Abstract
Lymphogranuloma venereum (LGV) is a systemic sexually transmitted infection caused by Chlamydia trachomatis serovars L1 to L3. The current LGV cases in Europe are mainly characterized by an anorectal syndrome, spreading within men who have sex with men (MSM). Whole-genome sequencing of LGV strains is crucial to the study of bacterial genomic variants and to improve strategies for contact tracing and prevention. In this study, we described the whole genome of a C. trachomatis strain (LGV/17) responsible for a case of rectal LGV. LGV/17 strain was isolated in 2017 in Bologna (North of Italy) from a HIV-positive MSM, presenting a symptomatic proctitis. After the propagation in LLC-MK2 cells, the strain underwent whole-genome sequencing by means of two platforms. Sequence type was determined using the tool MLST 2.0, whereas the genovariant was characterized by an ompA sequence evaluation. A phylogenetic tree was generated by comparing the LGV/17 sequence with a series of L2 genomes, downloaded from the NCBI website. LGV/17 belonged to sequence type ST44 and to the genovariant L2f. Nine ORFs encoding for polymorphic membrane proteins A-I and eight encoding for glycoproteins Pgp1-8 were detected in the chromosome and in the plasmid, respectively. LGV/17 was closely related to other L2f strains, even in the light of a not-negligible variability. The LGV/17 strain showed a genomic structure similar to reference sequences and was phylogenetically related to isolates from disparate parts of the world, indicative of the long-distance dynamics of transmission.
1. Introduction
Lymphogranuloma venereum (LGV) is a systemic sexually transmitted infection (STI) caused by Chlamydia trachomatis serovars L1 to L3 [1,2].
Since 2003, several outbreaks of LGV have spread across Europe and other high-income countries, mainly among HIV-positive men who have sex with men (MSM) [3,4]. The current LGV cases are essentially characterized by a proctitis, manifesting with anorectal ulceration, tenesmus, anal pain and mucous or bloody discharge (anorectal syndrome) [5,6]. However, several cases of LGV have recently been detected in asymptomatic subjects (up to 30–40% of cases), as well as in lower-risk groups of MSM (e.g., HIV-negative patients) [7].
The current LGV epidemic in Western Europe is caused by the L2 biovar of C. trachomatis with a predominance of the L2b genovariant [8]. Nevertheless, many countries have been reporting increasing cases due to other L2 variants, such as L2f or the hybrid L2-L2b/D-Da [2,9,10,11].
Laboratory diagnosis of LGV is mainly based on nucleic acid amplification techniques (NAAT) able to detect C. trachomatis and differentiate LGV from non-LGV serovars (e.g., assays based on ompA or pmpH genes) [12,13]. Other diagnostic procedures, such as microbial cultures, have a limited use and present critical issues. Indeed, Chlamydia cultures are impaired by the obligate intracellular nature of the microorganism, requiring the use of cell lines and trained personnel, incurring high costs and long time [14].
Beyond the technical difficulties, chlamydial culture has a critical role, allowing researchers to obtain the viable bacterial strain for further phenotypic and genotypic studies, such as whole genome sequencing (WGS). In this context, some culture-independent methods have been recently developed to sequence the whole genome of C. trachomatis directly from clinical samples [10].
WGS-based studies of chlamydial strains can be useful for different reasons, such as: (i) to deepen virulence and pathogenicity factors; (ii) to discover and monitor the presence of new genomic variants; (iii) to decipher the spread of clonal complexes during outbreaks; (iv) to improve strategies for contact tracing and the prevention of LGV spread; (v) to study LGV genomic evolution over the years in different countries.
Considering that limited WGS data of LGV strains are available, herein we sequenced the whole genome of a C. trachomatis strain (LGV/17) responsible for a case of rectal LGV, isolated from a symptomatic MSM in 2017 in the North of Italy.
2. Materials and Methods
2.1. Strain Isolation and LGV Diagnosis
A C. trachomatis strain, named LGV/17, was isolated in May 2017 from a rectal swab submitted to the Microbiology Unit of IRCSS Sant’Orsola-Malpighi Hospital of Bologna (Italy) [15]. This strain was recovered during routine diagnostic procedures from a 37-year-old HIV-positive MSM attending the STI Outpatient Clinic of the hospital and presenting with an anorectal syndrome.
