Abstract
Staphylococcus aureus is a serious human pathogen that employs a number of virulence factors as part of its pathogenesis. The purpose of the present study was to explore marine bacteria as a source of compounds that modulate virulence gene expression in S. aureus. During the global marine Galathea 3 expedition, a strain collection was established comprising bacteria that express antimicrobial activity against Vibrio anguillarum and/or Staphylococcus aureus. Within this collection we searched colony material, culture supernatants, and cell extracts for virulence modulating activity showing that 68 out of 83 marine bacteria (affiliated with the Vibrionaceae and Pseudoalteromonas sp.) influenced expression of S. aureus hla encoding α-hemolysin toxin and/or spa encoding Protein A. The isolate that upon initial screening showed the highest degree of interference (crude ethyl acetate extract) was a Vibrio nigripulchritudo. Extraction, purification and structural elucidation revealed a novel siderophore, designated nigribactin, which induces spa transcription. The effect of nigribactin on spa expression is likely to be independent from its siderophore activity, as another potent siderophore, enterobactin, failed to influence S. aureus virulence gene expression. This study shows that marine microorganisms produce compounds with potential use in therapeutic strategies targeting virulence rather than viability of human pathogens.
1. Introduction
The marine environment has proven to be a reservoir of microorganisms producing compounds with interesting biomedical properties [1]. Examples of such compounds include thiopeptides from a sponge-associated Bacillus cereus strain with antibacterial activity against multiple drug resistant strains of staphylococci and enterococci [2]; andrimid, a broad spectrum antibiotic produced by Vibrio coralliilyticus [3], and the antibiotic holomycin that interferes with RNA synthesis purified from Photobacterium halotolerans [3]. While new antibacterial compounds may prove efficient in treating infectious diseases, human pathogens have a profound ability to acquire resistance resulting in serious health care problems. These include methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus as well as extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae [4,5]. To address the therapeutic failures associated with antibiotic resistance, other strategies, including anti-virulence therapies, are being considered. Antivirulence agents inhibit the production or activity of disease-causing factors of the infecting organism, and thereby disarm the pathogen of its virulence traits [6]. As quorum-sensing (QS) signalling systems are central regulators of virulence gene expression in many pathogens while being absent in humans, they represent highly promising targets for the development of anti-virulence therapeutics, possibly in combination with traditional antibiotics [7,8,9]. Several quorum sensing inhibitors (QSIs) targeting QS systems in Gram-negative pathogens have been identified, including ajoene from garlic that reduces the infective ability of Pseudomonas aeruginosa in a pulmonary infectious mouse model [10].
In Gram-positive bacteria, QS is commonly mediated by auto-inducing cyclic peptides. One example is the agr quorum sensing system in Staphylococcus aureus [11]. S. aureus causes a variety of infections ranging from mild skin infections to life-threatening bacteremia and endocarditis, with many strains being resistant to a number of antibiotics [12]. Pathogenesis of S. aureus is attributed to a multitude of virulence factors, of which a major part is controlled by agr [11]. The agr QS system is composed by an external signal, an autoinducing cyclic peptide that upon binding to the membrane-bound sensor histidine kinase (AgrC) activates the response regulator AgrA and induces virulence gene expression via a regulatory RNA, RNAIII [13,14]. Activation of agr results in expression of extracellular virulence factors including the key toxin, α-hemolysin, while cell surface-associated virulence factors, such as Protein A, are repressed [14]. Previously, we identified a putative QSI compound produced by the marine bacterium Photobacterium halotolerans that dramatically reduces hla and RNAIII expression while increasing spa production [15]. To address how abundant such compounds are in the marine environment we have screened a collection of marine bacteria for compounds that modulate S. aureus virulence gene expression.
2. Results and Discussion
2.1. Modulation of S. aureus Virulence Gene Expression by Marine Bacteria
As part of the global marine Galathea 3 expedition, a collection of bacterial strains belonging to the genera Vibrio, Ruegeria, and Pseudoalteromonas was established based on antibacterial activity against Vibrio anguillarum and/or Staphylococcus aureus [16]. Using a reporter fusion assay [17] we screened culture extracts, supernatants, and colony material of 83 strains from this collection for the ability to inhibit expression of S. aureus hla (α-hemolysin) as well as interference with the S. aureus agr quorum sensing system reported as decreased hla and increased spa expression (Figure 1, Table 1).
