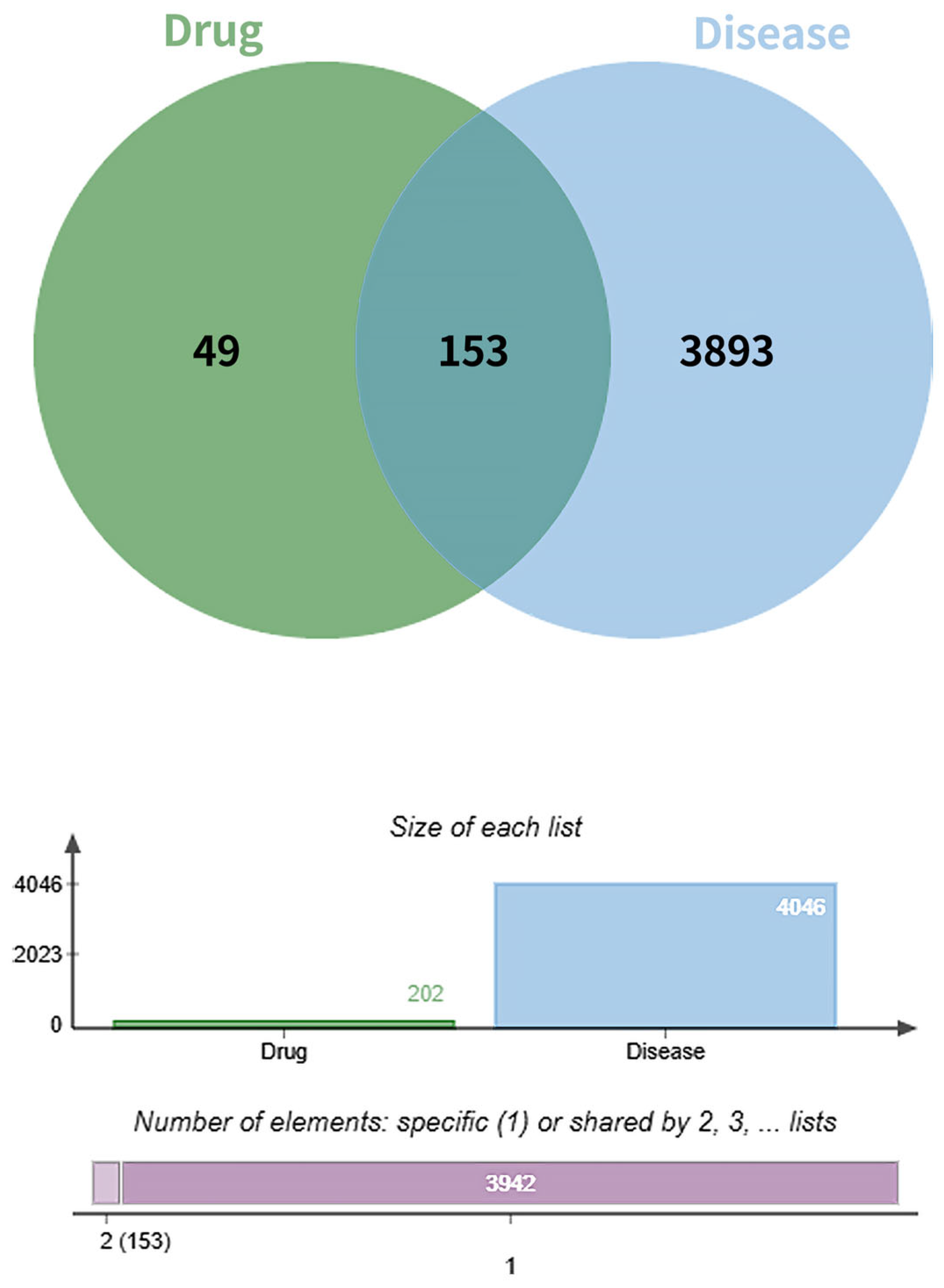
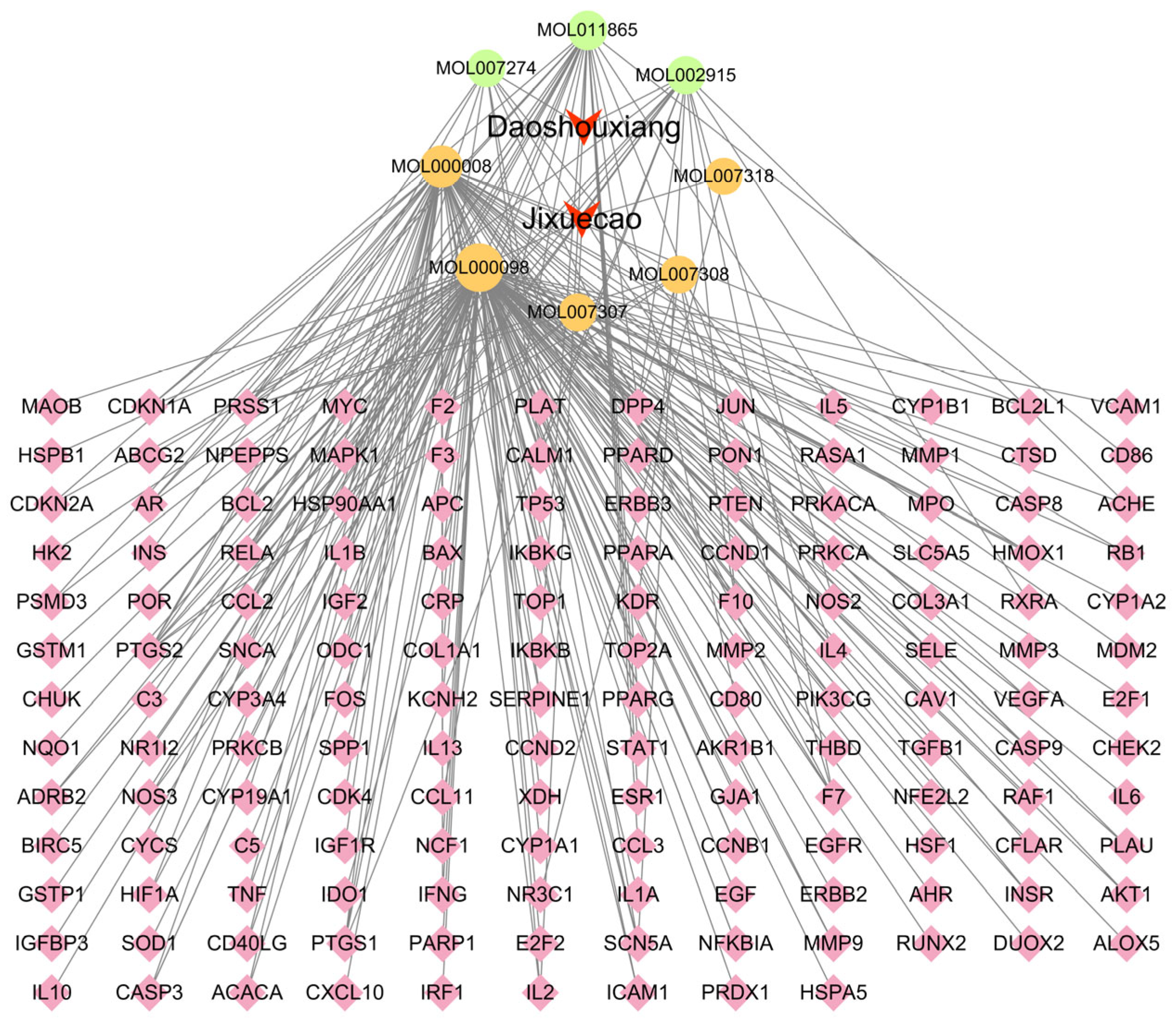
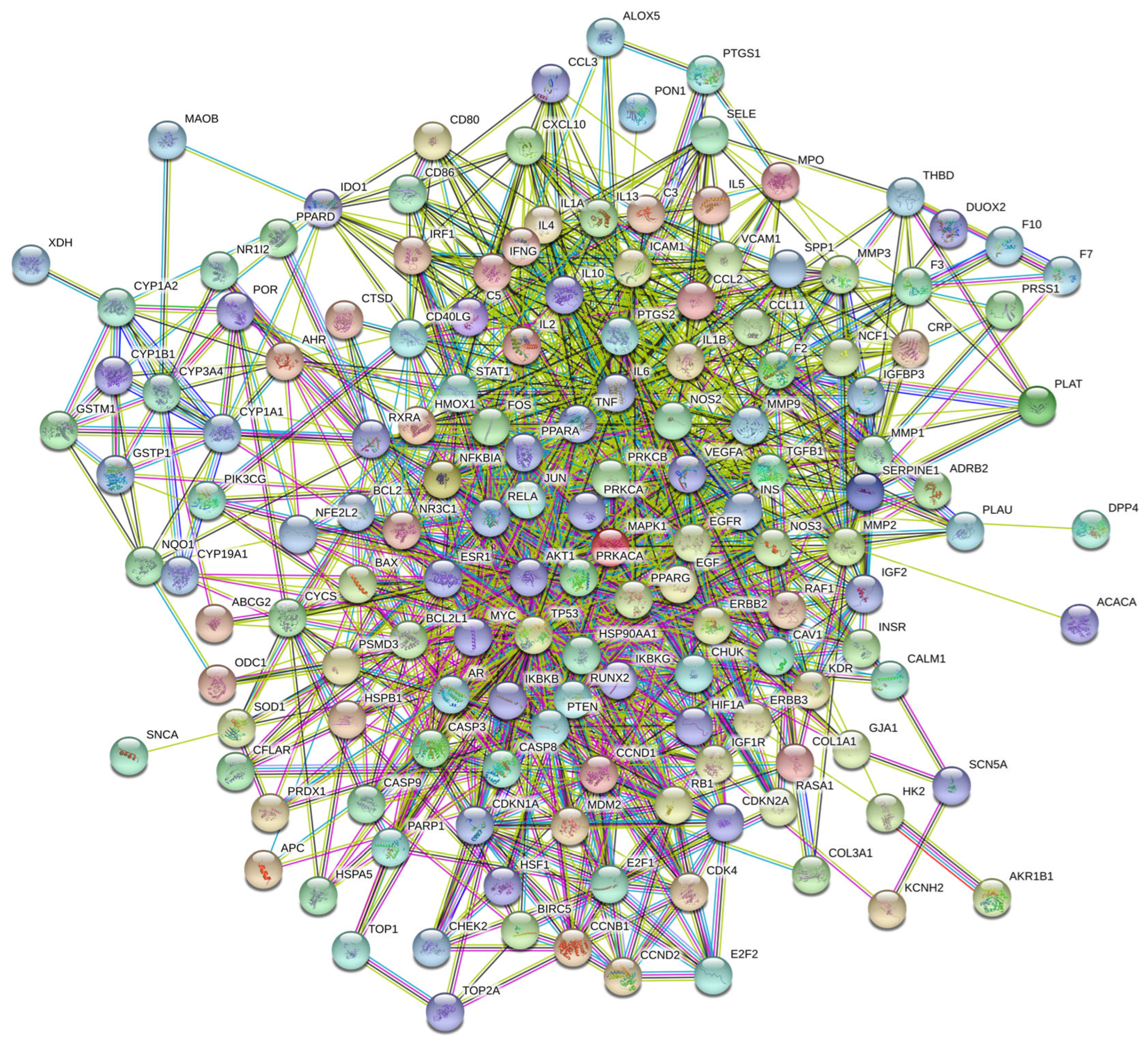
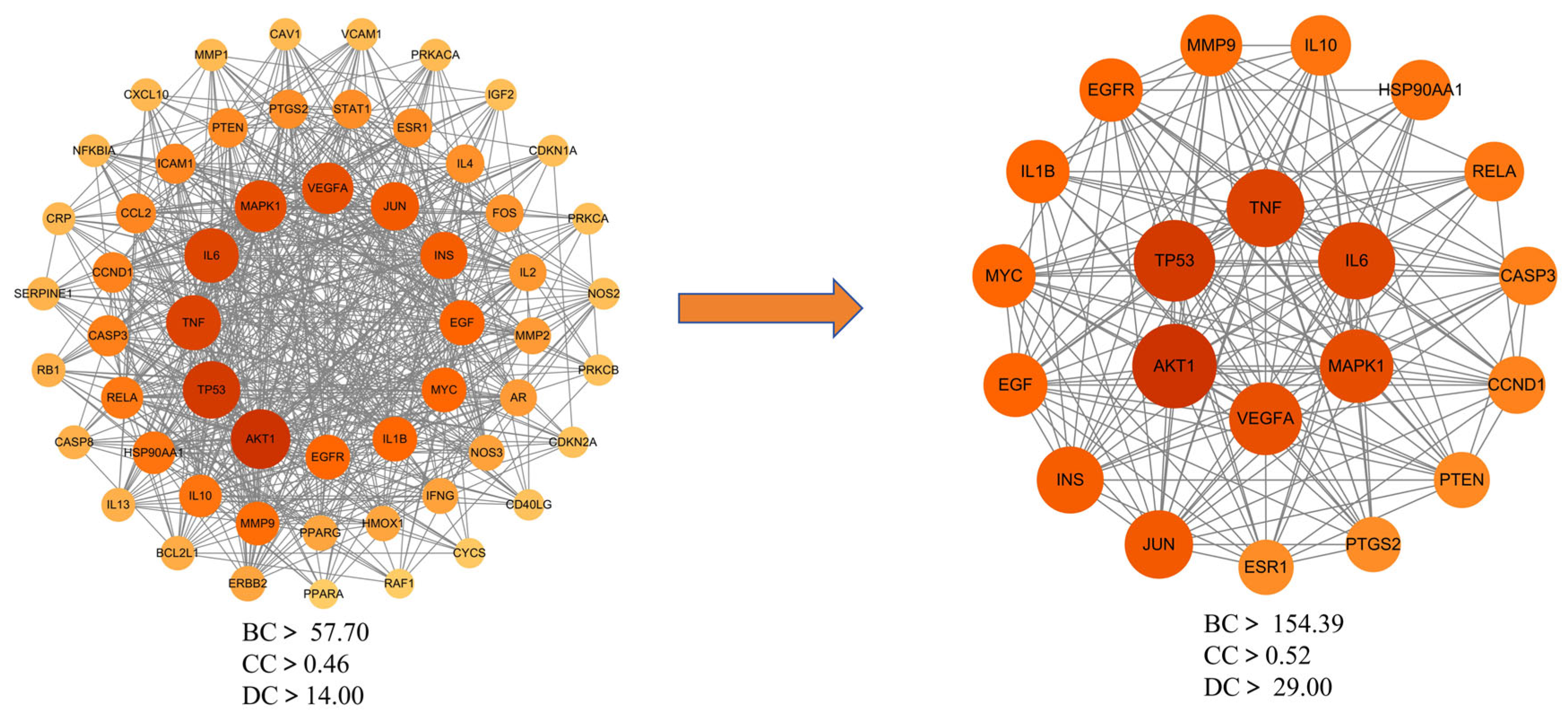
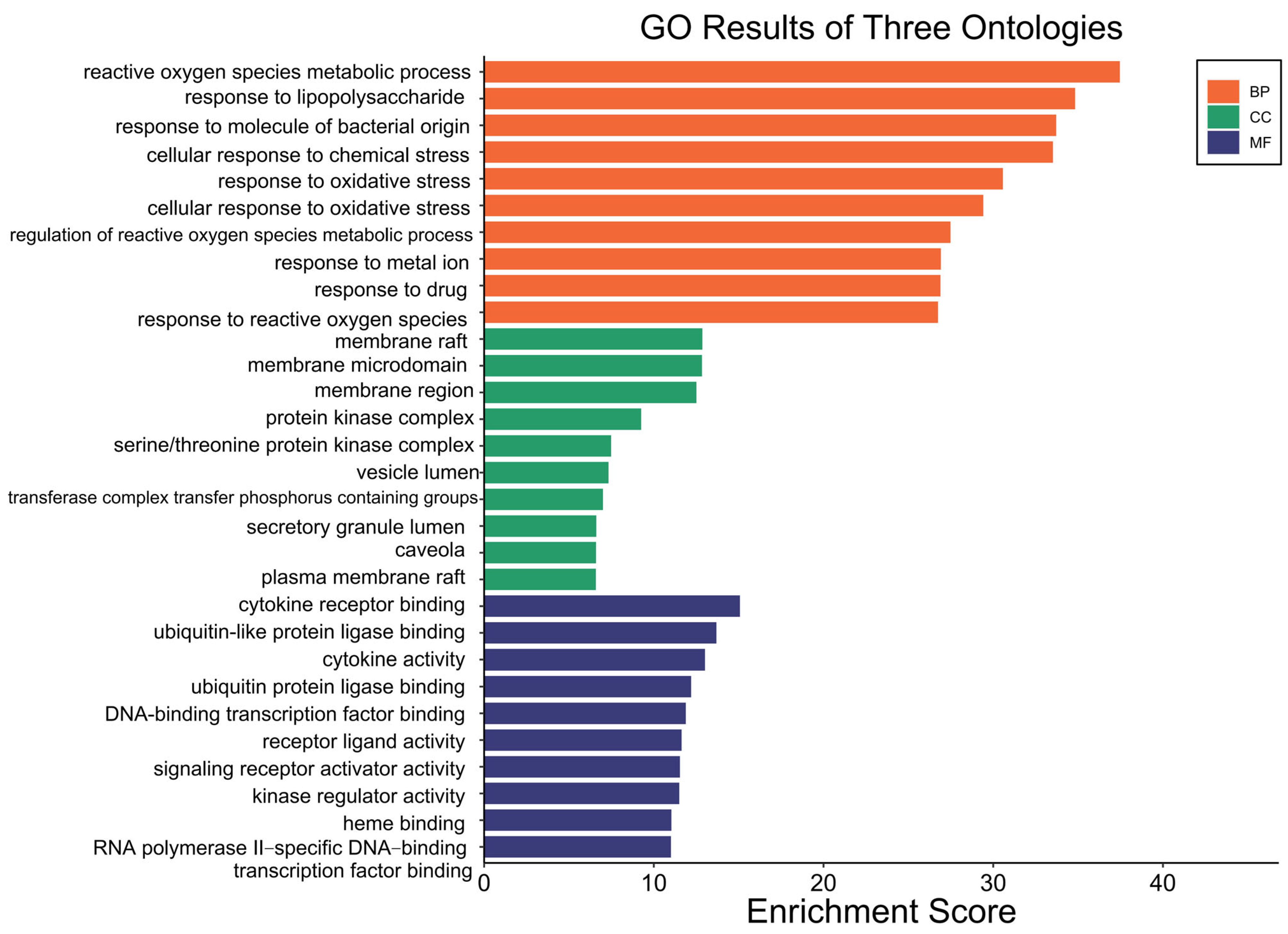
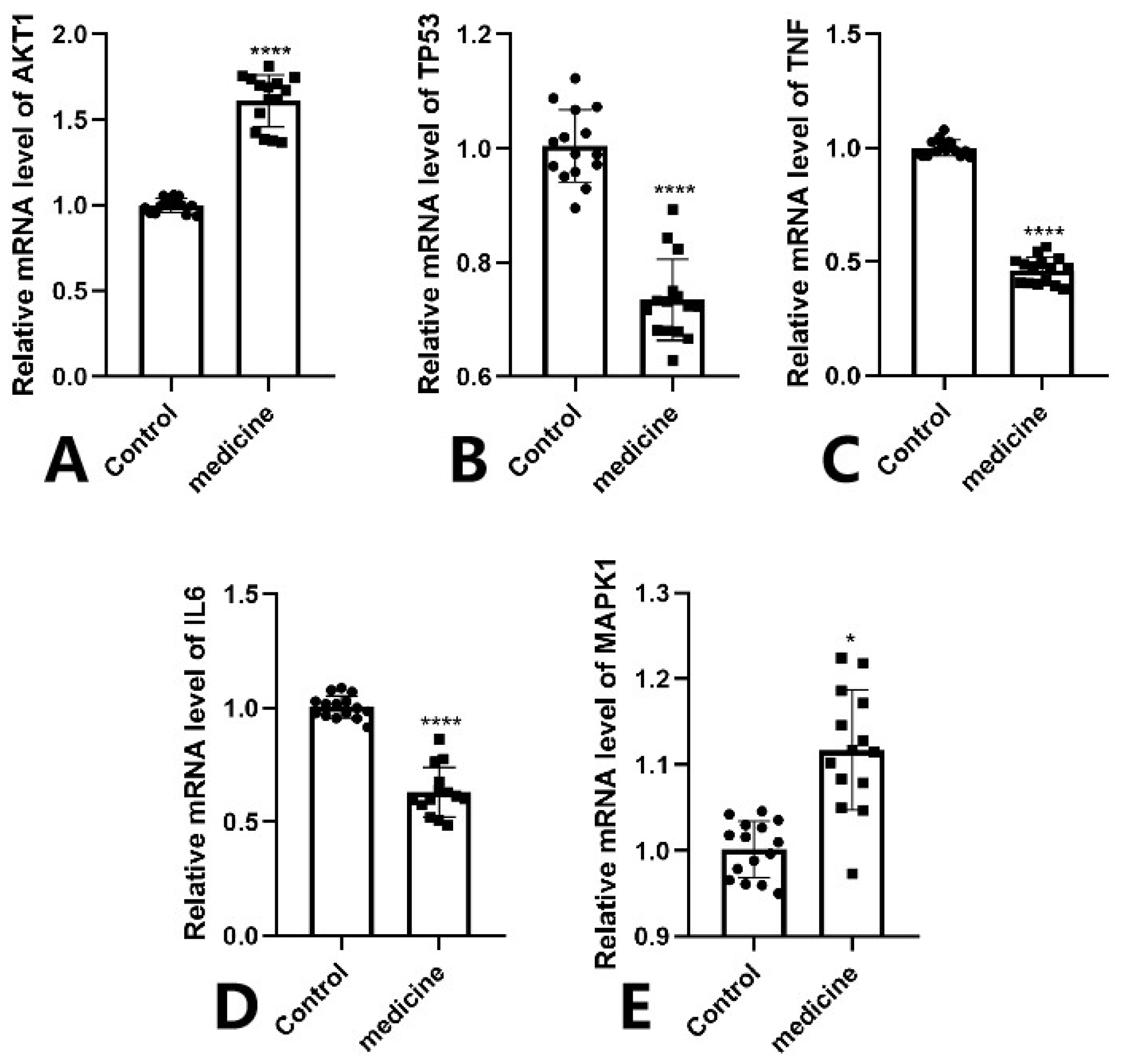
3.7. Molecular Docking Validation Between Key Targets and Active Compounds
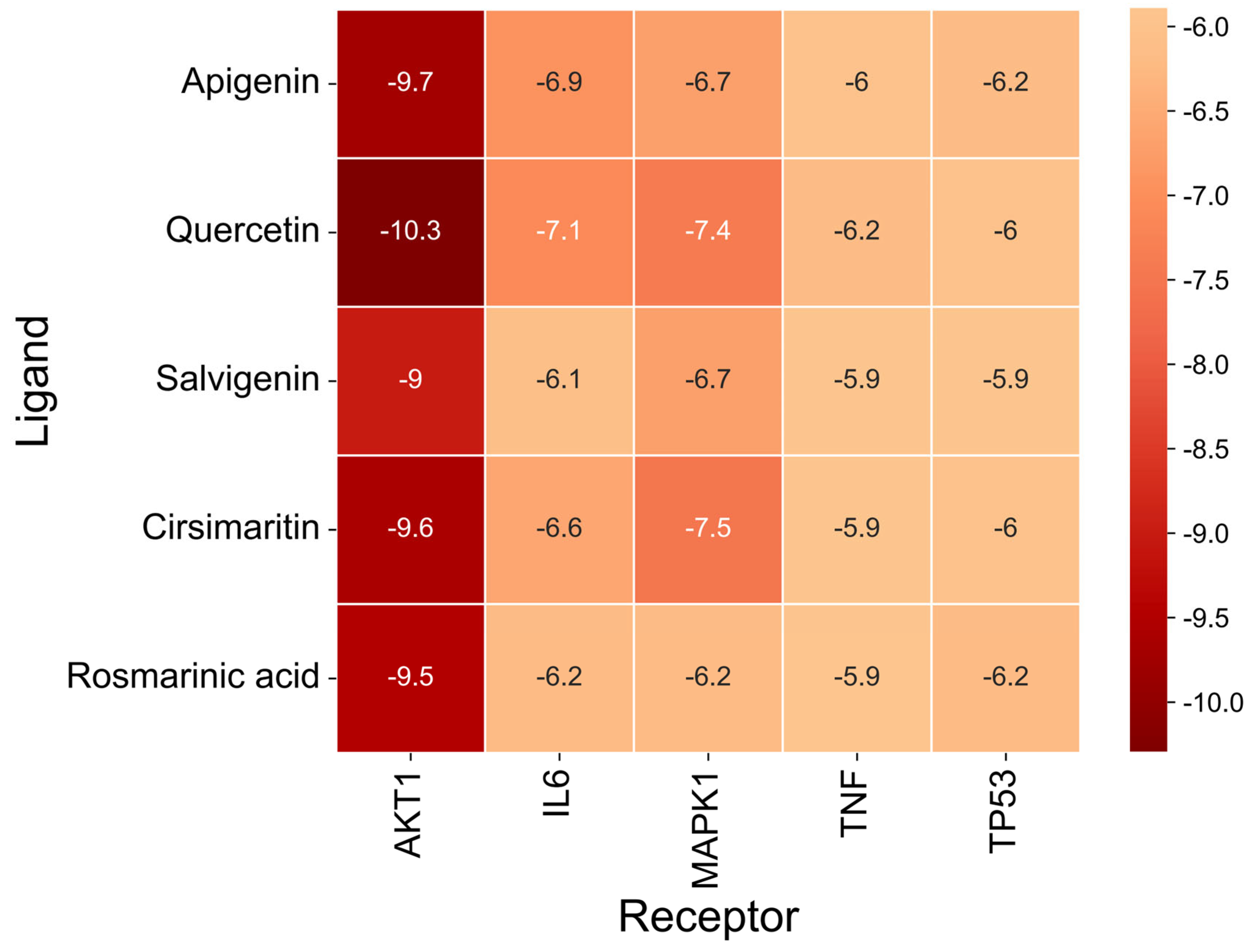