The patient underwent a clinical visit, and a rectal swab was collected in a sucrose-phosphate-glutamine (SPG) medium. A part of the sample was processed by a commercial NAAT (Versant CT/GC DNA 1.0 Assay; Siemens Healthcare Diagnostics, Terrytown, USA), revealing a positive result for C. trachomatis [16]. Subsequently, a molecular genotyping, based on an ompA gene semi-nested PCR and RFLP analysis, identified a C. trachomatis L2 serovar [17].
The remaining part of the rectal swab was likewise inoculated in LLC-MK2 cells (ATCC® CCL-7). The presence of C. trachomatis was confirmed by the demonstration of chlamydial inclusions by direct immunofluorescence, as previously described [18]. After the isolation, the C. trachomatis strain was propagated for ∼1/2 weeks in LLC-MK2 cells (ATCC® CCL-7TM). C. trachomatis elementary bodies (EBs) were purified from cell debris and reticulate bodies by Renografin density gradient centrifugation, as described elsewhere [19].
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Sant‘Orsola-Malpighi Hospital, Bologna (78/2017/U/Tess). Informed consent was obtained from the subject involved in the study.
2.2. Whole Genome Sequencing and Subsequent Analyses
Genomic DNA was extracted from C. trachomatis EBs using a DNeasy Blood & Tissue Kit (Qiagen, Basel, Switzerland), following the manufacturer’s instructions, and further cleaned up with AMPure XP magnetic beads (Beckman Coulter, Milan, Italy). Whole-genome sequencing was performed using both Illumina iSeq 100 (Illumina, San Diego, CA, USA) and Oxford Nanopore MinION (Oxford Nanopore Technologies, Oxford, UK) systems, as previously described [20]. Briefly, short-read sequences were obtained using the Illumina iSeq 100 platform (iSeq Reagent Kit v2; Illumina, San Diego, CA, USA), using the iSeq Reagent kit v2 with 2 × 150 paired-end reads after Illumina DNA Prep paired-end library preparation. Long-read sequences were obtained by the Oxford Nanopore MinION (Oxford Nanopore Technologies, Oxford, UK) platform using a one-direction (1D) library prepared with a rapid sequencing kit (SQK-RAD004) and sequenced on a MinION Mk1C with SpotON flow cell R9.4 with the stand-alone version of MinKNOW v1.15.2 software with local base-calling.
A read quality evaluation was performed with FastQC 0.11.9 and seqtk 1.3. Prior to genome assembly, a filtering step was conducted to remove any contaminant human reads. First, the reference human genome GRCh38 (available at https://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/001/405/GCA_000001405.15_GRCh38/ accessed on 1 November 2022) was downloaded from the NCBI website and indexed with the Burrows–Wheeler Aligner (BWA) 0.7.17 using the index command. Next, Illumina and Nanopore reads were mapped to the reference human genome with default parameters using BWA and Minimap2 2.17, respectively. Last, Samtools 1.6 was used to sort the .sam alignment files with the sort command, extract the unmapped reads from the files using the view command with the -f parameter, and convert the resulting files to a .fastq format. Filtered reads were used for de novo genome assembly with a hybrid approach using Unicycler 0.5.0. Assembly quality was evaluated using Quast 5.0.2 and Bandage 0.8.1.
The genome nucleotide sequence was annotated using RASTtk 2.0, and virulence genes were detected by aligning the genome against the Virulence Factor Database’s (VFDB’s) full protein sequence dataset with diamond 2.0.15. Sequence type was determined using the tool MLST 2.0 (https://cge.food.dtu.dk/services/MLST/ accessed on 5 December 2022) from the Center for Genomic Epidemiology (CGE) website. The plasmid nucleotide sequence was aligned to the NCBI non-redundant (nr) database using blastn 2.12.0 in order to identify potential high-homology plasmids.
ParSNP 1.7.4 was launched to generate a phylogenetic tree based on core genome single nucleotide polymorphisms (cgSNPs). Chlamydia trachomatis L2 genomes were downloaded from the NCBI website and used for the phylogenetic analysis. Strain 434/Bu (GenBank accession number GCA_000068585) was used as a reference. The phylogenetic tree was reviewed and manually annotated with metadata from the NCBI database using iTOL v6.
