, from its HR-ESI-MS (m/z 206.0426 [M + Na]+, calcd. for C8H9NO4Na 206.0424) and NMR data (Table 3). The 1H and 13C NMR spectra displayed two sp2 methine groups (δH 9.07, H-1′, 1H, s, δC 144.3, C-1′), (δH 6.40, H-3, 1H, s, δC 109.5, C-3), one methylene group (δH 4.32, H-4′, 2H, s, δC 59.4, C-4′), one methyl group (δH 2.10, 3′-CH3, 3H, s, δC 23.3, 3′-CH3), and two exchangeable protons (δH 9.25, 2′-NH, 1H, s), (δH 5.73, 4′-OH, 1H, s), which were assigned by HSQC spectrum. The hydroxymethyl substituted 3-ene-butyrolactone moiety was evidenced by 1H-1H COSY correlation of H-3/H-4′ and HMBC correlations of H-3/C-2, C-1, C-4, C-4′, H-4′/C-4. The singlet of H-3 and H-4′ in 1H NMR spectrum confirmed that they were only located at position 3 and 4′, respectively. Correlations of 3′-CH3/C-3′, 2′-NH/C-3′, C-1′ and H-1′/C-3′, C-2, C-1 indicated that a N-vinylacetamide group was connected to the 3-ene-butyrolactone moiety. Consequently, the structure of compound 4 was determined to be 2-(N-vinylacetamide)-4-hydroxymethyl-3-ene-butyrolactone (Figure 5).

Table 3.
NMR spectroscopic data for compound 4.
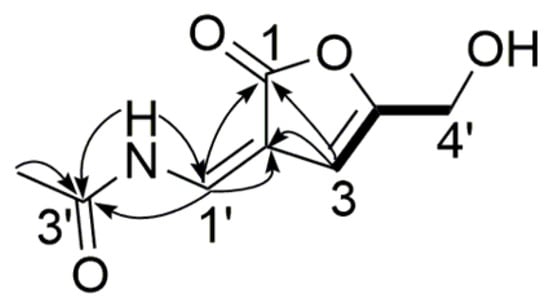
Figure 5.
Key HMBC (  ) and 1H-1H COSY (
) and 1H-1H COSY (  ) correlations of compound 4.
) correlations of compound 4.
Compound 5 was obtained as a white powder, and its molecular formula was assigned as C43H79NO9, with five degrees of unsaturation, from its HR-ESI-MS (m/z 754.5827 [M + H]+, calcd. for C43H80NO9 754.5828) and NMR data (Table 4). The 1H NMR spectrum displayed two sets of ortho-coupled protons (δH 5.45, H-4, 1H, dd, 14.8, 6.0 Hz), (δH 5.72, H-5, 1H, d, 14.8 Hz), (δH 5.49, H-3′, 1H, dd, 15.8, 4.2 Hz), (δH 5.85, H-4′, 1H, dt, 15.8, 5.8 Hz), one sp2 proton (δH 5.08, H-7, 1H, t, 6.0 Hz), three methyl groups (δH 0.89, H-17, H-19′; 3H each, t, 6.8 Hz), (δH 1.57, H-18, 3H, s), one exchangeable proton (δH 7.39, NH, 1H, s), and several oxidized sp3 methine or methylene groups. The 13C NMR spectrum showed seven sp2 carbon atoms and nine oxidized sp3 carbon atoms. The overlap signals (δH 1.26, 36H, δC 29.4–32.1) indicated the existence of long aliphatic carbon chains in the structure. The NMR data were exactly similar to those of chrysogeside D reported in the literature [], which suggested that they were analogs. The difference between the two compounds was that compound 5 had only one methylene group between C-5 and C-7. Correlations of H-5/H-4, H-5/H-6, H-6/H-7, H-7/H-18, H-9/H-10 in 1H-1H COSY spectrum, together with correlations of H-7/C-6, H-9/C-6, C-8, C-10, C-18, H-18/C-6, C-8, C-9 in HMBC spectrum confirmed that there was only one methylene group between C-5 and C-7. The methanolysis products of compound 5 were further analyzed by HR-ESI-MS in order to determine the lengths of the aliphatic carbon chains. When the fragments m/z 217.0678 [M + Na]+ (calcd. for C7H14O6Na 217.0683), and the negative mode m/z 229.0488 [M + Cl]− (calcd. for C7H14ClO6 229.0484), along with the 13C NMR data were compared with those of chrysogeside D, it indicated that compound 5 had the same methyl d-glucopyranoside moiety as chrysogeside D. The NMR signals of the anomeric proton and carbon at δH 4.37/δC 103.1 suggested the β-configuration of the glucoside []. Besides, the same C-19 fatty acid fragment m/z 335.2558 [M + Na]+ (calcd. for C19H36O3Na 335.2557) and its methyl ester m/z 349.2713 [M + Na]+ (calcd. for C20H38O3Na 349.2713) were also detected in the HR-ESI-MS spectrum. The observation of a 15.8 Hz coupling between H-3′ and H-4′ allowed assignment of the E configuration to this double bond. Furthermore, a C-18 unsaturated fatty acid fragment m/z 298.2738 [M + H]+ (calcd. for C18H36NO2 298.2741) and its methyl ester m/z 312.2896 [M + H]+ (calcd. for C19H38NO2 312.2897) were detected. The E configuration of the C-4, C-5 double bond was assigned on the basis of a large coupling (15.8 Hz) between H-4 and H-5. The 13C chemical shifts at C-1, C-2, C-3, C-1′, and C-2′ were very similar to the related cerebrosides reported in the literature [], so the absolute configuration of C-2, C-3, and C-2′ was determined to be S, R, R, respectively. Consequently, the structure of compound 5 was assigned as (2R,3E)-2-hydroxy-N-[(2S,3R,4E,7E)-1-β-d-glucopyranosyloxy-3-hydroxy-8-methylheptadec-4,7-dien-2-yl]nonadec-3-enamide (Figure 6), and this compound has been named chrysogeside F.