Binding energy is considered a critical parameter for evaluating the stability and affinity of interactions between small-molecule ligands and protein targets. In this study, molecular docking was performed to assess the binding interactions between five core protein targets—AKT1, TP53, TNF, IL6, and MAPK1—and the top five bioactive compounds of Fespixon cream: quercetin, apigenin, rosmarinic acid, salvigenin, and cirsimaritin. The ligands quercetin, apigenin, rosmarinic acid, salvigenin, and cirsimaritin were optimized to their minimum energy state prior to docking. The 2D structures were retrieved from the PubChem database and converted to 3D conformations using ChemBio3D Ultra 14.0. The MM2 force field was applied to minimize energy and optimize molecular geometry to ensure physiochemical relevance during docking.
The docking results (
Figure 7 and
Table 9) showed that all compound–target pairs exhibited minimum binding energies lower than −5.0 kcal/mol, suggesting favorable and stable interactions. Among them, quercetin showed the strongest binding affinity with AKT1, with a minimum binding energy of −10.3 kcal/mol, indicating that this compound–target interaction may play a central role in the pharmacological mechanism of Fespixon cream for diabetic foot ulcer treatment. The binding conformations were visualized using PyMOL software (
Figure 8). Furthermore, the most potent active compound identified in our study—quercetin—was individually docked with the remaining four top-ranked core targets identified through PPI network construction and hub gene screening: TP53, TNF, IL6, and MAPK1. The corresponding binding conformations were also visualized using PyMOL (
Figure 9,
Figure 10,
Figure 11 and
Figure 12).