Characterization of LGV genovariants was performed through an evaluation of the ompA gene variant, as previously described [2]. Briefly, BLAST analysis was performed by aligning the whole-genome of the LGV/17 strain against the LGV ompA gene variants available in GenBank representing the different genovariants (i.e., L2a, L2, L2b, L2c, L2d, L2e, L2, L2g, L2bV1, L2bV2, L2bV3, L2bV4, and L2-L2b/D-Da).
2.3. Data Availability
The complete genome sequence of the C. trachomatis strain LGV/17 was deposited in the NCBI database and is freely accessible at BioSample, accession number SAMN31133397.
3. Results
We generated a high-quality genome assembly of the clinical isolate LGV/17 using Illumina and Nanopore sequencing technologies. Illumina sequencing produced a total of 1,711,892 paired-end reads with an average Phred quality score of 35. Nanopore sequencing generated 102,458 reads ranging from 109 to 69,834 bp, with an average length of 3432 nucleotides and a mean Phred score of 20. After removing reads that mapped to the human genome, a total of 397,163 (23.2%) Illumina reads and 51,862 (50.6%) Nanopore reads were selected for the C. trachomatis genome assembly. The resulting de novo assembly produced a complete and circular genome with a total length of 1,045,668 bp, which consisted of a 1,038,168 bp chromosome and a 7500 bp plasmid. The mean coverage was 280×, and the G+C content was 41.3%. The genome was identified as sequence type ST44, and Blast analysis of the ompA gene variants showed that it belonged to the genovariant L2f (100% identity and coverage to ompA of the reference C. trachomatis isolate 128C/07 (EU676181)).
Functional annotation of the genome identified 953 protein-encoding sequences and 37 tRNAs on the chromosome (Figure 1). As shown in Table 1, we identified nine ORFs involved in virulence mechanisms, clustered in three genomic locations. These encoded for polymorphic membrane proteins (Pmps) A-I, autotransporter adhesins involved in the initial phase of C. trachomatis infection [21]. Eight ORFs encoding for glycoproteins Pgp1-8 were also found in the plasmid (Figure 1). The plasmid nucleotide sequence showed high homology to the plasmid described in 1988 by Comanducci et al. and believed to be required for C. trachomatis growth within mammalian cells (Query Cover 100%, Identity 99.98%) (GenBank Accession Number: X07547.1) [22].
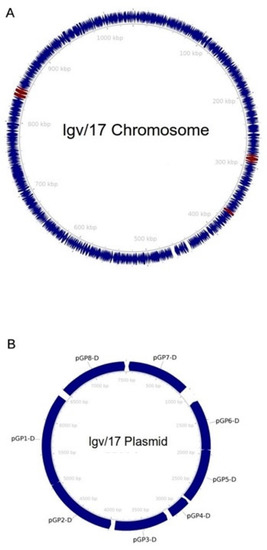
Figure 1.
Graphic representation of the LGV/17 genome. The chromosome is shown in panel (A), while the plasmid is in panel (B). Coding sequences are shown as navy-blue arrows. Maroon arrows indicate genes related to virulence mechanisms.

Table 1.
List of genes encoding for Pmps. Respective locations on the LGV/17 strain chromosome are shown.
Phylogenetic analysis (Figure 2) indicated that, even in light of the significant variability, the LGV/17 strain is closely related to the other strains identified over the years as the L2f genovariant in other countries.
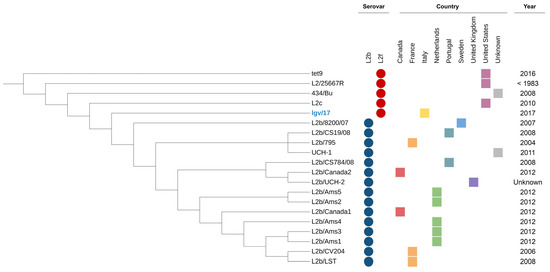
Figure 2.
Phylogenetic tree annotated with metadata from each strain. Serovars are indicated by colored circles, while the countries are represented by colored squares.