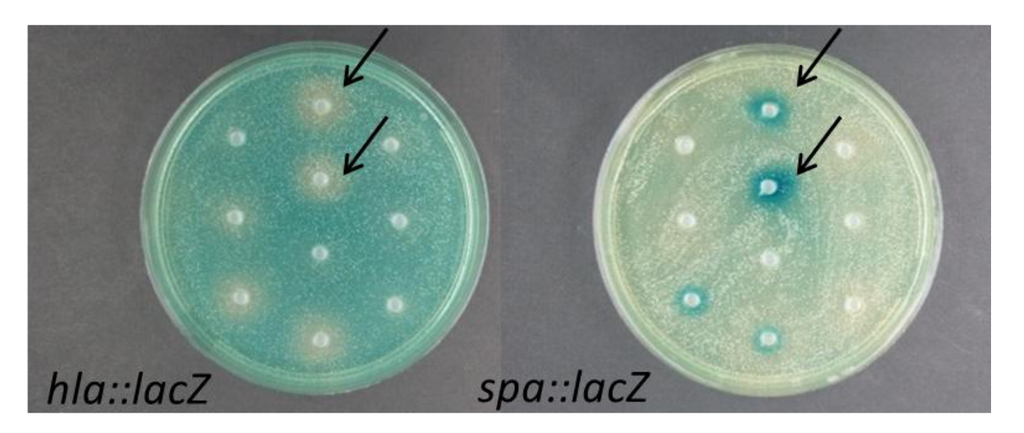
Figure 1.
Screening of crude EtOAc extracts of Vibrio nigripulchritudo isolates S2600 (top arrow) and S2604 (bottom arrow) obtained from the Solomon Sea [16] in the S. aureus hla::lacZ and spa::lacZ S. aureus reporter fusion assays.
Colony material from almost all tested Vibrionaceae strains reduced hla expression, while ethylacetate (EtOAc) extracts of 17 strains and only a single culture supernatant showed this activity (Table 1). Nine extracts and colony material of 8 strains showed both repression of hla and induction of spa transcription.

Table 1.
Screening of marine bacterial material, extracts and culture supernatants for interference with S. aureus virulence gene expression. The number of isolates displaying down-regulation of hla and combined down-regulation of hla with up-regulation of spa are listed in the table.
| Genus/family | Number of strains tested | hla interference | hla/spa interference | ||||
|---|---|---|---|---|---|---|---|
| Colony material | Extract | Supernatant | Colony material | Extract | Supernatant | ||
| Pseudoalteromonas | 41 | 37 | 15 | 0 | 19 | 0 | 0 |
| Vibrionaceae | 37 | 30 | 15 | 1 | 8 | 9 | 0 |
| Ruegeria | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
Colony material of 37 out of 41 Pseudoalteromonas strains reduced hla expression, and this activity was retained in 15 of the EtOAc extracts. When agr interference was monitored as the combination of hla repression and spa induction, none of the EtOAc extracts proved positive whereas colony material from 19 strains did. The active species covered P. phenolica, P. rubra, P. ruthenica, and P. luteoviolacea. Comparison with previous work on antibiotic activity of the strains tested here [16] showed that growth inhibition is often independent from modulation of virulence gene expression (see supplementary data). None of the five tested Ruegeria strains affected virulence gene expression.
The present study adds to recent work of marine bacteria as sources of QS inhibitors and modulators of virulence gene expression. A marine Bacillus species was found to interfere with QS-controlled virulence factor production and biofilm formation in Pseudomonas aeruginosa PAO1 and violacein pigment production in Chromobacterium violaceum [18]. Ability to interfere with QS in P. aeruginosa was also seen in marine microorganisms isolated around the Great Barrier Reef. Of 284 tested extracts, 64 (23%) were active in a general, LuxR-derived QS screen, and of these 36 (56%) were also active in a specific P. aeruginosa QS screen [9]. Thus, marine bacteria seem to be common producers of compounds targeting virulence gene expression in both Gram-positive and Gram-negative bacteria possibly through modulation of QS systems.
2.2. Interference of Virulence Gene Expression by Vibrio nigripulchritudo
One strain, Vibrio nigripulchritudo S2604, displayed particularly prominent reduction of hla expression while increasing the expression of spa (Figure 1). The activity was expressed both under shaken and stagnant growth conditions. Under stagnant conditions the activity was enhanced when substituting glucose with melibiose (data not shown). In addition to V. nigripulchritudo S2604, we examined the five remaining isolates of V. nigripulchritudo grown in the presence of melibiose without aeration to determine if the ability to modulate S. aureus virulence gene expression was unique to strain S2604 or a general property of V. nigripulchritudo (Figure 2). To address whether V. nigripulchritudo strains directly influence agr quorum sensing the extracts were screened for effect on expression of the regulatory RNAIII molecule, one of the key effector molecules of the agr quorum sensing system [19]. Here, we observed that extracts of some V. nigripulchritudo strains reduced RNAIII expression whereas for other extracts the RNAIII expression was only marginally affected (Figure 2), indicating that the different V. nigripulchritudo strains produce a variety of QS-modulating compounds.
