«Salivaomics» of Different Molecular Biological Subtypes of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Group Description
2.2. Determination of the Expression of the Receptors for Estrogen, Progesterone and HER2
2.3. Saliva Collection and Analysis
2.4. Statistical Analysis
3. Results
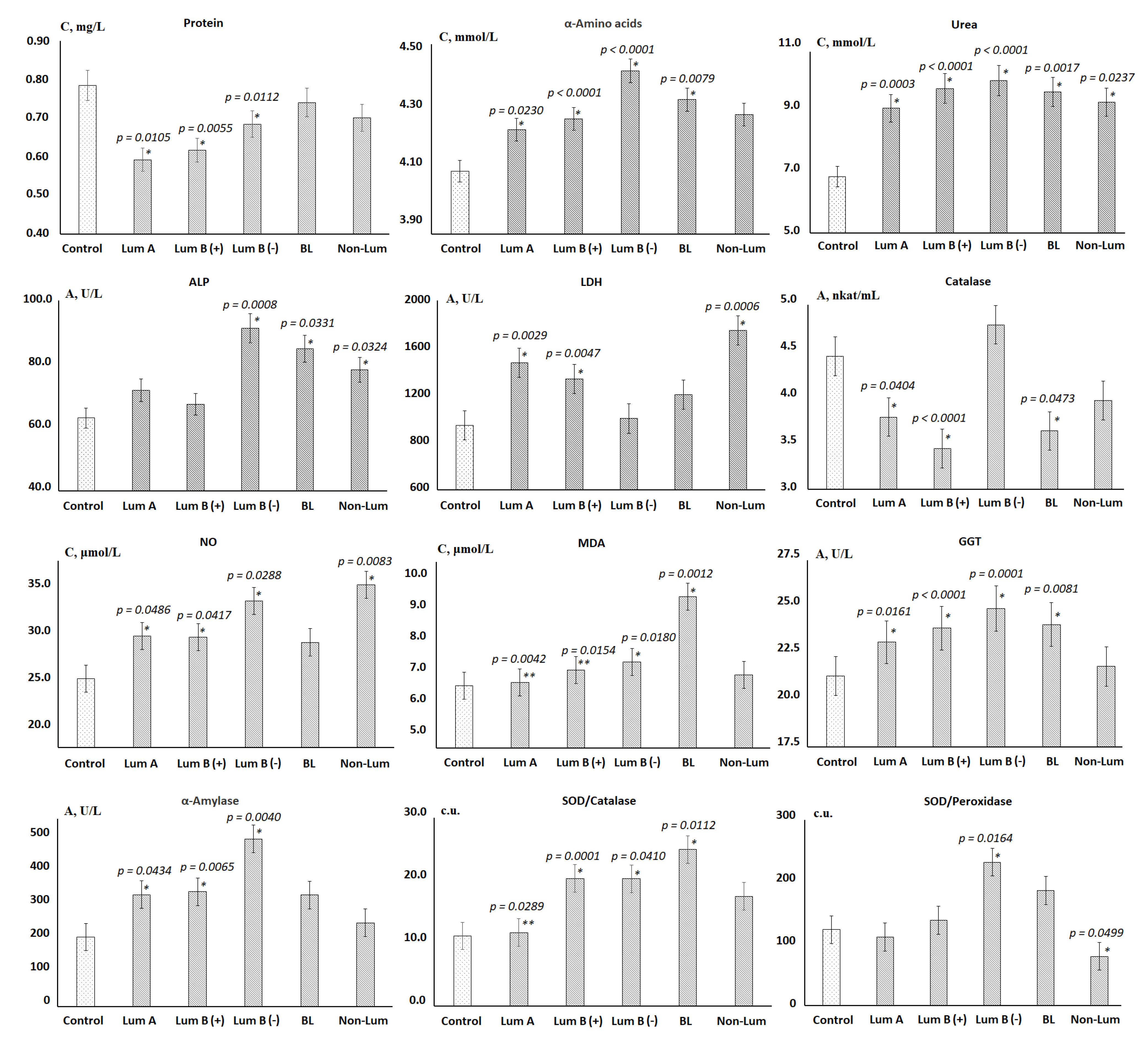
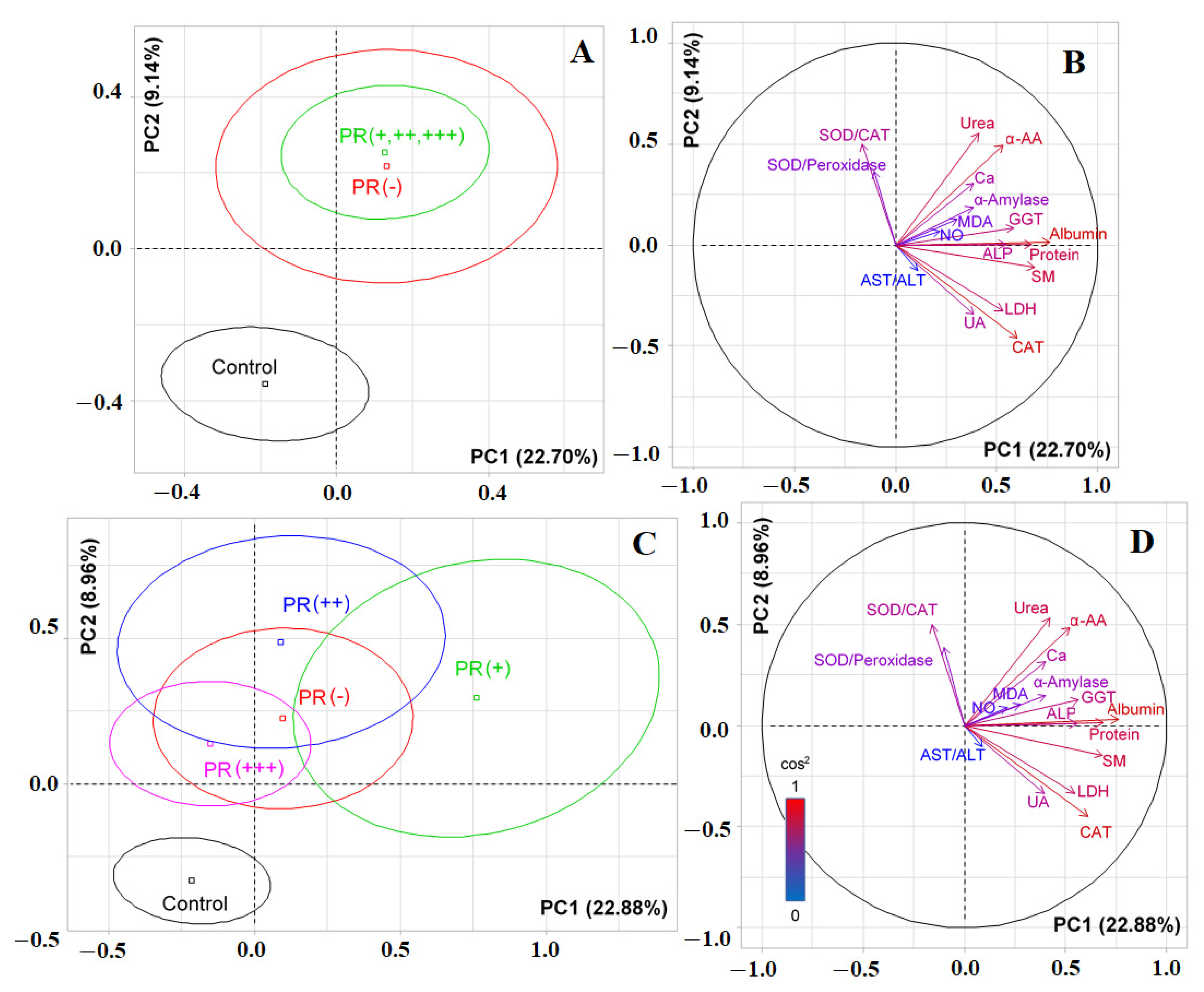
3.1. Changes in the Biochemical Composition of the Saliva of Patients with Breast Cancer, Depending on Its Molecular Biological Subtype
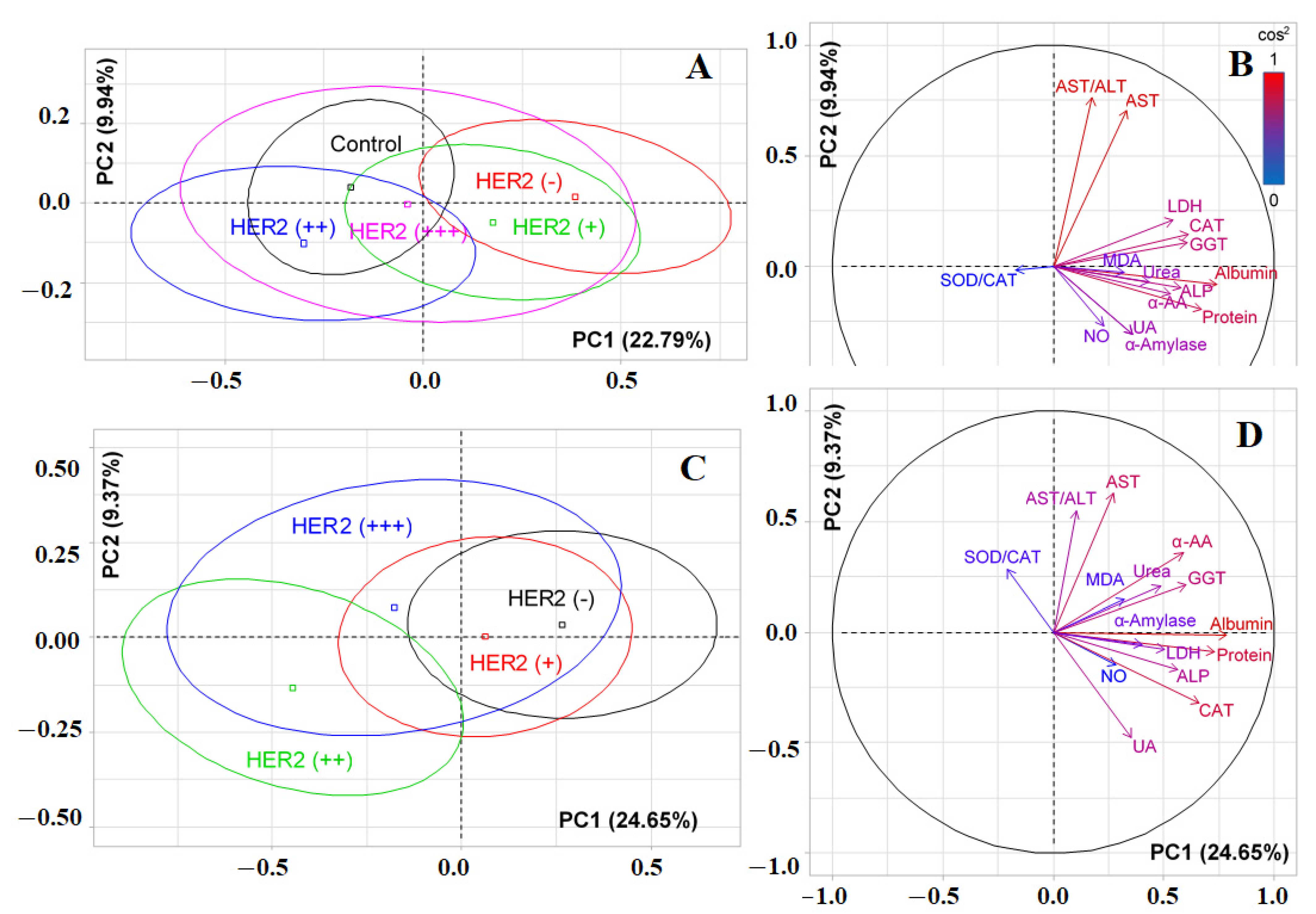
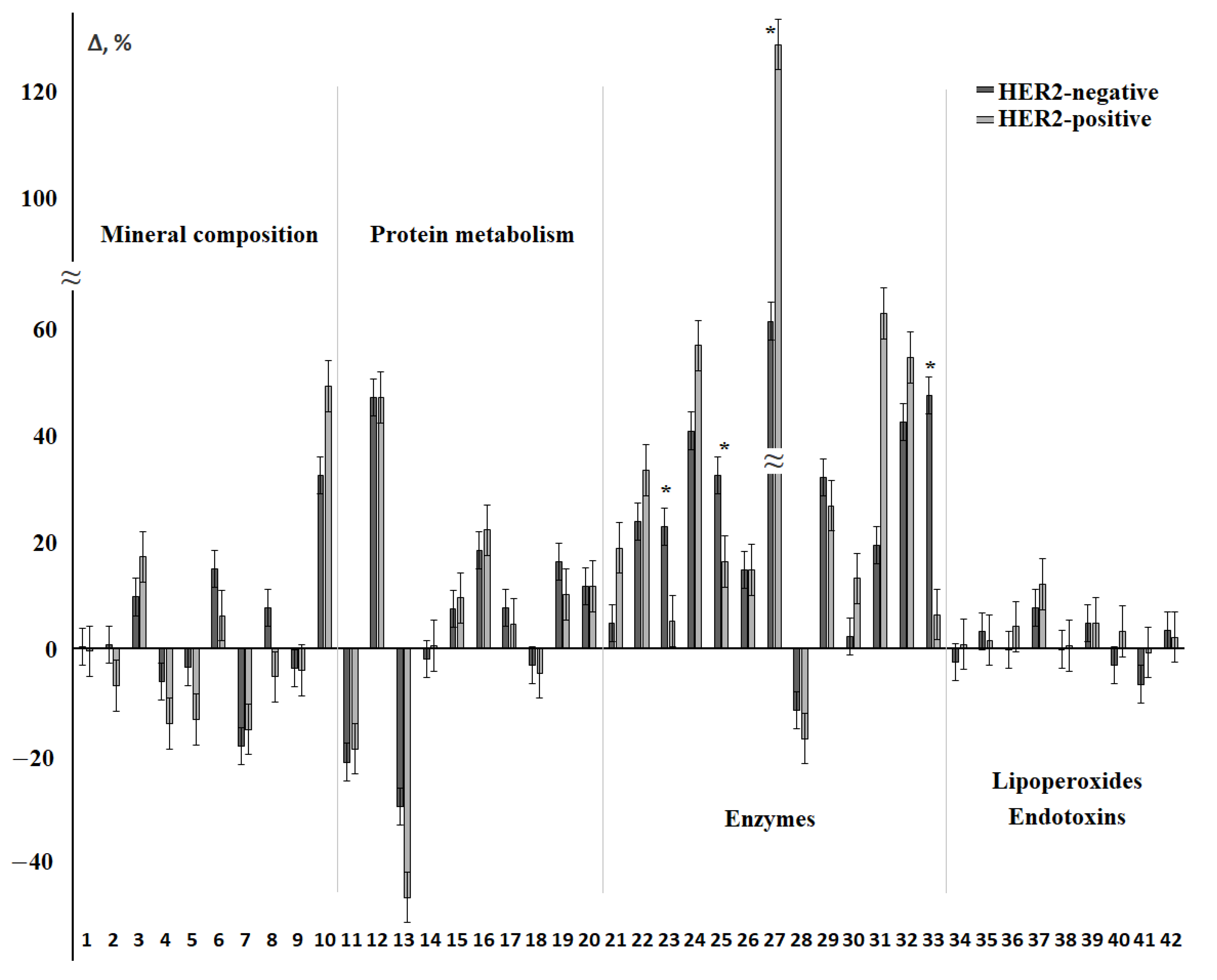
3.2. Changes in the Salivary Biochemical Composition of Breast Cancer Patients Depending on the HER2 Status
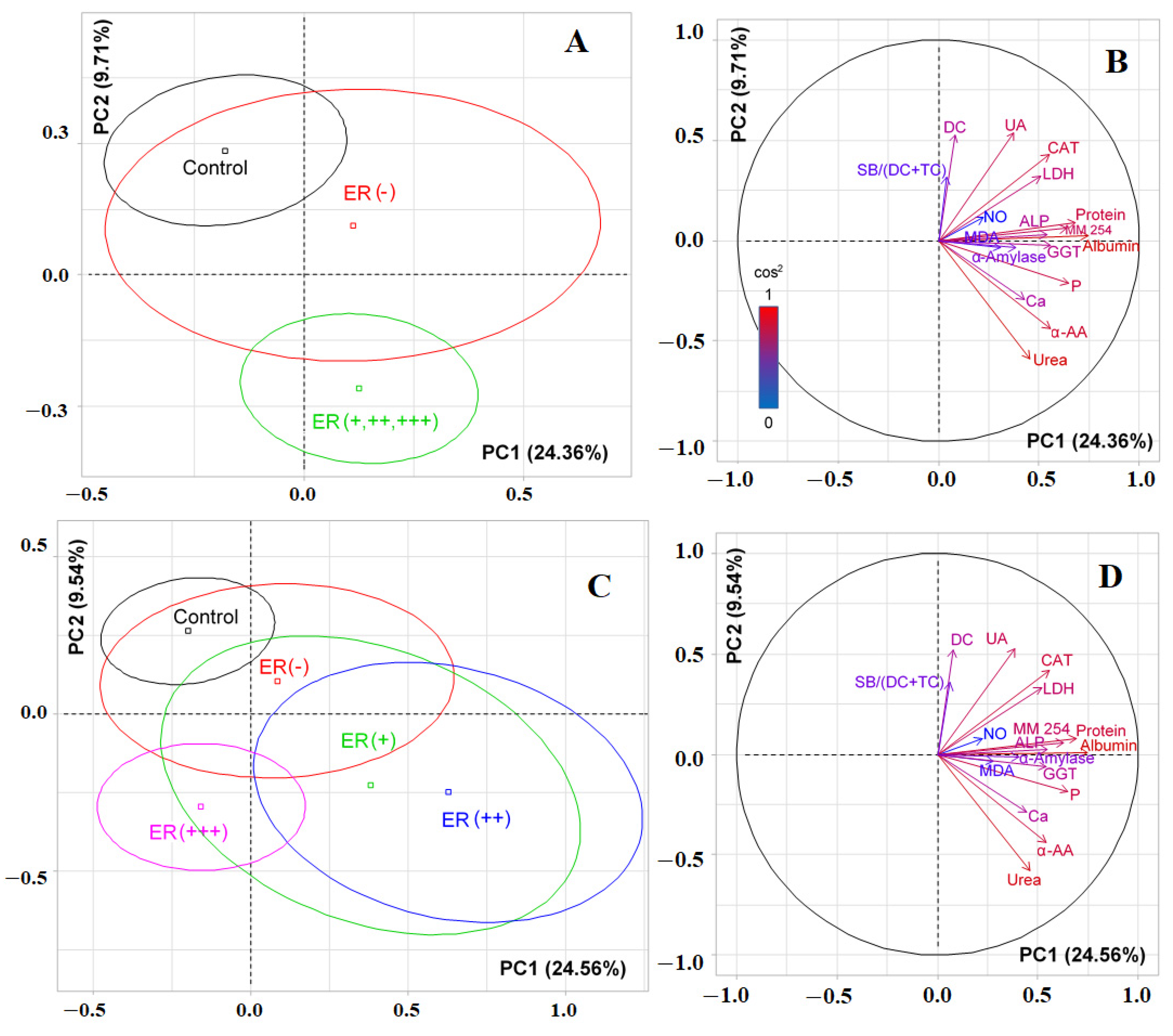
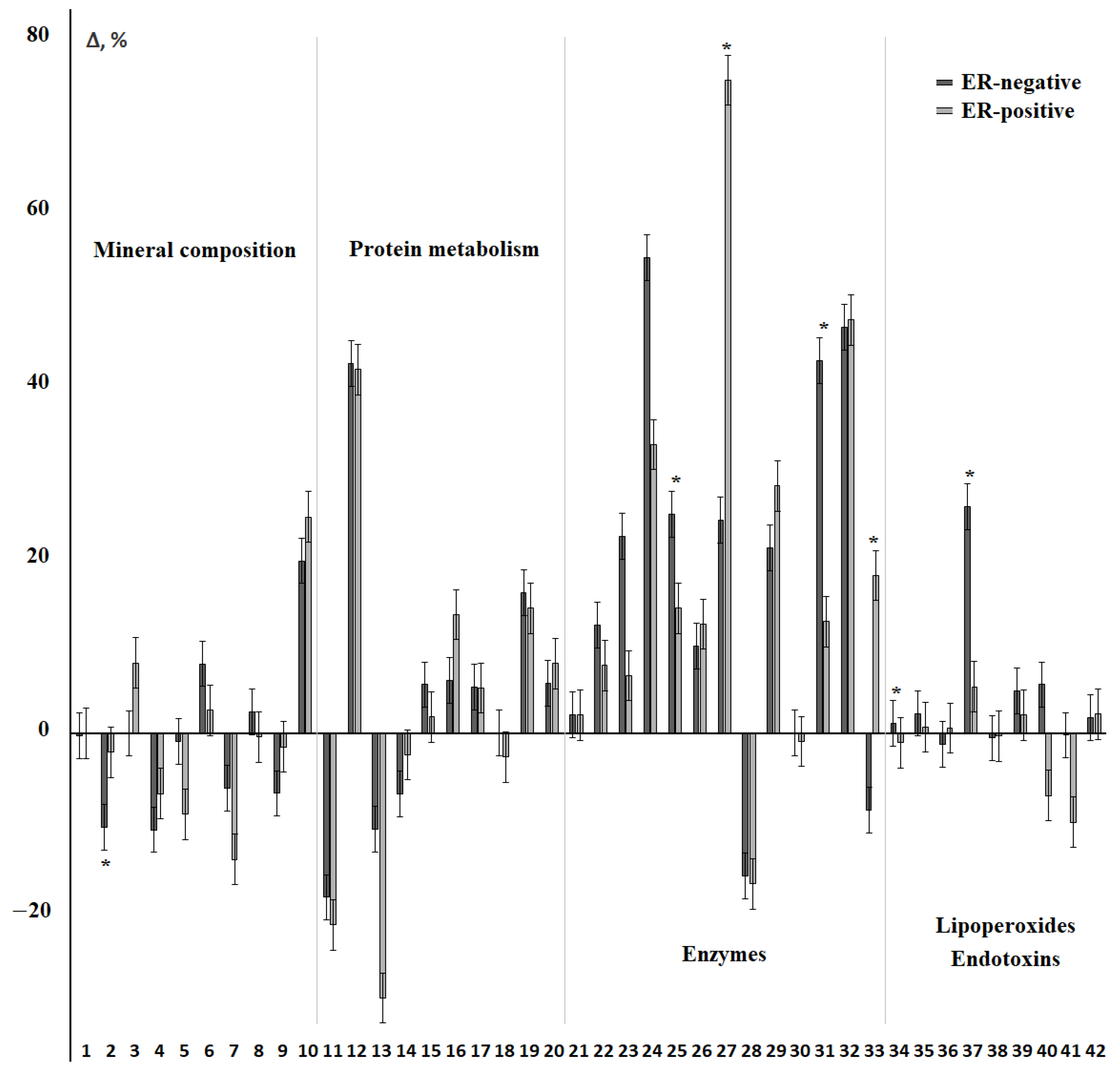