Table 4.
NMR spectroscopic data for compound 5.
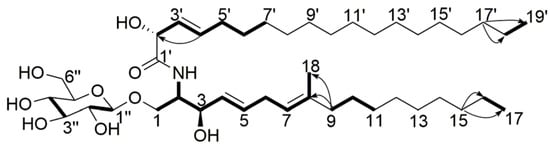
Figure 6.
Key HMBC (  ) and 1H-1H COSY (
) and 1H-1H COSY (  ) correlations of compound 5.
) correlations of compound 5.
Compound 6 was identified as alternarienonic acid by comparison with data in reference []. Compound 7 was identified as talaroflavone by comparison with data in reference []. Compound 8 was identified as alternariol by comparison with data in reference []. Compound 9 was identified as alternariol 5-O-methyl ether by comparison with data in reference []. Compound 10 was identified as 4′-epialtenuene by comparison with data in reference []. Compound 11 was identified as altenuene by comparison with data in reference []. Compound 12 was identified as 1(2)-linolyl-2(1)-palmityl-glycero-O-4′-(N,N,N-trimethyl) homoserine by comparison with data in reference []. Compound 13 was identified as 1,2-dilinolylglycero-O-4′-(N,N,N-trimethyl) homoserine by comparison with data in reference []. Compound 14 was identified as 5,8-epidioxy-5α,8α-ergosta-6, 22E-dien-3β-ol by comparison with data in reference []. Compound 15 was identified as 5,8-epidioxy-5α,8α-ergosta-6,9,22E-dien-3β-ol by comparison with data in reference []. Compound 16 was identified as (22E,24R)-24-methyl-5α-cholesta-7,22-diene-3β,5,6β-triol by comparison with data in reference []. Compound 17 was identified as altenusin by comparison with data in reference []. Compound 18 was identified as tentoxin by comparison with data in reference []. Compound 19 was identified as tricycloalternarene A by comparison with data in reference []. Compound 20 was identified as 2,5-dimethyl-7-hydroxychromone by comparison with data in reference []. Compound 21 was identified as 7-hydroxy-2-hydroxymethyl-5-methyl-4H-chromen-4-one by comparison with data in reference []. Compound 22 was identified as β-adenosine by comparison with data in reference []. Compound 23 was identified as uridine by comparison with data in reference []. Compound 24 was identified as nicotinamide by comparison with data in reference [].
2.2. Inhibitory Activity against BRD4 Protein
The inhibitory activity of compounds 1–24 against BRD4 protein was evaluated using the BRD4 bromodomain 1 (BD1) inhibitor screening assay kit and the positive control was JQ1 (Table 5). Among these compounds, compound 2 exhibited a potent inhibition rate of 88.1%. Compound 1 showed a moderate inhibition rate of 57.7%. The inhibition rates of the other compounds were all below 35.0% when tested at a concentration of 10 µM.

Table 5.
Inhibition rates of compounds 1–24 against BRD4 protein a.
3. Discussion
As epigenetic readers of the histone code, BET family proteins (including BRD2, BRD3, BRD4, and BRDT) play an important role in a number of human diseases. Among them, BRD4 is the most extensively studied member. It contains two highly conserved N-terminal bromodomains (BD1 and BD2) that recognize acetylated lysine residues, an extraterminal domain and a C-terminal domain which has little sequence homology to other BET family members []. BD1 and BD2 interact with acetylated chromatin and non-histone proteins to regulate transcription, DNA replication, cell cycle progression, and other cellular activities []. The C-terminal domain interact with the transcription elongation factor P-TEFb as well as RNA polymerase II and has been implicated in promoting gene transcription []. As literature reports, disrupting the protein-protein interactions between BRD4 and acetyl-lysine can effectively block cell proliferation in cancer [,], cytokine production in acute inflammation [], as well as alleviate HIV-1 latency []. BRD4 is thus considered as a promising therapeutic target for multiple diseases and targeting BRD4 has attracted significant interest of researchers in drug discovery. Although a number of divers BRD4 inhibitors have been reported, very few of them exhibit excellent selectivity among BET family members or sub-bromodomains []. Thus, more potent and specific BRD4 inhibitors are still urgently needed.