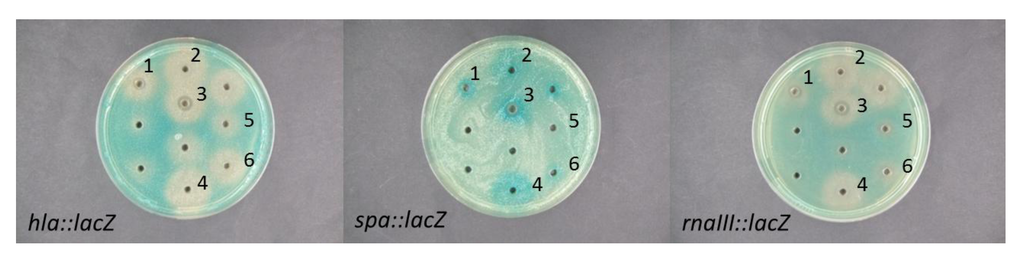
Figure 2.
Effect of EtOAc extracts of Vibrio nigripulchritudo isolates (1: S2604, 2: S2600, 3: S2601, 4: S2603, 5: S1072, 6: S2156) grown stagnant with 0.4% melibiose on expression of hla, spa and rnaIII S. aureus reporter fusions. Clearing zones around the wells represents restricted growth and antimicrobial activity of the tested compound.
2.3. Nigribactin, a Novel Siderophore from Vibrio nigripulchritudo S2604, Enhances Spa Transcription
Dereplication and fractionation by explorative solid-phase extraction (E-SPE) [20] of EtOAc extract obtained from V. nigripulchritudo S2604 indicated the presence of a novel, uncharged, apolar compound. Fractionation of a large scale S2604 extract followed by NMR revealed that a novel compound, designated nigribactin, is responsible for the spa enhancing activity (Figure 3). Surprisingly nigribactin did not modulate expression of hla and RNAIII, indicating that several compounds in the original extract influence virulence gene expression (Figure 3).
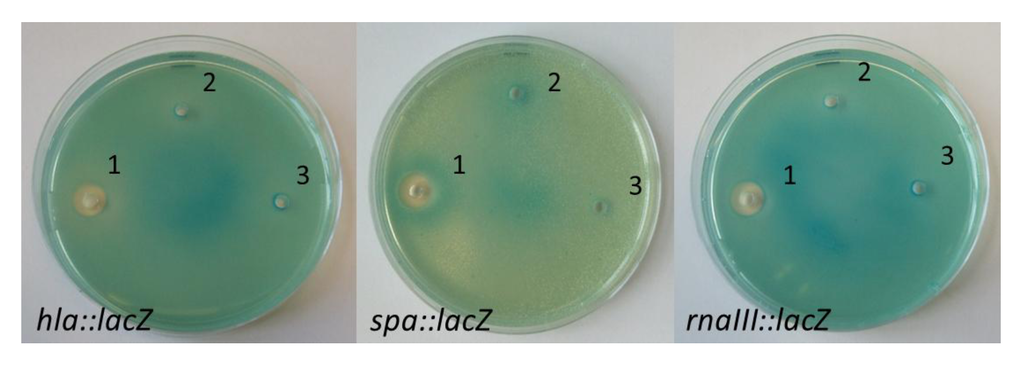
Figure 3.
Screening of purified nigribactin for its effect on S. aureus hla, spa and RNAIII expression. 1: 2 mg·mL−1 nigribactin dissolved in DMSO; 2: DMSO; 3: sterile water.
At high concentrations nigribactin inhibits growth of S. aureus as observed by lack of growth closest to the well (Figure 3) with a minimal inhibitory concentration during growth in liquid medium of >10 μg·mL−1 (data not shown). However, in the plate assay (Figure 3) the spa-inducing activity was observed further from the well where only sub-lethal concentrations of nigribactin are present. The ability of nigribactin to enhance spa transcription was confirmed by Northern blot analysis showing a substantial increase in spa transcription in the exponential growth phase (Figure 4).