3.3. Changes in the Salivary Biochemical Composition of Breast Cancer Patients Depending on the ER Status
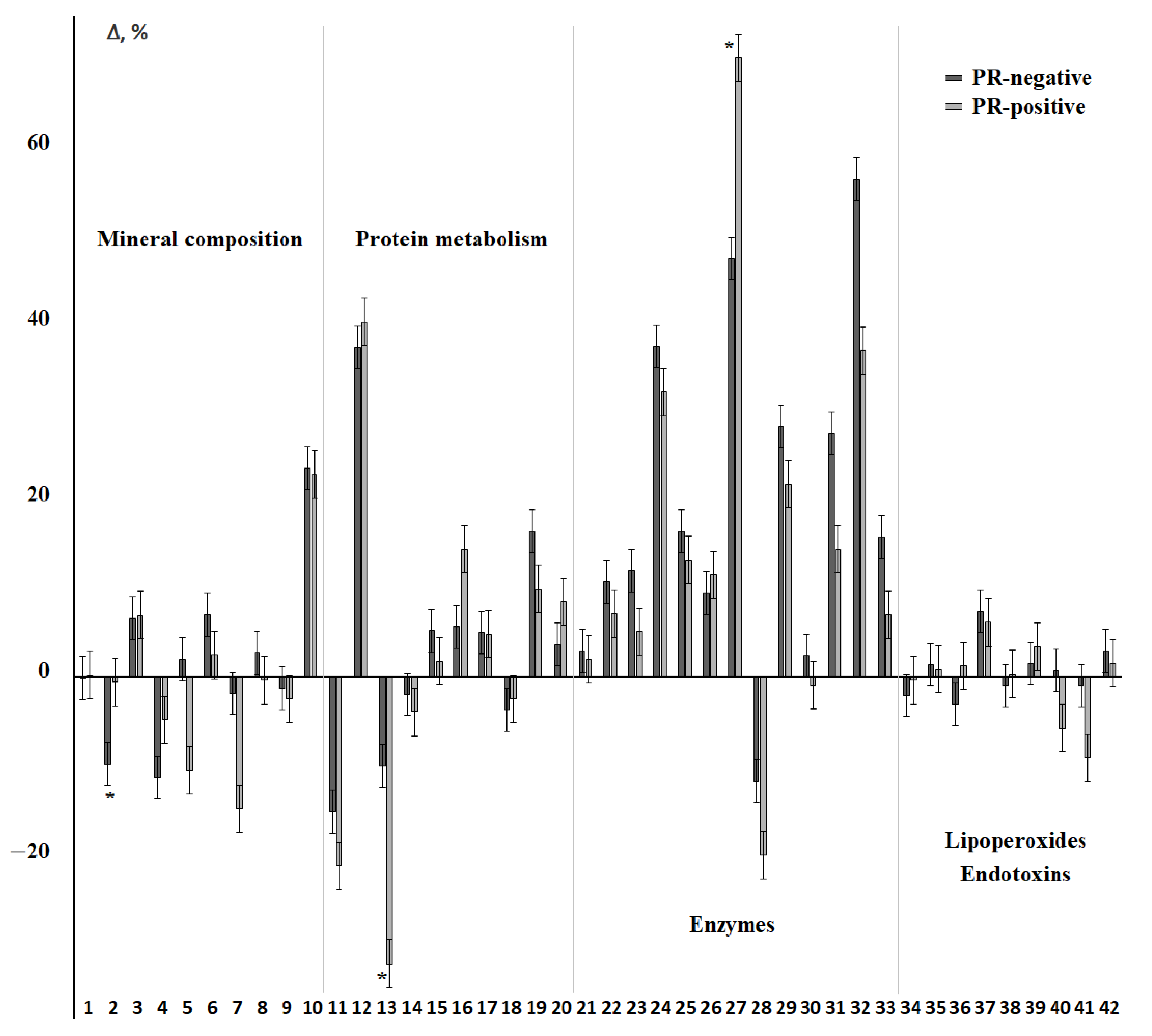
3.4. Changes in the Salivary Biochemical Composition of Breast Cancer Patients Depending on the PR Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017. A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed]
- Veronesi, U.; Boyle, P. Breast Cancer. In International Encyclopedia of Public Health, 2nd ed.; Reference Module in Biomedical Sciences; Academic Press: Cambridge, MA, USA, 2017; pp. 272–280. [Google Scholar]
- Wark, P.A.; Peto, J. Cancer Epidemiology, 2nd ed.; Reference Module in Biomedical Sciences International Encyclopedia of Public Health; Academic Press: Oxford, UK, 2017; pp. 339–346. [Google Scholar]