4. Discussion
To the best of our knowledge, this is the first report about the WGS of an LGV-associated C. trachomatis strain isolated from Italy. This work expands the existing literature on C. trachomatis genomic data, which could potentially be used to help in detecting, monitoring, and controlling LGV spread and outbreaks. Moreover, these data could be useful for the study of the genomic evolution of LGV over time and across different countries, as well as for deepening the pathogenesis and virulence of C. trachomatis biovars.
At first, we found that the LGV/17 strain belonged to L2f genovar. As previously reported [2], L2f was found to be the most common genovar in the urban areas of Bologna (North of Italy) in the years 2016–2017 and 2017–2018, accounting for about 50% of all LGV cases detected during these periods. In agreement with Marangoni et al., we hereby confirm the association between L2f genovar and the presence of rectal signs and symptoms (i.e., tenesmus, rectal discharge, anal pain), as well as other STIs, such as HIV [2].
Multi-locus sequence typing (MLST) has become a useful tool for studying the genetic diversity of important public health pathogens, leading to improvements in the monitoring and molecular epidemiology of C. trachomatis infections [23,24]. Several MLST-like systems able to identify LGV strains have been previously proposed, but none of them has been broadly adopted by the scientific community [25,26,27]. In silico extraction of MLST profiles using the Chlamydiales scheme [25], currently available at PubMLST (https://pubmlst.org/ accessed on 5 December 2022), showed that the LGV/17 strain belongs to sequence type 44 (ST44), being indistinguishable from the parent-like L2b/UCH-1/proctitis lineage [10].
Compared to MLST, the WGS approach has a higher discriminatory power, allowing timely characterization of circulating types and variants, which can help to disclose transmission chains, guide therapies, and identify emerging public-health harm [10]. However, WGS-based typing is still not applicable for routine surveillance of C. trachomatis because of several limitations, such as costs and technical difficulties.
By means of a chromosome functional annotation, we identified nine ORFs encoding for the polymorphic membrane proteins (Pmps) A-I, clustered in three genomic locations. Pmps are a group of membrane-bound surface-exposed proteins that have been characterized as autotransporter adhesins, crucial for the initial phases of chlamydial infection [21]. These proteins are differently regulated in response to stress (e.g., beta-lactams) and also play an important role as potent antigenic proteins involved in the immunopathogenesis of chlamydial infections [28]. Interestingly, the expression of Pmps is variably regulated in different chlamydial serovars and may induce differential immune responses with specific serovars [29].
Pmps are transcribed in vitro in various C. trachomatis strains including L2 and are all translocated to the bacterial surface [30,31]. However, it has been demonstrated that Pmps exhibit variable transcription patterns: for example, pmpD gene from the C. trachomatis serovar L2 upregulated when reticulate bodies (RBs) differentiated into elementary bodies (EBs) (i.e., about 16–24 h post-infection) [31].
When looking at the LGV/17 plasmid, eight ORFs encoding for glycoproteins Pgp1-8 were detected. In C. trachomatis serovar L2, the plasmid is known to regulate the expression of more than 20 genes at the transcription level [32,33], and plasmid-free organisms displayed reduced expression levels of glycogen synthesis genes, lacking glycogen accumulation in inclusions [33].
The significant role of plasmid glycoproteins Pgp1-8 has been recognized by different studies [32]. In fact, it has been shown that Pgp1, -2, -6, and -8 are critical for plasmid maintenance, that Pgp4 is a transcriptional regulator of chlamydial gene expression and glycogen synthesis, while Pgp3, -5, and -7 are necessary for chlamydial growth in vitro [34,35].
Finally, we conducted a phylogenetic analysis by comparing the entire LGV/17 sequence with a series of C. trachomatis L2 genomes downloaded from the NCBI website. As expected, our strain was closely related to other L2f strains, despite a not-negligible variability. These data potentially indicate a ‘genetic drift’ of L2 strains over the years and in different regions/areas of the world. The city where the LGV/17 strain was isolated is a high-density urban area (i.e., Bologna) with a significant exchange of people coming from different European/non-European countries. Thus, the creation of different sexual networks and dynamics of transmission could lead to a complex epidemiological scenario, where different LGV genovariants co-exist, co-evolve, and spread within the population.