In the course of our search for new BRD4 inhibitors as well as lead compounds from marine-derived fungi, a strain of Alternaria sp. NH-F6 was isolated from deep-sea sediment samples collected from South China Sea. The ethyl acetate extracts of the fermentation products yielded two new perylenequinones (1–2), one new alternaric acid (3), 2-(N-vinylacetamide)-4-hydroxymethyl-3-ene-butyrolactone (4), one new cerebroside (5), together with 19 known compounds including alternarienonic acid (6), talaroflavone (7), alternariol (8), alternariol 5-O-methyl ether (9), 4′-epialtenuene (10), altenuene (11), diacylglycerotrimethyl homoserine lipids (12–13), 5,8-epidioxy-5α,8α-ergosta-6,22E-dien-3β-ol (14), 5,8-epidioxy-5α,8α-ergosta-6,9,22E-dien-3β-ol (15), (22E,24R)-24-methyl-5α-cholesta-7,22-diene-3β,5,6β-triol (16), altenusin (17), tentoxin (18), tricycloalternarene A (19), 2,5-dimethyl-7-hydroxychromone (20), 7-hydroxy-2-hydroxymethyl-5-methyl-4H-chromen-4-one (21), β-adenosine (22), uridine (23), and nicotinamide (24). These metabolites were mainly classified into quinones, glycerides, steroids, pyranones, nitrogen-containing compounds, terpenoids, and phenolics, and the structural types were almost the same as those reported in Alternaria species []. Altertoxins have been reported in the literature from several Alternaria species []. These compounds have similar structures to compounds 1 and 2 and they usually have epoxides in the structures. The epoxides in the altertoxins could be opened under mild conditions to form compounds like 1 and 2.
BRD4 inhibitor screening assay showed that compound 2 was a potent inhibitor with an inhibition rate of 88.1% at a concentration of 10 µM. Compound 1 showed a moderate inhibition rate of 57.7% when tested at the same concentration. This indicated that they might also possess significant antitumor, antiviral, or anti-inflammatory activities. The discovery of new BRD4 inhibitors may provide reference for drug design and make a contribution to the treatment of cancer or other diseases. Currently, more than a dozen of BRD4 inhibitors have progressed into human clinical trials for the treatment of cancer or other diseases []. These molecules usually have a unique head moiety that can form critical hydrogen bonds with residues of the BRD4 binding pocket. Among the hydrogen bond, they often contain a small hydrophobic group such as a methyl group []. Since compounds 1 and 2 are structurally different from these molecules, the exact mechanism of its inhibitory effect against BRD4 protein will be explored in our future research.
4. Experimental Section
4.1. General
HR-ESI-MS spectra were obtained using Agilent-6230Q-TOF mass spectrometer (Agilent Technologies China Co., Ltd., Shanghai, China). Ultra-high purity helium was used as the collision gas and high-purity nitrogen as the nebulizing gas. 1D- and 2D-NMR spectra were recorded on Bruker Ascend 600 and 400 MHz spectrometers (Bruker Technologies Beijing Co., Ltd., Beijing, China) using tetramethylsilane as the internal standard. Infrared ray (IR) spectra were recorded on Bruker Vector 22 (Bruker Technologies Beijing Co., Ltd., Beijing, China) and optical rotations were measured by JASCO P1010 (JASCO China Co., Ltd., Shanghai, China). High performance liquid chromatography (HPLC) analyses were performed on Shimadzu LC-20 AT prominence liquid chromatograph (Shimadzu China Co., Ltd., Shanghai, China). Preparative HPLC using a LC-3000 system with photodiode array detector purchased from Beijing Chuangxintongheng Science & Technology Co., Ltd. (Beijing, China), was used to purify compounds isolated from atmospheric silica gel column chromatography. A Pursuit XRs 10 C18 column (250 × 21.2 mm) was used for preparative HPLC. Preparative medium performance liquid chromatography (MPLC) was carried out on an octadecylsilyl C18 column (25 mm × 500 mm). Time-resolved fluorescence resonance energy transfer (TR-FRET) signals were measured by multifunctional microplate reader SpectraMax M5. BRD4 (BD1) inhibitor screening assay kit was purchased from Cisbio Bioassays (Codolet, France). Silica gel (100–200 mesh) was purchased from Qingdao Haiyang Chemical Group Co., Ltd., (Qingdao, China). Sephadex LH-20 was purchased from GE Healthcare (Beijing, China). Dichloromethane and methanol were purchased from Tianjin Damao Chemical Reagents Co., Ltd. (Tianjin, China). Deionized water was prepared using a Milli-Q system (Millipore, Bedford, MA, USA).