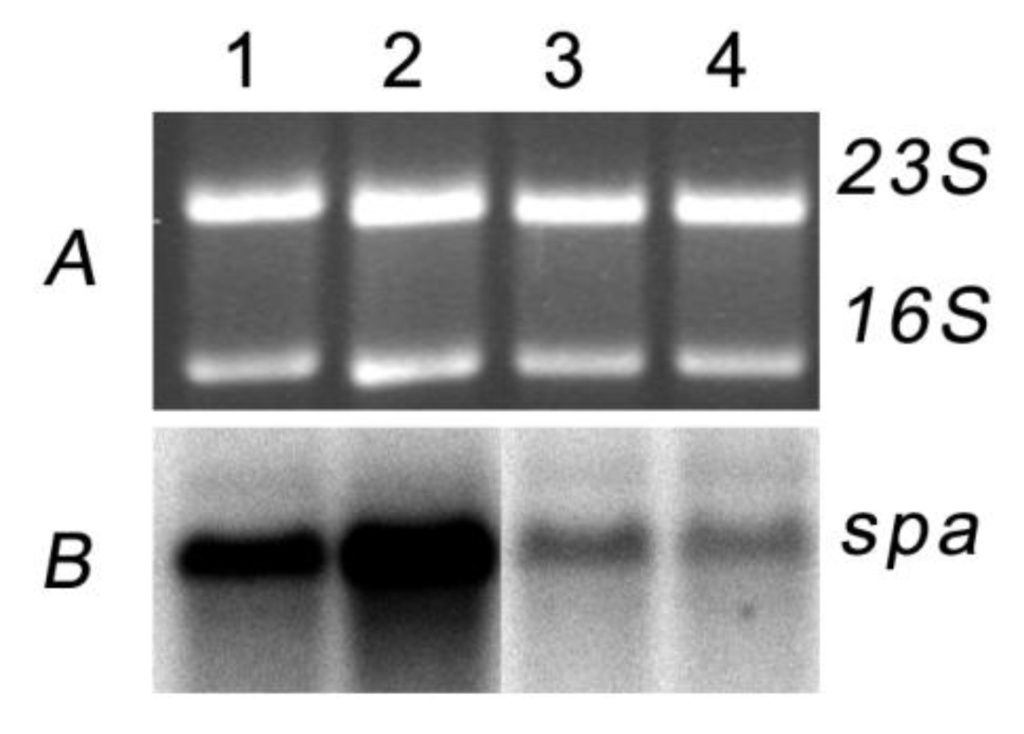
Figure 4.
Transcription of spa is induced in the presence of nigribactin at low optical density but not in transition to stationary phase. Equal amounts of RNA from S. aureus NCTC8325-4 sampled 30 min (1, 2) and 90 min (3, 4) after addition of DMSO (1, 3) or 5µg·mL−1 nigribactin (2, 4) both added at OD600= 0.4 reacted with a spa specific probe.
Nigribactin (C30H32N4O9, calc monoisotopic mass 592.2169 Da) showed to be a catechol hydroxyphenyloxalone with a norspermidine backbone, giving it high structural similarity to siderophores from Vibrio such as vibriobactin and fluvibactin [21]. Siderophores are low molecular weight iron chelators typically produced in response to low-iron stress [22]. The structure of nigribactin was established by comparison of 1D and 2D NMR data recorded in DMSO-d6 (13C data given in Section 3.3) to data for fluvibactin from Vibrio fluvialis [21]. Analysis of the NMR data revealed that the nigribactin structure differs from that of fluvibactin only by containing one less methyl group in the 5-membered oxazoline ring and one less hydroxyl group (Figure 5).
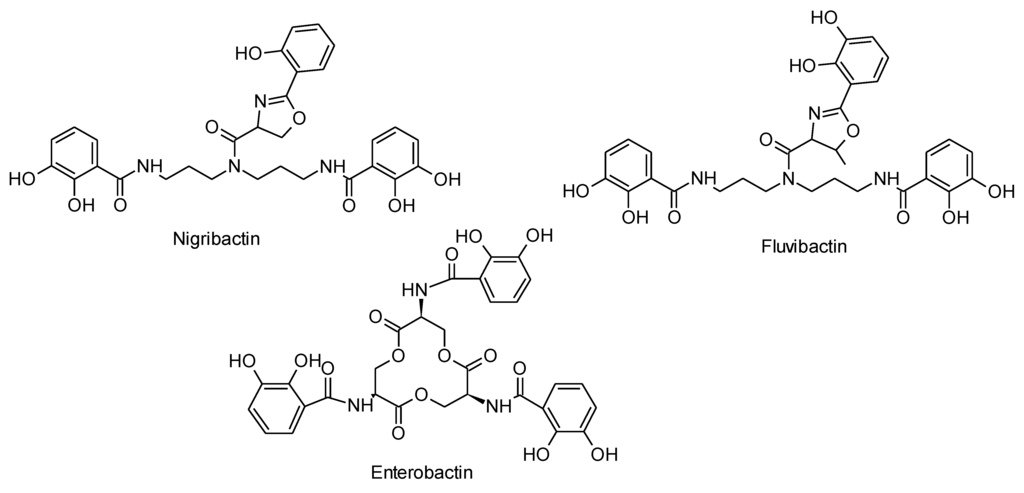
Figure 5.