5. Conclusions
In conclusion, we described the whole-genome of a L2f C. trachomatis strain responsible for a rectal LGV in an Italian HIV-positive MSM. The LGV/17 strain showed a genomic structure similar to reference sequences and it was phylogenetically related to isolates from disparate parts of the world, indicative of long-distance dynamics of transmission. Further perspectives include: (i) the isolation and collection of other LGV strains from Italy, to better understand C. trachomatis genomic evolution; (ii) in depth studies of the in vitro behavior of the LGV/17 strain (e.g., interactions with different cellular types).
Author Contributions
Conceptualization, C.F. and A.M.; methodology, P.G. and S.A.; software, P.G. and S.A.; validation, T.L., C.F. and A.M.; formal analysis, V.G., M.E.D. and S.A.; investigation, V.G., M.E.D. and S.A.; resources, T.L. and P.G.; data curation, C.F., P.G. and S.A.; writing—original draft preparation, C.F., A.M. and P.G.; supervision, T.L., C.F. and A.M.; project administration, C.F. and P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Sant‘Orsola-Malpighi Hospital, Bologna (78/2017/U/Tess).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The complete genome sequence of C. trachomatis strain LGV/17 is freely available at BioSample accession number SAMN31133397.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Stary, G.; Meyer, T.; Bangert, C.; Kohrgruber, N.; Gmeinhart, B.; Kirnbauer, R.; Jantschitsch, C.; Rieger, A.; Stary, A.; Geusau, A. New Chlamydia trachomatis L2 strains identified in a recent outbreak of lymphogranuloma venereum in Vienna, Austria. Sex Transm. Dis. 2008, 35, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, A.; Foschi, C.; Tartari, F.; Gaspari, V.; Re, M.C. Lymphogranuloma venereum genovariants in men having sex with men in Italy. Sex Transm. Infect. 2021, 97, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Childs, T.; Simms, I.; Alexander, S.; Eastick, K.; Hughes, G.; Field, N. Rapid increase in lymphogranuloma venereum in men who have sex with men, United Kingdom, 2003 to September 2015. Euro Surveill. 2015, 20, 30076. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, N.H.; de Vries, H.J. Lymphogranuloma venereum among men who have sex with men. An epidemiological and clinical review. Expert. Rev. Anti. Infect. Ther. 2014, 12, 697–704. [Google Scholar] [CrossRef]
- Savage, E.J.; van de Laar, M.J.; Gallay, A.; van der Sande, M.; Hamouda, O.; Sasse, A.; Hoffmann, S.; Diez, M.; Borrego, M.J.; Lowndes, C.M.; et al. European Surveillance of Sexually Transmitted Infections (ESSTI) network. Lymphogranuloma venereum in Europe, 2003–2008. Euro Surveill. 2009, 14, 19428. [Google Scholar] [CrossRef]
- Foschi, C.; Marangoni, A.; D’Antuono, A.; Nardini, P.; Compri, M.; Bellavista, S.; Filippini, A.; Bacchi Reggiani, M.L.; Cevenini, R. Prevalence and predictors of Lymphogranuloma venereum in a high risk population attending a STD outpatients clinic in Italy. BMC Res. Notes 2014, 7, 225. [Google Scholar] [CrossRef]
- Hughes, Y.; Chen, M.Y.; Fairley, C.K.; Hocking, J.S.; Williamson, D.; Ong, J.J.; De Petra, V.; Chow, E.P.F. Universal lymphogranuloma venereum (LGV) testing of rectal chlamydia in men who have sex with men and detection of asymptomatic LGV. Sex. Transm. Infect. 2022, 98, 582–585. [Google Scholar] [CrossRef]