4.2. Fungus Material
Alternaria sp. NH-F6 was isolated by the first author using the standard agar plate dilution method from the marine sediments collected from Sansha City (16°83′ N, 112°48′ E), South China Sea, in October 2015. Sediments were sampled at a depth of 3927 m. The fungus was identified by observing the morphological characteristics and analysis of the 26s ribosomal deoxyribose nucleic acid (rDNA) regions (GenBank accession number: KY 378939.1). The strain was deposited in BeNa Culture Collection (BNCC 151307), Beijing, China.
4.3. Fermentation and Isolation
Alternaria sp. NH-F6 was maintained on potato dextrose agar (PDA) medium at 28 °C for seven days. Spores of the strain were inoculated into 500 mL Erlenmeyer flasks containing 250 mL potato dextrose broth (PDB) medium (200 g potatoes, 20 g glucose, 25 g baysalt, and 1000 mL distilled water) using a sterile inoculation loop and incubated at 28 °C on a rotary shaker at 180 rpm for three days. Large scale solid fermentation was carried out in 200 bottles of 500 mL Erlenmeyer flasks each containing 122 g rice medium (40 g rice, 2 g baysalt, and 80 mL distilled water). Each flask was inoculated with 15.0 mL culture medium and incubated at 28 °C for 15 days.
When the fermentation was finished, the fermentation products were extracted with equal volume of ethyl acetate twice to yield 36.0 g crude extract. This extract was subjected to silica gel column chromatography (6 × 80 cm, 250 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (1:0, 80:1, 40:1, 20:1, 5:1, 1:1, 0:1, v/v, each 2.5 L) to give 11 fractions (Fr. A–K).
Fr. B (0.95 g) was subsequently subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (150:1, 100:1, 80:1, 40:1, 0:1, v/v, each 200 mL) to give six fractions (Fr. B-1 to Fr. B-6). Fr. B-4 (33.2 mg) was dissolved in CH2Cl2 and kept standing for two days to precipitate compound 9 (0.9 mg). Fr. B-5 (347.4 mg) was subjected to silica gel column chromatography (1.5 × 40 cm, 10 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (100:1, 60:1, 30:1, 15:1, 0:1, v/v, each 100 mL) to give four fractions (Fr. B-5-1 to Fr. B-5-4). Fr. B-5-2 (52.0 mg) was fractionated by preparative HPLC using 95% CH3OH-H2O as the mobile phase (flow rate 10 mL/min, λ = 210 nm) to give compound 15 (6.3 mg, tR = 27 min) and compound 14 (19.7 mg, tR = 33 min). Fr. B-6 (103.5 mg) was subjected to silica gel column chromatography (1.5 × 40 cm, 10 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (80:1, 40:1, 20:1, 15:1, 0:1, v/v, each 100 mL) to give four fractions (Fr. B-6-1 to Fr. B-6-4). Fr. B-6-3 (66.4 mg) was purified on preparative HPLC using 45% CH3OH-H2O as the mobile phase (flow rate 10 mL/min, λ = 210 nm) to give compound 20 (4.7 mg, tR = 33 min).
Fr. D (0.6 g) was subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (80:1, 50:1, 20:1, 0:1, v/v, each 200 mL) to give three fractions (Fr. D-1 to Fr. D-3). Fr. D-2 (254.2 mg) was fractionated by preparative MPLC using a 90 min gradient from 10% CH3OH-H2O to 100% CH3OH-H2O (flow rate 10 mL/min, λ = 210 nm). Fr. D-2-1 (tR = 38–48 min) and Fr. D-2-2 (tR = 66.5–73 min) were recycled and dried at 45 °C under vacuum condition, respectively. Fr. D-2-1 (55.6 mg) was subsequently purified on preparative HPLC (28% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 7 (5.3 mg, tR = 40 min). Fr. D-2-2 (42.8 mg) was also purified on preparative HPLC (48% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 18 (15.7 mg, tR = 48 min) and compound 19 (7.3 mg, tR = 52 min).
Fr. F (1.21 g) was subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (50:1, 30:1, 10:1, 5:1, 0:1, v/v, each 200 mL) to give five fractions (Fr. F-1 to Fr. F-5). Fr. F-3 (124.1 mg) was fractionated by preparative MPLC using a 90 min gradient from 10% CH3OH-H2O to 100% CH3OH-H2O (flow rate 10 mL/min, λ = 210 nm). Fr. F-3-1 (tR = 19–25 min) and Fr. F-3-2 (tR = 55–70 min) were recycled and dried at 45 °C under vacuum condition, respectively. Fr. F-3-1 (12.0 mg) was subsequently purified on preparative HPLC (20% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 4 (4.0 mg, tR = 32 min). Fr. F-3-2 (73.2 mg) was also purified on preparative HPLC (38% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 11 (28.2 mg, tR = 74 min) and compound 10 (9.2 mg, tR = 81 min). Fr. F-4 (46.0 mg) was purified on Sephadex LH-20 column chromatography (3 × 40 cm, 50 g gel) and eluted with CH3OH to give compound 6 (9.0 mg).