Structures of the bacterial siderophores nigribactin (this study), fluvibactin [21] and enterobactin [23].
The structural similarity of nigribactin to known siderophores prompted us to address if nigribactin is a siderophore. We confirmed prominent iron-chelating activity of nigribactin by examining dilutions of purified nigribactin using the colometric CAS assay [24] (Figure 6A). However, the siderophore activity of nigribactin appears not to be responsible for the effect on spa expression as neither another catechol siderophore, enterobactin [25] (Figure 5), nor 2,2-dipyridyl, an iron chelating compound, induced spa transcription (Figure 6B).
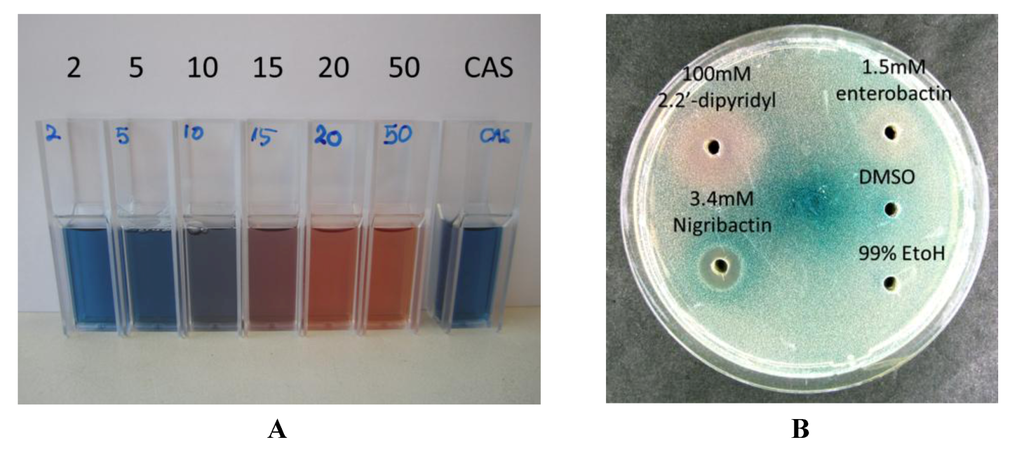
Figure 6.
(A) Confirmation of siderophore activity of nigribactin by addition at 2, 5, 10, 15, 20, and 50 µM to vials carrying Chrome azurol S (CAS);(B) Test of enterobactin (dissolved in DMSO) and 2,2′-dipyridyl (dissolved in 99% EtOH) against the S. aureus 8325-4 spa::lacZ reporter fusion [26] incorporated into an agar plate.
3. Experimental Section
3.1. Bacterial Strains
Of 512 marine bacterial strains isolated during the global Galathea 3 expedition [16], 83 strains were selected for the present study. The screening assay applied in this study is described by [17] using S. aureus strains carrying different gene reporter fusions, including S. aureus 8325-4 hla::lacZ [26], S. aureus 8325-4 spa::lacZ [26] and S. aureus 8325-4 rnaIII::lacZ [27,28]. S. aureus strain 8325-4 [29] was used for Northern blot analyses.
3.2. Bacterial Growth Conditions, Culture Extraction and Virulence Gene Expression Assay
Marine bacteria were grown in 30 mL sea salt solution (SSS; Sigma S9883; 40 g·L−1) with 0.4% glucose and 0.3% casamino acids for three days at 25 °C with (200 rpm) and without (0 rpm) aeration. Culture supernatants were prepared by sterile filtration. Cultures were extracted with an equal volume of EtOAc, transferring the organic phase to a new vial, and evaporating under nitrogen gas until dryness. Fractionation by explorative solid-phase extraction (E-SPE) was performed according to [20]. Dry extracts and fractions were redissolved in 300 µL 80% EtOH for biological testing as described in [17]. For screening of colony material, marine bacteria were grown on Marine Agar 2216 (Difco 212185) for 24 h at 25 °C, and a lump of colony material was placed on top of agar plates containing S. aureus [17] but without wells in the plates, and incubated for 48 h at 30 °C. By using a combination of reporter strains looking for both up- and -down regulation, we were able to detect and exclude strains being natural producers of β-galactosidase.
3.3. Northern Blot Analysis
S. aureus 8325-4 was grown in TSB at 37 °C at 200 rpm. Nigribactin was added at OD600 = 0.4 and samples for RNA extraction were taken after 30 and 90 min. Northern blot analysis using a probe targeting spa was performed as described previously [30]. Probes were made using the primers spa forward (5′-GGG GGT GTA GGT ATT GCA TCT G-3′) and spa reverse (5′-GGG GCT CCT GAA GGA TCG TC-3′).