- Christerson, L.; de Vries, H.J.; de Barbeyrac, B.; Gaydos, C.A.; Henrich, B.; Hoffmann, S.; Schachter, J.; Thorvaldsen, J.; Vall-Mayans, M.; Klint, M.; et al. Typing of lymphogranuloma venereum Chlamydia trachomatis strains. Emerg. Infect. Dis. 2010, 16, 1777–1779. [Google Scholar] [CrossRef]
- Isaksson, J.; Carlsson, O.; Airell, Å.; Strömdahl, S.; Bratt, G.; Herrmann, B. Lymphogranuloma venereum rates increased and Chlamydia trachomatis genotypes changed among men who have sex with men in Sweden 2004-2016. J. Med. Microbiol. 2017, 66, 1684–1687. [Google Scholar] [CrossRef]
- Borges, V.; Cordeiro, D.; Salas, A.I.; Lodhia, Z.; Correia, C.; Isidro, J.; Fernandes, C.; Rodrigues, A.M.; Azevedo, J.; Alves, J.; et al. Chlamydia trachomatis: When the virulence-associated genome backbone imports a prevalence-associated major antigen signature. Microb. Genom. 2019, 5, e000313. [Google Scholar] [CrossRef]
- Rodríguez-Domínguez, M.; Puerta, T.; Menéndez, B.; González-Alba, J.M.; Rodríguez, C.; Hellín, T.; Vera, M.; González-Sainz, F.J.; Clavo, P.; Villa, M.; et al. Clinical and epidemiological characterization of a lymphogranuloma venereum outbreak in Madrid, Spain: Co-circulation of two variants. Clin. Microbiol. Infect. 2014, 20, 219–225. [Google Scholar] [CrossRef]
- Morré, S.A.; Spaargaren, J.; Fennema, J.S.; de Vries, H.J.; Coutinho, R.A.; Peña, A.S. Real-time polymerase chain reaction to diagnose lymphogranuloma venereum. Emerg. Infect. Dis. 2005, 11, 1311–1312. [Google Scholar] [CrossRef]
- Petrovay, F.; Balla, E.; Németh, I.; Gönczöl, É. Genotyping of Chlamydia trachomatis from the endocervical specimens of high-risk women in Hungary. J. Med. Microbiol. 2009, 58 Pt 6, 760–764. [Google Scholar] [CrossRef] [PubMed]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef]
- Foschi, C.; Bortolotti, M.; Polito, L.; Marangoni, A.; Zalambani, C.; Liparulo, I.; Fato, R.; Bolognesi, A. Insights into penicillin-induced Chlamydia trachomatis persistence. Microb. Pathog. 2020, 142, 104035. [Google Scholar] [CrossRef]
- Marangoni, A.; Foschi, C.; Nardini, P.; Compri, M.; Cevenini, R. Evaluation of the Versant CT/GC DNA 1.0 assay (kPCR) for the detection of extra-genital Chlamydia trachomatis and Neisseria gonorrhoeae infections. PLoS ONE 2015, 10, e0120979. [Google Scholar] [CrossRef]
- Gallo Vaulet, L.; Entrocassi, C.; Corominas, A.I.; Rodriguez Fermepin, M. Distribution study of Chlamydia trachomatis genotypes in symptomatic patients in Buenos Aires, Argentina: Association between genotype E and neonatal conjunctivitis. BMC Res. Notes 2010, 3, 34. [Google Scholar] [CrossRef]
- Marangoni, A.; Fiorino, E.; Gilardi, F.; Aldini, R.; Scotti, E.; Nardini, P.; Foschi, C.; Donati, M.; Montagnani, M.; Cevenini, M.; et al. Chlamydia pneumoniae acute liver infection affects hepatic cholesterol and triglyceride metabolism in mice. Atherosclerosis 2015, 41, 471–479. [Google Scholar] [CrossRef]
- Marziali, G.; Marangoni, A.; Foschi, C.; Re, M.C.; Calonghi, N. Effect of Sugars on Chlamydia trachomatis Infectivity. Pathogens 2020, 9, 298. [Google Scholar] [CrossRef]
- Gaibani, P.; Amadesi, S.; Lazzarotto, T.; Ambretti, S. Genome characterization of a Klebsiella pneumoniae co-producing OXA-181 and KPC-121 resistant to ceftazidime/avibactam, meropenem/vaborbactam, imipenem/relebactam and cefiderocol isolated from a critically ill patient. J. Glob. Antimicrob. Resist. 2022, 30, 262–264. [Google Scholar] [CrossRef]