Fr. G (1.25 g) was subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (60:1, 30:1, 20:1, 0:1, v/v, each 200 mL) to give five fractions (Fr. G-1 to Fr. G-5). Fr. G-4 (460.0 mg) was fractionated by preparative MPLC using a 90 min gradient from 10% CH3OH-H2O to 100% CH3OH-H2O (flow rate 10 mL/min, λ = 210 nm). Fr. G-4-1 (tR = 20–25 min), Fr. G-4-2 (tR = 30–35 min) and Fr. G-4-3 (tR = 37–45 min) were recycled and dried at 45 °C under vacuum condition, respectively. Fr. G-4-1 (4.7 mg) was purified on Sephadex LH-20 column chromatography (1.5 × 40 cm, 25 g gel) and eluted with CH3OH to give compound 3 (2.4 mg). Fr. G-4-2 (42.6 mg) was purified on Sephadex LH-20 column chromatography (3 × 40 cm, 50 g gel) and eluted with CH3OH to give compound 17 (39.3 mg). Fr. G-4-3 (11.3 mg) was purified on preparative HPLC using 30% CH3OH-H2O as the mobile phase (flow rate 10 mL/min, λ = 210 nm) to give compound 21 (2.4 mg, tR = 59 min). Fr. G-5 (10.2 mg) was also purified on preparative HPLC (55% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 8 (1.2 mg, tR = 47 min).
Fr. H (2.0 g) was fractionated by silica gel column chromatography (3 × 40 cm, 30 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (30:1, 20:1, 10:1, 1:1, v/v, each 300 mL) to give seven fractions (Fr. H-1 to Fr. H-7). Fr. H-3 (161.0 mg) was fractionated by preparative MPLC using a 90 min gradient from 10% CH3OH-H2O to 100% CH3OH-H2O (flow rate 10 mL/min, λ = 210 nm) to give compound 16 (3.7 mg, tR = 72 min). Fr. H-4 (371.4 mg) was also fractionated by preparative MPLC using a 40 min gradient from 10% CH3OH-H2O to 100% CH3OH-H2O (flow rate 10 mL/min, λ = 210 nm). Fr. H-4-1 (tR = 9-22 min) and Fr. H-4-2 (tR = 37–48 min) were recycled and dried at 45 °C under vacuum condition, respectively. Fr. H-4-1 (63.5 mg) was purified on preparative HPLC using 40% CH3OH-H2O as the mobile phase (flow rate 10 mL/min, λ = 210 nm) to give compound 1 (4.0 mg, tR = 105 min). Fr. H-4-2 (57.3 mg) was subjected to silica gel column chromatography (1 × 20 cm, 3 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (30:1, 15:1, 8:1, v/v, each 30 mL) to give compound 2 (2.0 mg). Fr. H-7 (160.1 mg) was purified on preparative HPLC (98% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 5 (17.0 mg, tR = 48 min).
Fr. I (0.23 g) was subjected to silica gel column chromatography (1.5 × 40 cm, 10 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (10:1, 5:1, 0:1, v/v, each 100 mL) to give three fractions (Fr. I-1 to Fr. I-3). Fr. I-3 (9.4 mg) was purified on Sephadex LH-20 column chromatography (1.5 × 40 cm, 25 g gel) and eluted with CH3OH to give compound 23 (6.7 mg).
Fr. J (0.64 g) was dissolved in ethyl acetate and kept standing for two days to precipitate compound 22 (9.7 mg).
Fr. K (0.64 g) was subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (15:1, 10:1, 5:1, v/v, each 200 mL) to give four fractions (Fr. K-1 to Fr. K-4). Fr. K-4 (142.6 mg) was fractionated by preparative HPLC using 98% CH3OH-H2O as the mobile phase (flow rate 10 mL/min, λ = 210 nm) to give compound 13 (10.7 mg, tR = 62 min) and compound 12 (18.6 mg, tR = 78 min).
Fr. L (1.2 g) was subjected to silica gel column chromatography (3 × 40 cm, 20 g silica gel) and eluted with a gradient of CH2Cl2–CH3OH (40:1, 25:1, 5:1, v/v, each 200 mL) to give five fractions (Fr. L-1 to Fr. L-5). Fr. L-5 (25.2 mg) was purified on preparative HPLC (20% CH3OH-H2O, flow rate 10 mL/min, λ = 210 nm) to give compound 24 (1.1 mg, tR = 15 min).