3.4. Purification and Structural Elucidation of Nigribactin
Strain S2604 was grown in 2 L sea salt solution (Sigma S9883; 40 g·L−1) with 0.4% melibiose and 0.3% casamino acids for three days (0 rpm) at 25 °C. On day 3, the culture was extracted with 750 mL EtOAc for 24 h. The organic extract was dry loaded onto 10 g Sepra ZT C18 (Phenomenex, Torrance, CA) and dried before packing into a 60 g SNAP column (Biotage, Uppsala, Sweden) with 50 g pure resin in the base. Using an Isolera flash purification system (Biotage) the extract was subjected to a crude fractionation using an acetonitrile (MeCN)/H2O gradient (flow rate 40 mL·min−1) starting with 10% MeCN (2 column volumes (CV), isocratic), increasing to 100% MeCN (10 CV) before washing with 100% MeCN (2 CV). Fractions were automatically collected using UV detection (210 and 320 nm). The fractions inducing spa activity (120 mg) were pooled, evaporated, and redissolved in 1.2 mL EtOAc/methanol (MeOH; 1:3 v/v) for diol separation (Isolute diol, Biotage) on the Isolera system. A total of nine fractions (fraction size 12 mL) were collected from the diol column (10 g SNAP column) ranging from heptane, dichloromethane (DCM), EtOAc to pure MeOH, running under gravity. The fractions (28 mg total) with spa activity (25% DCM in heptane to 100% MeOH) were pooled and purified on a Luna II C18 column (250 × 10 mm, 5 μm) (Phenomenex) using a 45%–70% MeCN/H2O gradient (buffered with 20 mM formic acid, flow rate 4 mL·min−1) over 20 min on a Gilson 322 liquid chromatograph with a 215 liquid handler/injector (BioLab, Risskov, Denmark). All fractions were analysed by LC-UV-MS according to standard procedures [20] before pooling. This yielded 1.6 mg of nigribactin.
NMR spectra were recorded on a Varian Unity Inova 500 MHz spectrometer equipped with a 5 mm probe using standard pulse sequences. 13C data was confirmed on a Bruker Avance 800 MHz spectrometer at the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules. The NMR data used for the structural assignment of nigribactin (Figure 7) were acquired in DMSO-d6 (Table 2).
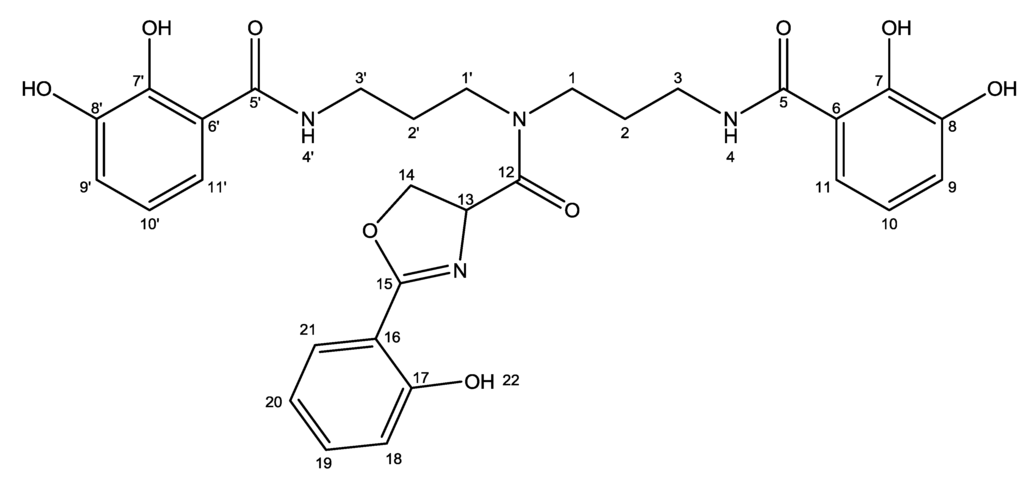
Figure 7.
Structure of nigribactin with numbered atoms as assigned in Table 2.

Table 2.