- Grabinski, V.F.; Brawley, O.W. Disparities in Breast Cancer. Obstet. Gynecol. Clin. N. Am. 2022, 49, 149–165. [Google Scholar] [CrossRef]
- Sullivan, C.L.; Butler, R.; Evans, J. Impact of a Breast Cancer Screening Algorithm on Early Detection. J. Nurse Pract. 2021, 17, 1133–1136. [Google Scholar] [CrossRef]
- Poddubnaya, I.V.; Kolyadina, I.V.; Kalashnikov, N.D.; Borisov, D.A.; Makarova, M.V. Population “portrait” of breast cancer in Russia: Analysis of Russian registry data. Sovrem. Onkol. 2015, 17, 25–29. [Google Scholar]
- Loi, S. The ESMO clinical practise guidelines for early breast cancer: Diagnosis, treatment and follow-up: On the winding road to personalized medicine. Ann. Oncol. 2019, 30, 1183–1184. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R.; Kiran, P. Tumor microenvironment pathways: Cross regulation in breast cancer metastasis. Genes Dis. 2022, 9, 310–324. [Google Scholar] [CrossRef]
- Kushlinskiy, N.Y.; Krasil’nikov, M.A. Biological Markers of Tumors: Fundamental and Clinical Studies; M: Izd-vo RAMN: Moscow, Russia, 2017; p. 632. [Google Scholar]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed]
- Nuyten, D.S.A.; Hastie, T.; Chi, J.-T.A.; Chang, H.Y.; van de Vijver, M.J. Combining biological gene expression signatures in predicting outcome in breast cancer: An alternative to supervised classification. Eur. J. Cancer 2008, 44, 2319–2329. [Google Scholar] [CrossRef][Green Version]
- Barzaman, K.; Karami, J.; Zarei, Z.; Hosseinzadeh, A.; Kazemi, M.H.; Moradi-Kalbolandi, S.; Safari, E.; Farahmand, L. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 2020, 84, 106535. [Google Scholar] [CrossRef]