The flow rate of atmospheric silica gel and Sephadex LH-20 column chromatography described above was regulated by gravity.
4.4. Methanolysis of Compound 5
Compound 5 (2 mg) was refluxed with 20% HCl–MeOH (5 mL) for 18 h. The products were dried at 42 °C under vacuum conditions. To analyze the products, the sample was directly driven into HR-ESI-MS by an autosampler after dissolving in methanol. The HR-ESI-MS ionization source parameters were set up as follows: the capillary voltage was set up at 4.0 kV. The drying gas temperature was maintained at 325 °C. The nebulizer pressure was set up at 20 psi and the drying gas flow was 5 L/min. The injection volume was 5 μL and the flow rate was set up at 200 μL/min. The sample temperature was maintained at 35 °C.
12β-Chloro-3,9α,11β,13β,16-pentahydroxy-8,9,10,11,12,13-hexahydro-6(7H)-one (1): red, amorphous powder; +46.0 (c 0.10, MeOH); UV (MeOH) λmax 200, 260, 287 nm; IR (film) νmax 3424, 1633, 1026 cm−1; 1H NMR and 13C NMR spectroscopic data, Table 1; HR-ESI-MS [M − H]− m/z 387.0641 (calcd. for C20H16ClO6, 387.0641).
3,11α,12β,13β,16-Pentahydroxy-11,12-dihydroperylen-6(13H)-one (2): red, amorphous powder; +9.0 (c 0.10, MeOH); UV (MeOH) λmax 199, 226, 257 nm; IR (film) νmax 3442, 1625, 1030 cm−1; 1H and 13C NMR spectroscopic data, Table 1; HR-ESI-MS [M + H]+ m/z 351.0862 (calcd. for C20H15O6, 351.0863).
Alternarienonic acid B (3): viscous, yellow, oil; +70.0 (c 0.10, MeOH); UV (MeOH) λmax 223, 248, 299 nm; IR (film) νmax 3367, 1701, 1586, cm−1; 1H and 13C NMR spectroscopic data, Table 2; HR-ESI-MS [M − H]− m/z 277.0724 (calcd. for C14H13O6 277.0718).
2-(N-Vinylacetamide)-4-hydroxymethyl-3-ene-butyrolactone (4): white, powder; UV (MeOH) λmax 227, 256 nm; IR (film) νmax 3480, 3361, 1650 cm−1; 1H and 13C NMR spectroscopic data, Table 3; HR-ESI-MS [M + Na]+ m/z 206.0426 (calcd. for C8H9NO4Na 206.0424).
Chrysogeside F (5): white, powder; −4.0 (c 0.10, MeOH); UV (MeOH) λmax 207 nm; IR (film) νmax 3367, 2921, 1071, cm−1; 1H and 13C NMR spectroscopic data, Table 4; HR-ESI-MS [M + H]+ m/z 754.5827 (calcd. for C43H80NO9 754.5828).
4.5. BRD4 Inhibitor Screening Assay
The commercially available assay kit (BRD4 BD1 TR-FRET assay kit) based on time-resolved fluorescence technology was used to evaluate the inhibition rates of BRD4 protein according to the supplier’s protocols. HTRF assays were performed in white 384 Well Shallow Well Standard Height Plates (Nunc™) with a total working volume of 20 μL. Compounds were diluted, with diluent and detection buffer supplied by TR-FRET assay kit (0.8% final DMSO), from a concentration stock of 10 mM in 100% DMSO for the primary screen. All HTRF reagents were reconstituted and diluted according to the supplier’s protocols. For each assay, 5 μL of Biotin-labeled peptide ligand was added to each well at a final concentration of 20.6 nM. 5 μL of His-tagged BRD4-BD1 was then added to each well at a final concentration of 11 nM. Afterwards, 2.5 μL of compounds (final concentration 10 μM) were added to the well and incubated at room temperature for 30 min. 5 μL of Anti-His-Tb cryptate was added at a final concentration of 0.73 nM. After that, streptavidin-XL665 was added to the well at a final concentration of 2.57 nM. The mix was incubated at room temperature for 1 h and HTRF signals were measured at 620 and 665 nm. Results were analyzed with a two-wavelength signal ratio: (intensity (665 nm)/intensity (620 nm)) × 104. The inhibition rate was calculated as follows: inhibition rate (%) = 1 − [(compound signal) − (min signal)]/[(max signal) − (min signal)] × 100, where “max signal” is the signal ratio with the compound vehicle alone (DMSO), and ‘min signal’ is the signal ratio without streptavidin-XL665. Results were expressed as the mean value of triplicate determinations and the positive control was JQ1.