NMR spectroscopic data (DMSO-d6) of nigribactin.
| Atom | δC (ppm) | δH (ppm) (multiplicity, J (Hz)) | HMBC |
|---|---|---|---|
| 1a | 43.2 | 3.38 (1H, m) | 1′, 2, 3, 12 |
| 1b | 43.2 | 3.36 (1H, m) | 1′, 2, 3, 12 |
| 2 | 26.8 | 1.77 (2H, m) | - |
| 3 | 36.3 | 3.26 (2H, m) | 1, 2, 5 |
| 4 | - | 8.74 | 5 |
| 5 | 169.4 | - | - |
| 6 | 114.8 | - | - |
| 7 | 149.4 | - | - |
| 8 | 145.9 | - | - |
| 9 | 118.6 | 6.88 (1H, d, 7.6) | 7, 8, 11 |
| 10 | 117.7 | 6.65 (1H, t, 7.6) | 6, 8 |
| 11 | 116.9 | 7.22 (1H, d, 8.0) | 5, 7, 9 |
| 1′a | 44.6 | 3.68 (1H, m) | 2′, 12 |
| 1′b | 44.6 | 3.56 (1H, m) | 2′, 12 |
| 2′ | 28.2 | 1.95 (2H, p, 7.2) | 1′, 3′ |
| 3′ | 36.3 | 3.38 (2H, m) | 1′, 2′, 5′ |
| 4′ | - | 8.83 | 5′ |
| 5′ | 169.6 | - | - |
| 6′ | 114.8 | - | - |
| 7′ | 149.4 | - | - |
| 8′ | 145.9 | - | - |
| 9′ | 118.6 | 6.88 (1H, d, 7.6) | 7′, 8′, 11′ |
| 10′ | 117.7 | 6.65 (1H, t, 7.6) | 6′, 8′ |
| 11′ | 116.9 | 7.26 (1H, d, 8.0) | 5′, 7′, 9′ |
| 12 | 168.3 | - | - |
| 13 | 64.2 | 5.36 (dd, 9.5, 6.7) | 12, 15 |
| 14a | 69.2 | 4.77 (1H, t, 7.5) | 12, 13, 15 |
| 14b | 69.2 | 4.54 (1H, t, 8.9) | 12, 15 |
| 15 | 165.3 | - | - |
| 16 | 109.6 | - | - |
| 17 | 158.6 | - | - |
| 18 | 116.4 | 6.97 (1H, d, 8.3) | 16, 17, 20 |
| 19 | 133.9 | 7.44 (1H, t, 7.9) | 17, 21 |
| 20 | 118.9 | 6.93 (1H, t, 7.6) | 16, 18 |
| 21 | 127.9 | 7.62 (1H, dd, 7.8, 1.1) | 17, 19 |
| 22 (–OH) | - | 11.7 | 16, 17, 18 |
| –OH | - | 12.7 | - |
| –OH | - | 12.6 | - |
| –OH | - | 9.12 | - |
| –OH | - | 9.09 | - |
4. Conclusions
This study shows that a substantial number of marine bacteria (80%), collected from various marine habitats worldwide, are able to influence S. aureus virulence gene expression. From Vibrio nigripulchritudo we isolated a new siderophore, nigribactin, which enhances the expression of spa encoding Protein A. While the crude extract of this bacterium also showed hla-repressing activity, we failed to isolate a single compound both repressing hla and inducing spa expression. However, since a large number of strains displayed this combination of activities such compounds are likely to be abundant. From a biological perspective, it is intriguing that bacteria from marine habitats produce compounds that influence virulence gene expression of a pathogen normally associated with warm-blooded animals. Thus, our study shows that marine bacteria are a source of compounds that affect virulence gene expression in S. aureus and ultimately, such compounds may aid in the treatment of infectious diseases.
Supplementary Files
Acknowledgments
This work was supported by funding from the Programme Committee for Food, Health and Welfare under the Danish Strategic Research Council. This is Galathea 3 contribution no. P98. We thank Charlotte H. Gotfredsen for acquisition of NMR data, Lisette K. Nielsen for assistance with nigribactin purification, Jette Melchiorsen for siderophore testing, and Lene Maj Petersen for NMR proofreading. We thank the Danish Instrument Center for NMR Spectroscopy of Biological Macromolecules at the Carlsberg Laboratory for NMR time.