- Vasilevsky, S.; Stojanov, M.; Greub, G.; Baud, D. Chlamydial polymorphic membrane proteins: Regulation, function and potential vaccine candidates. Virulence 2016, 7, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Comanducci, M.; Ricci, S.; Ratti, G. The structure of a plasmid of Chlamydia trachomatis believed to be required for growth within mammalian cells. Mol. Microbiol. 1988, 2, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Patiño, L.H.; Camargo, M.; Muñoz, M.; Ríos-Chaparro, D.I.; Patarroyo, M.A.; Ramírez, J.D. Unveiling the Multilocus Sequence Typing (MLST) Schemes and Core Genome Phylogenies for Genotyping Chlamydia trachomatis. Front. Microbiol. 2018, 9, 1854. [Google Scholar] [CrossRef] [PubMed]
- Feodorova, V.A.; Saltykov, Y.V.; Kolosova, A.A.; Rubanik, L.V.; Poleshchuk, N.N.; Motin, V.L. Emergence of Novel Chlamydia trachomatis Sequence Types among Chlamydia Patients in the Republic of Belarus. Microorganisms 2022, 10, 478. [Google Scholar] [CrossRef]
- Pannekoek, Y.; Morelli, G.; Kusecek, B.; Morré, S.A.; Ossewaarde, J.M.; Langerak, A.A.; van der Ende, A. Multi locus sequence typing of Chlamydiales: Clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 2008, 8, 42. [Google Scholar] [CrossRef]
- Dean, D.; Bruno, W.J.; Wan, R.; Gomes, J.P.; Devignot, S.; Mehari, T.; de Vries, H.J.; Morré, S.A.; Myers, G.; Read, T.D.; et al. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis. 2009, 15, 1385–1394. [Google Scholar] [CrossRef]
- Bom, R.J.; Christerson, L.; Schim van der Loeff, M.F.; Coutinho, R.A.; Herrmann, B.; Bruisten, S.M. Evaluation of high-resolution typing methods for Chlamydia trachomatis in samples from heterosexual couples. J. Clin. Microbiol. 2011, 49, 2844–2853. [Google Scholar] [CrossRef]
- Tan, C.; Hsia, R.C.; Shou, H.; Haggerty, C.L.; Ness, R.B.; Gaydos, C.A.; Dean, D.; Scurlock, A.M.; Wilson, D.P.; Bavoil, P.M. Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect. Immun. 2009, 77, 3218–3226. [Google Scholar] [CrossRef]
- Gomes, J.P.; Hsia, R.C.; Mead, S.; Borrego, M.J.; Dean, D. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect. 2005, 7, 410–420. [Google Scholar] [CrossRef]
- Tan, C.; Hsia, R.C.; Shou, H.; Carrasco, J.A.; Rank, R.G.; Bavoil, P.M. Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell Microbiol. 2010, 12, 174–187. [Google Scholar] [CrossRef]
- Kiselev, A.O.; Stamm, W.E.; Yates, J.R.; Lampe, M.F. Expression, processing, and localization of PmpD of Chlamydia trachomatis Serovar L2 during the chlamydial developmental cycle. PLoS ONE 2007, 2, e568. [Google Scholar] [CrossRef]
- Gong, S.; Yang, Z.; Lei, L.; Shen, L.; Zhong, G. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J. Bacteriol. 2013, 195, 3819–3826. [Google Scholar] [CrossRef]
- Carlson, J.H.; Whitmire, W.M.; Crane, D.D.; Wicke, L.; Virtaneva, K.; Sturdevant, D.E.; Kupko, J.J., III; Porcella, S.F.; Martinez-Orengo, N.; Heinzen, R.A.; et al. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 2008, 76, 2273–2283. [Google Scholar] [CrossRef]
- Song, L.; Carlson, J.H.; Whitmire, W.M.; Kari, L.; Virtaneva, K.; Sturdevant, D.E.; Watkins, H.; Zhou, B.; Sturdevant, G.L.; Porcella, S.F.; et al. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 2013, 81, 636–644. [Google Scholar] [CrossRef]
- Wang, Y.; Cutcliffe, L.T.; Skilton, R.J.; Persson, K.; Bjartling, C.; Clarke, I.N. Transformation of a plasmid-free, genital tract isolate of Chlamydia trachomatis with a plasmid vector carrying a deletion in CDS6 revealed that this gene regulates inclusion phenotype. Pathog. Dis. 2013, 67, 100–103. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).