- Luond, F.; Tiede, S.; Christofori, G. Breast cancer as an example of tumour heterogeneity and tumour cell plasticity during malignant progression. Br. J. Cancer 2021, 125, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Bertos, N.R.; Park, M. Breast cancer-one term, many entities? J. Clin. Investig. 2011, 121, 3789–3796. [Google Scholar] [CrossRef] [PubMed]
- Kolyadina, I.V.; Poddubnaya, I.V.; Frank, G.A.; Komov, D.V.; Ozhereliev, A.S.; Karseladze, A.I.; Ermilova, V.D.; Vishnevskaya, Y.V.; Makarenko, N.P.; Kerimov, R.A.; et al. The Prognostic Significance of Receptor Status in Patients with Early Breast Cancer. Mod. Technol. Med. 2012, 4, 48–53. [Google Scholar]
- Rapado-González, Ó.; Majem, B.; Muinelo-Romay, L.; López-López, R.; Suarez-Cunqueiro, M.M. Cancer Salivary Biomarkers for Tumours Distant to the Oral Cavity. Int. J. Mol. Sci. 2016, 17, 1531. [Google Scholar] [CrossRef]
- Bel’skaya, L.V. Possibilities of using saliva for the diagnosis of cancer. Klin. Lab. Diagn. (Russ. Clinical Lab. Diagn.) 2019, 64, 333–336. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.W.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. BBA-Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef]
- Roblegg, E.; Coughran, A.; Sirjani, D. Saliva: An all-rounder of our body. Eur. J. Pharm. Biopharm. 2019, 142, 133–141. [Google Scholar] [CrossRef]