References
- Rahman, H.; Austin, B.; Mitchell, W.J.; Morris, P.C.; Jamieson, D.J.; Adams, D.R.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar] [CrossRef]
- Nagai, K.; Kamigiri, K.; Arao, N.; Suzumura, K.; Kawano, Y.; Yamaoka, M.; Zhang, H.; Watanabe, M.; Suzuki, K. YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. I. taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J. Antibiot. 2003, 56, 123–128. [Google Scholar] [CrossRef]
- Wietz, M.; Mansson, M.; Gotfredsen, C.H.; Larsen, T.O.; Gram, L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 2010, 8, 2946–2960. [Google Scholar] [CrossRef]
- Hidron, A.I.; Edwards, J.R.; Patel, J.; Horan, T.C.; Sievert, D.M.; Pollock, D.A.; Fridkin, S.K. NHSN annual update: Antimicrobial-Resistant pathogens associated with healthcare-associated infections: Annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008, 29, 996–1011. [Google Scholar] [CrossRef]
- Moran, G.J.; Krishnadasan, A.; Gorwitz, R.J.; Fosheim, G.E.; McDougal, L.K.; Carey, R.B.; Talan, D.A. EMERGEncy ID Net Study Group. Methicillin-Resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 2006, 355, 666–674. [Google Scholar]
- Shoham, M. Antivirulence agents against MRSA. Future Med. Chem. 2011, 3, 775–777. [Google Scholar]
- Gotoh, Y.; Eguchi, Y.; Watanabe, T.; Okamoto, S.; Doi, A.; Utsumi, R. Two-Component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010, 13, 232–239. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Givskov, M. Quorum-Sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar]
- Skindersoe, M.E.; Ettinger-Epstein, P.; Rasmussen, T.B.; Bjarnsholt, T.; De Nys, R.; Givskov, M. Quorum sensing antagonism from marine organisms. Mar. Biotechnol. 2008, 10, 56–63. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar]
- Novick, R.P.; Projan, S.J.; Kornblum, J.; Ross, H.F.; Ji, G.; Kreiswirth, B.; Vandenesch, F.; Moghazeh, S. The agr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 1995, 248, 446–458. [Google Scholar] [CrossRef]
- Stefani, S.; Goglio, A. Methicillin-Resistant Staphylococcus aureus: Related infections and antibiotic resistance. Int. J. Infect. Dis. 2010, 14, 19–22. [Google Scholar] [CrossRef]
- Novick, R.P.; Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef]
- Mansson, M.; Nielsen, A.; Kjærulff, L.; Gotfredsen, C.H.; Wietz, M.; Ingmer, H.; Gram, L.; Larsen, T.O. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine Photobacterium. Mar. Drugs 2011, 9, 2537–2552. [Google Scholar] [CrossRef]
- Gram, L.; Melchiorsen, J.; Bruhn, J.B. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol. 2010, 12, 439–451. [Google Scholar] [CrossRef]
- Nielsen, A.; Nielsen, K.F.; Frees, D.; Larsen, T.O.; Ingmer, H. Method for screening compounds that influence virulence gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 509–512. [Google Scholar] [CrossRef]
- Musthafa, K.S.; Saroja, V.; Pandian, S.K.; Ravi, A.V. Antipathogenic potential of marine Bacillus sp. SS4 on N-acyl-homoserine-lactone-mediated virulence factors production in Pseudomonas aeruginosa (PAO1). J. Biosci. 2011, 36, 55–67. [Google Scholar] [CrossRef]
- George, E.A.; Muir, T.W. Molecular mechanisms of agr quorum sensing in virulent Staphylococci. Chembiochem 2007, 8, 847–855. [Google Scholar] [CrossRef]
- Mansson, M.; Phipps, R.K.; Gram, L.; Munro, M.H.; Larsen, T.O.; Nielsen, K.F. Explorative solid-phase extraction (E-SPE) for accelerated microbial natural product discovery, dereplication, and purification. J. Nat. Prod. 2010, 73, 1126–1132. [Google Scholar] [CrossRef]
- Yamamoto, S.; Okujo, N.; Fujita, Y.; Saito, M.; Yoshida, T.; Shinoda, S. Structures of two polyamine-containing catecholate siderophores from Vibrio fluvialis. J. Biochem. 1993, 113, 538–544. [Google Scholar]
- Payne, S.M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993, 1, 66–69. [Google Scholar]
- Abergel, R.J.; Warner, J.A.; Shuh, D.K.; Raymond, K.N. Enterobactin protonation and iron release: Structural characterization of the salicylate coordination shift in ferric enterobactin. J. Am. Chem. Soc. 2006, 128, 8920–8931. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef]
- Chan, P.F.; Foster, S.J. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 1998, 144, 2469–2479. [Google Scholar] [CrossRef]
- Horsburgh, M.J.; Aish, J.L.; White, I.J.; Shaw, L.; Lithgow, J.K.; Foster, S.J. SigmaB modulates virulence determinant expression and stress resistance: Characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002, 184, 5457–5467. [Google Scholar]
- Fairhead, H. University of Sheffield, Sheffield, UK. Personal communication, 1998.
- Novick, R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 1967, 33, 155–166. [Google Scholar] [CrossRef]
- Jelsbak, L.; Ingmer, H.; Valihrach, L.; Cohn, M.T.; Christiansen, M.H.; Kallipolitis, B.H.; Frees, D. The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One 2010, 5, e12752. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).