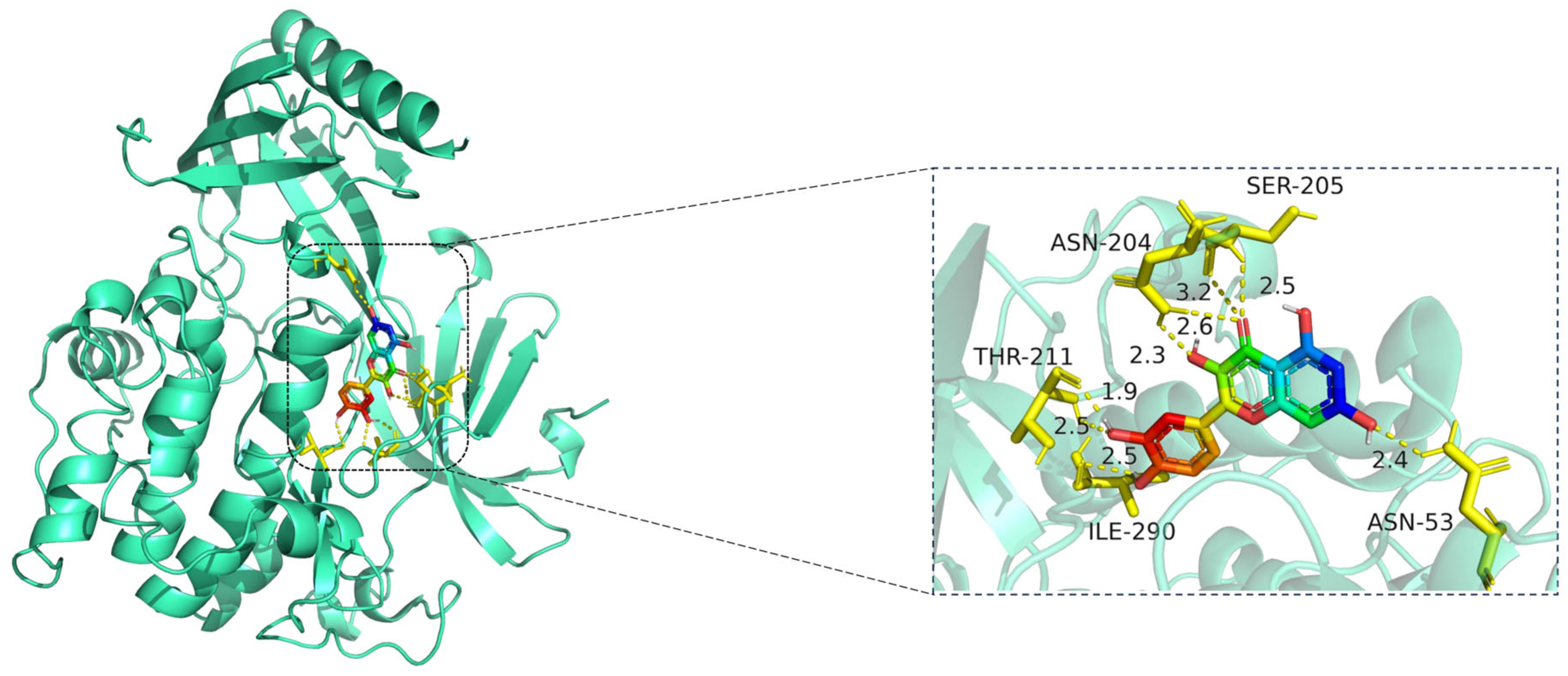
The docking visualization shows that quercetin fits well into the ATP-binding pocket of AKT1 and is stabilized by a network of hydrogen bonds and polar interactions involving key amino acid residues (
Figure 8):
ASN-204 forms two hydrogen bonds with quercetin at distances of 2.6 Å and 3.2 Å, stabilizing the chromone ring.
SER-205 forms a hydrogen bond at 2.5 Å, likely contributing to polar anchoring.
THR-211 exhibits strong dual interactions with distances of 1.9 Å and 2.3 Å, suggesting close spatial proximity and strong electrostatic contribution.
ILE-290 interacts via hydrogen bonding (2.5 Å) and likely hydrophobic stabilization with the aromatic rings of quercetin.
ASN-53 contributes a polar interaction at 2.4 Å, located at the periphery of the binding pocket.
These residues collectively form a polar and hydrophobic microenvironment, tightly stabilizing the ligand. Notably, THR-211 and ASN-204 are located in the AKT1 hinge region and may influence kinase activity regulation, suggesting that quercetin could act as a competitive or allosteric modulator. The hydrogen bond distances (1.9–3.2 Å) fall within the optimal interaction range, reinforcing the high-affinity binding observed in the docking score (−10.3 kcal/mol) [
15].