Supplementary Materials
The following are available online at www.mdpi.com/1660-3397/15/3/76/s1, Figures S1–S38: HR-ESI-MS, UV, 1D, and 2D NMR spectra of compounds 1–5, Table S1: 1H and 13C NMR data of compound 6 (MeOH-d4), Table S2: abbreviations used in the article.
Author Contributions
Hui Ding performed the experiments for the strain’s isolation and fermentation, compounds’ isolation, structure elucidation, biological activity evaluation and prepared the manuscript; Dashan Zhang participated part of the strain’s fermentation and compounds’ isolation works; Biao Zhou participated part of the compounds’ isolation work; Zhongjun Ma supervised the research work and revised the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2012, 29, 144–222. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.G.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2010, 27, 165–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.H.; Li, X.M.; Li, C.S.; Ji, N.Y.; Wang, B.G. Secondary metabolites from Penicillium pinophilum SD-272, a marine sediment-derived fungus. Mar. Drugs 2013, 11, 2230–2238. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Quan, C.; Hou, X.; Fan, S. Potential pharmacological resources: Natural bioactive compounds from marine-derived fungi. Mar. Drugs 2016, 14, 76. [Google Scholar] [CrossRef]
- Vidler, L.R.; Brown, N.; Knapp, S.; Hoelder, S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J. Med. Chem. 2012, 55, 7346–7359. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Bamborough, P.; Diallo, H.; Goodacre, J.D.; Gordon, L.; Lewis, A.; Seal, J.T.; Wilson, D.M.; Woodrow, M.D.; Chung, C. Fragment-based discovery of bromodomain inhibitors part 2: Optimization of phenylisoxazole sulfonamides. J. Med. Chem. 2012, 55, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Conway, S.J. Bromodomains: Are readers right for epigenetic therapy? ACS Med. Chem. Lett. 2012, 3, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Sanchez, R.; Zhou, M.M. Scaling the druggability landscape of human bromodomains, a new class of drug targets. J. Med. Chem. 2012, 55, 7342–7345. [Google Scholar] [CrossRef] [PubMed]
- Okuno, T.; Natsume, I.; Sawai, K.; Sawamura, K.; Furusaki, A.; Matsumoto, T. Structure of antifungal and phytotoxic pigments produced by Alternaria sps. Tetrahedron Lett. 1983, 24, 5653–5656. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Bai, J.; Wang, Y.; Zhang, L.; Wu, X.; Hua, H. A new perylenequinone from a halotolerant fungus, Alternaria sp. M6. Chin. J. Nat. Med. 2012, 10, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Edrada-Ebel, R.; Indriani, I.D.; Wray, V.; Müller, W.E.; Totzke, F.; Zirrgiebel, U.; Schächtele, C.; Kubbutat, M.H.; Lin, W.H.; et al. Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detectionin its host plant Polygonum senegalense. J. Nat. Prod. 2008, 71, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wang, Y.; Sun, K.; Liu, P.; Yin, X.; Zhu, W. Cerebrosides and 2-pyridone alkaloids from the halotolerant fungus Penicillium chrysogenum grown in a hypersaline medium. J. Nat. Prod. 2011, 74, 1298–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.L.; Wang, Y.; Tao, H.W.; Peng, X.P.; Liu, P.P.; Zhu, W.M. Cerebrosides of the halotolerant fungus Alternaria raphani isolated from a sea salt field. J. Nat. Prod. 2009, 72, 1695–1698. [Google Scholar] [CrossRef] [PubMed]
- Keusgen, M.; Yu, C.M.; Curtis, J.M.; Brewer, D.; Ayer, S.W. A cerebroside from the marine fungus Microsphaeropsis olivacea (Bonord.) Höhn. Biochem. Syst. Ecol. 1996, 24, 465–468. [Google Scholar] [CrossRef]
- Naganuma, M.; Nishida, M.; Kuramochi, K.; Sugawara, F.; Yoshida, H.; Mizushina, Y. 1-Deoxyrubralactone, a novel specific inhibitor of families X and Y of eukaryotic DNA polymerases from a fungal strain derived from sea algae. Bioorg. Med. Chem. 2008, 16, 2939–2944. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhang, Q.; Gao, Y.; Tang, J.; Zhang, A.; Gao, J. Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antibacterial, antioxidant, and cytotoxic activities. J. Agric. Food Chem. 2014, 62, 3584–3590. [Google Scholar] [CrossRef] [PubMed]
- He, J.W.; Chen, G.D.; Gao, H.; Yang, F.; Li, X.X.; Peng, T.; Guo, L.D.; Yao, X.S. Heptaketides with antiviral activity from three endolichenic fungal strains Nigrospora sp., Alternaria sp. and Phialophora sp. Fitoterapia 2012, 83, 1087–1091. [Google Scholar] [CrossRef] [PubMed]