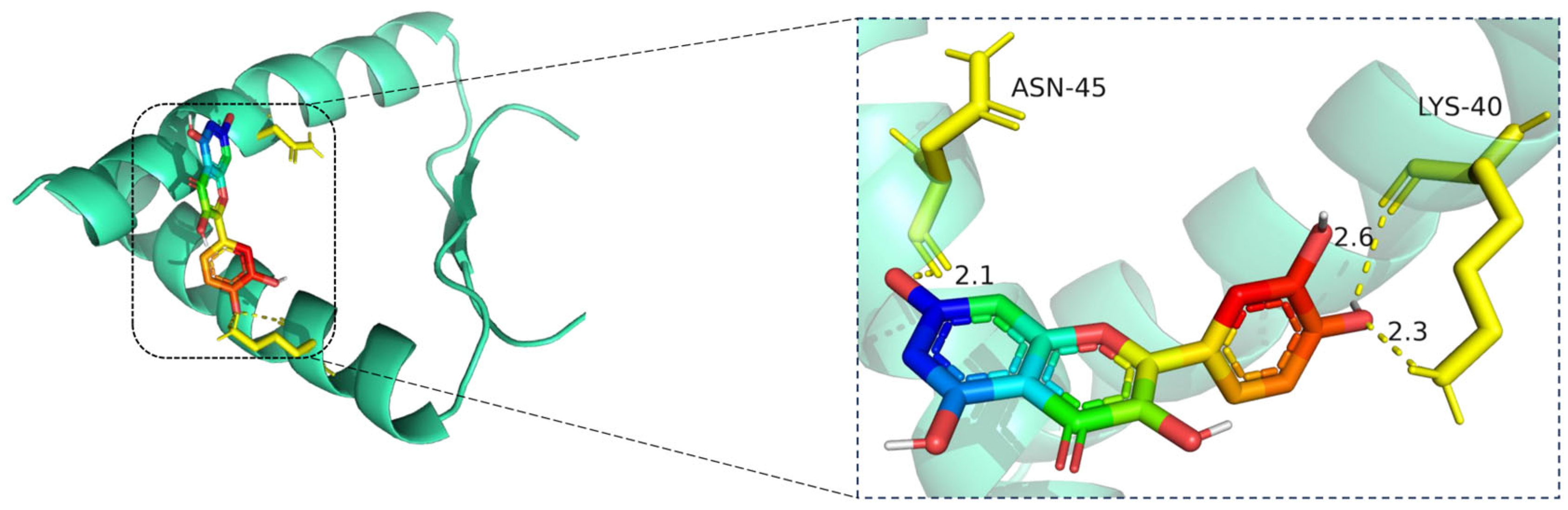
The docking visualization shows that quercetin fits well into the binding pocket of TP53 and is stabilized by a set of hydrogen bonds involving key amino acid residues (
Figure 9):
ASN-45 forms a strong hydrogen bond with the hydroxyl group of quercetin at a distance of 2.1 Å, contributing to anchoring the chromone scaffold.
LYS-40 establishes two hydrogen bonds with quercetin, at distances of 2.6 Å and 2.3 Å, respectively, indicating electrostatic interactions with both the phenolic and ketone moieties of the ligand.
These interactions create a polar environment that stabilizes the quercetin conformation within the TP53 pocket. The involved residues (ASN-45 and LYS-40) are located near the DNA-binding domain of TP53, implying a potential regulatory influence on its transcriptional activity. The hydrogen bond distances ranging from 2.1 to 2.6 Å fall within the favorable interaction range, suggesting high binding affinity consistent with the docking results.
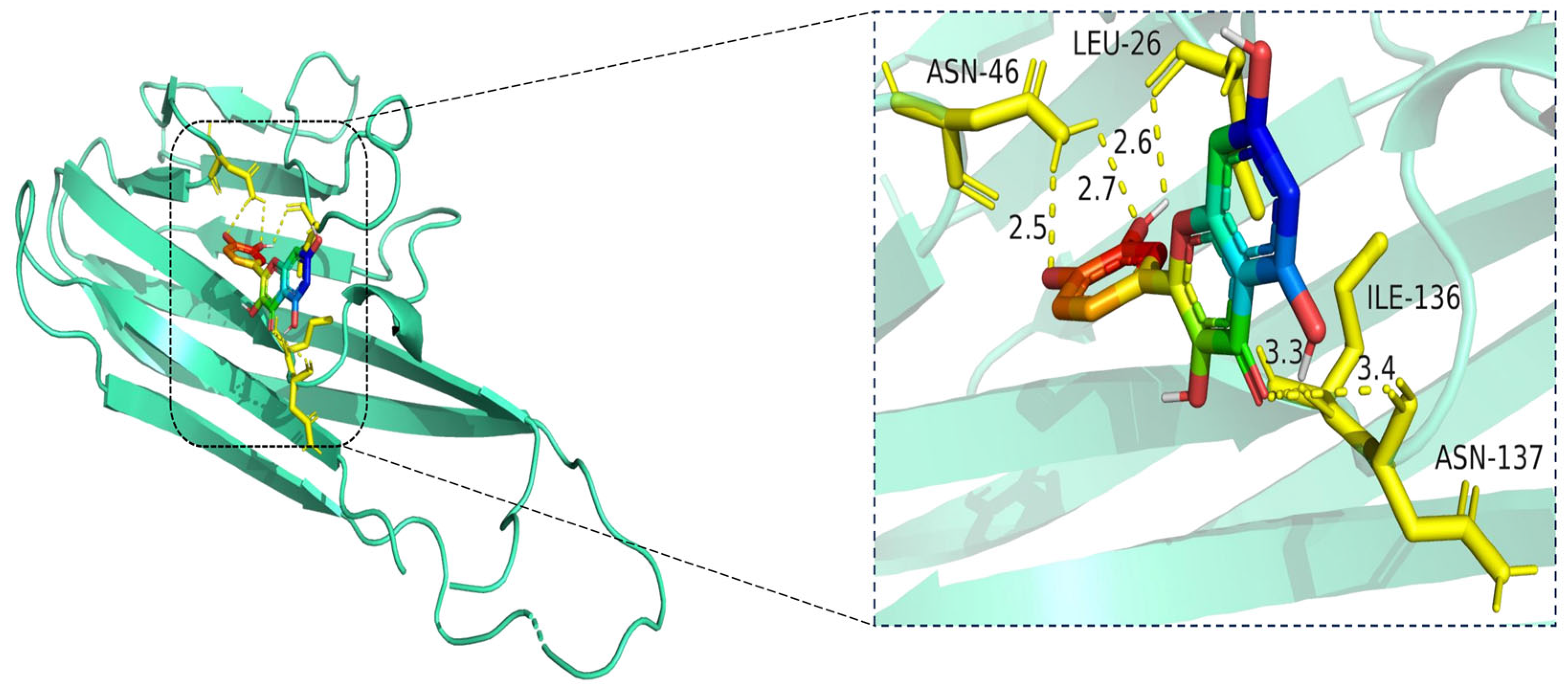
The docking visualization shows that quercetin fits well into the binding pocket of TNF-α and is stabilized by multiple hydrogen bonds and van der Waals interactions involving key amino acid residues (
Figure 10):
ASN-46 forms a hydrogen bond with the hydroxyl group of quercetin at a distance of 2.5 Å, contributing to anchoring the ligand within the hydrophilic region.
LEU-26 establishes two hydrogen bonds at 2.6 Å and 2.7 Å, likely stabilizing the planar aromatic structure of quercetin.
ILE-136 and ASN-137 contribute to hydrophobic and polar contacts, respectively, with distances of 3.3 Å and 3.4 Å, indicating weaker but spatially close interactions that help stabilize the complex.
Collectively, these interactions create a moderately hydrophilic binding environment, stabilizing quercetin within the TNF-α trimer interface. The interaction distances fall within the favorable range for hydrogen bonding (2.5–3.4 Å), supporting the predicted binding affinity.
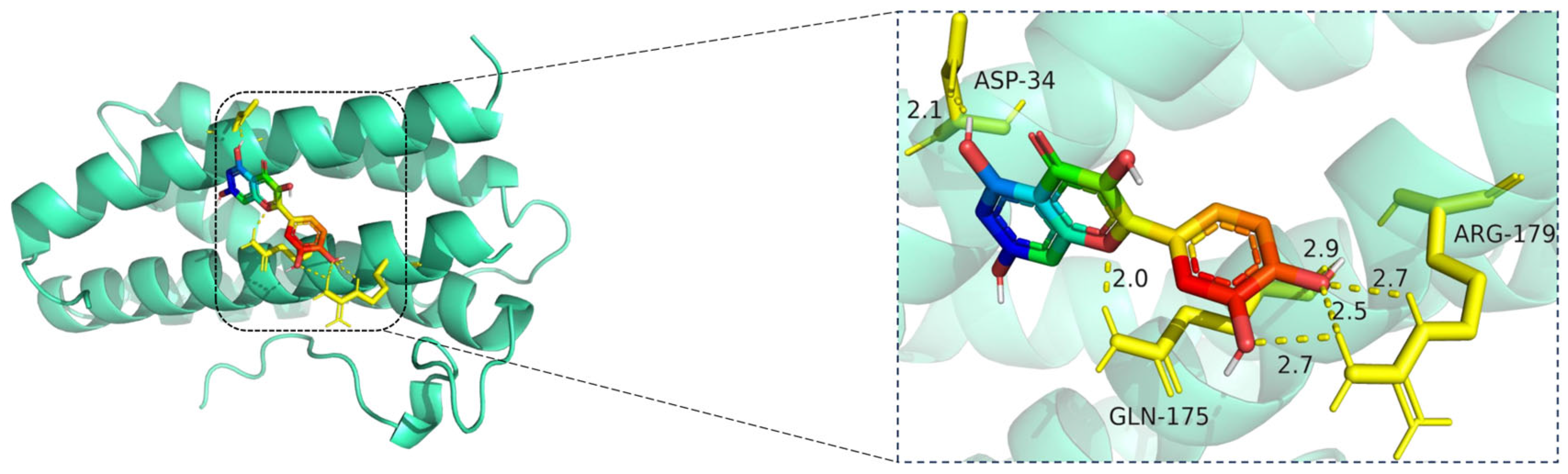
The docking visualization reveals that quercetin binds favorably to the IL6 receptor interface, stabilized by multiple hydrogen bonds with key residues (
Figure 11):
ASP-34 forms a hydrogen bond at 2.1 Å with the hydroxyl group of quercetin, providing polar anchoring at the entrance of the binding pocket.
GLN-175 contributes significantly to ligand stabilization via three hydrogen bonds at 2.0 Å, 2.5 Å, and 2.7 Å, indicating strong spatial complementarity and charge-based interactions.
ARG-179 forms two hydrogen bonds at 2.7 Å and 2.9 Å with the chromone ring and phenolic moiety, reinforcing the electrostatic stability of the complex.
Together, these residues form a dense hydrogen bonding network, suggesting that quercetin has a strong and specific interaction with IL6. The bond lengths (ranging from 2.0 to 2.9 Å) fall well within optimal hydrogen bonding distances.
The docking visualization shows that quercetin fits stably within the ATP-binding site of MAPK1 and is anchored through several key hydrogen bonds and polar interactions (
Figure 12):
TYR-316 forms a hydrogen bond with the hydroxyl group of quercetin at a distance of 2.3 Å, stabilizing the phenolic moiety.
ASN-82 interacts via a hydrogen bond at 2.5 Å, while ARG-135 forms another hydrogen bond at 2.6 Å, both contributing to polar stabilization within the binding pocket.
GLN-132 is positioned at 3.4 Å, suggesting weaker polar interactions with the chromone core of quercetin.
Together, these residues generate a polar microenvironment at the MAPK1 catalytic site, enhancing ligand binding. The interaction distances fall within optimal hydrogen bond range (2.3–3.4 Å), supporting the docking prediction.