- Kiet, T.T.; Schlegel, B.; Gräfe, U. New homoserine lipids from Xerocomus langbianensis. J. Basic Microbiol. 2002, 42, 133–136. [Google Scholar] [CrossRef]
- Ioannou, E.; Abdel-Razik, A.F.; Zervou, M.; Christofidis, D.; Alexi, X.; Vagias, C.; Alexis, M.N.; Roussis, V. 5α,8α-Epidioxysterols from the gorgonian Eunicella cavolini and the ascidian Trididemnum inarmatum: Isolation and evaluation of their antiproliferative activity. Steroids 2009, 74, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Piccialli, V.; Sica, D. Four new trihydroxylated sterols from the sponge Spongionella gracilis. J. Nat. Prod. 1987, 50, 915–920. [Google Scholar] [CrossRef]
- Singh, S.B.; Jayasuriya, H.; Dewey, R.; Polishook, J.D.; Dombrowski, A.W.; Zink, D.L.; Guan, Z.; Collado, J.; Platas, G.; Pelaez, F.; et al. Isolation, structure, and HIV-1-integrase inhibitory activity of structurally diverse fungal metabolites. J. Ind. Microbiol. Biotechnol. 2003, 30, 721–731. [Google Scholar] [PubMed]
- Kim, E.L.; Li, J.L.; Xiao, B.; Hong, J.; Yoo, E.S.; Yoon, W.D.; Jung, J.H. A new cyclic tetrapeptide from the jellyfish-derived fungus Phoma sp. Chem. Pharm. Bull. 2012, 60, 1590–1593. [Google Scholar] [PubMed]
- Yuan, L.; Zhao, P.; Ma, J.; Li, G.; Shen, Y. Tricycloalternarenes A–E: Five new mixed terpenoids from the endophytic fungal strain Alternaria alternata Ly83. Helv. Chim. Acta 2008, 91, 1588–1594. [Google Scholar] [CrossRef]
- Phaopongthai, J.; Wiyakrutta, S.; Meksuriyen, D.; Sriubolmas, N.; Suwanborirux, K. Azole-synergistic anti-candidal activity of altenusin, a biphenyl metabolite of the endophytic fungus Alternaria alternata isolated from Terminalia chebula Retz. J. Microbiol. 2013, 51, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Shiojima, K.; Nakajima, H.; Hamasaki, T. Structure and biological activity of plant growth regulators produced by Penicillium sp. No. 31f. Biosci. Biotechnol. Biochem. 1992, 56, 1138–1139. [Google Scholar] [CrossRef] [PubMed]
- Domondon, D.L.; He, W.; Kimpe, N.D.; Hofte, M.; Pöppe, J. β-adenosine, a bioactive compound in grass chaff stimulating mushroom production. Phytochemistry 2004, 65, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, L.; Kang, K.; Shan, C.; Wei, Y.; Zheng, C.; Guan, H. Secondary metabolites from green algae Ulva pertusa. Chem. Nat. Compd. 2010, 46, 828–830. [Google Scholar] [CrossRef]
- Teng, X.; Zhuang, Y.; Wang, Y.; Liu, P.; Xu, Z.; Zhu, W. Secondary metabolites from Penicillium sp. gxwz406 symbiotic with the gorgonian Echinogorgia flora. Chin. J. Mar. Drugs 2010, 29, 11–15. [Google Scholar]
- Zeng, L.; Zhou, M.M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002, 513, 124–128. [Google Scholar] [CrossRef]
- Wu, S.Y.; Chiang, C.M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 2007, 282, 13141–13145. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Kanno, Y.; Siegel, R.M.; Jang, M.K.; Lenardo, M.J.; Ozato, K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 2004, 13, 33–43. [Google Scholar] [CrossRef]
- Dey, A.; Nishiyama, A.; Karpova, T.; McNally, J.; Ozato, K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 2009, 20, 4899–4909. [Google Scholar] [CrossRef] [PubMed]
- Nicodeme, E.; Jeffrey, K.L.; Schaefer, U.; Beinke, S.; Dewell, S.; Chung, C.W.; Chandwani, R.; Marazzi, I.; Wilson, P.; Coste, H.; et al. Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Boehm, D.; Conrad, R.J.; Ott, M. Bromodomain proteins in HIV infection. Viruses 2013, 5, 1571–1586. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Philpott, M.; Muller, S.; Schulze, J.; Badock, V.; Eberspacher, U.; Moosmayer, D.; Bader, B.; Schmees, N.; Fernandez-Montalvan, A.; et al. Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1. J. Biol. Chem. 2014, 289, 9304–9319. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Fu, L.; Peng, Y.; Zhou, L. Metabolites from Alternaria fungi and their bioactivities. Molecules 2013, 18, 5891–5935. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, P.; Chen, H.; Wold, E.A.; Tian, B.; Brasier, A.R.; Zhou, J. Drug discovery targeting bromodomain-containing protein 4 (BRD4). J. Med. Chem. 2017, 14. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).