- Tong, P.; Yuan, C.; Sun, X.; Yue, Q.; Wang, X.; Zheng, S. Identification of salivary peptidomic biomarkers in chronic kidney disease patients undergoing hemodialysis. Clin. Chim. Acta 2019, 489, 154–161. [Google Scholar] [CrossRef]
- Khurshid, Z.; Warsi, I.; Moin, S.F.; Slowey, P.D.; Latif, M.; Zohaib, S.; Zafar, M.S. Biochemical analysis of oral fluids for disease detection. Adv. Clin. Chem. 2021, 100, 205–253. [Google Scholar]
- Navazesh, M.; Dincer, S. Salivary Bioscience and Cancer; Salivary Bioscience; Springer Nature: Cham, Switzerland, 2020; pp. 449–467. [Google Scholar]
- Bonne, N.J.; Wong, D.T. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 2012, 4, 82. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.T.W. Salivaomics. J. Am. Dent. Assoc. 2012, 143, 19S–24S. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Nonaka, T.; Ye, Q.; Wei, F.; Wong, D.T.W. Salivaomics, Saliva-Exosomics, and Saliva Liquid Biopsy; Salivary Bioscience; Springer Nature: Cham, Switzerland, 2020; pp. 157–175. [Google Scholar]
- Mockus, M.; Prebil, L.; Ereman, R.; Dollbaum, C.; Powell, M.; Yau, C.; Benz, C.C. First Pregnancy Characteristics, Postmenopausal Breast Density, and Salivary Sex Hormone Levels in a Population at High Risk for Breast Cancer. BBA Clin. 2015, 3, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Oakman, C.; Tenori, L.; Biganzoli, L.; Santarpia, L.; Cappadona, S.; Luchinat, C.; Di Leo, A. Uncovering the metabolomic fingerprint of breast cancer. Int. J. Biochem. Cell Biol. 2011, 43, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Porto-Mascarenhas, E.C.; Assad, D.X.; Chardin, H.; Gozal, D.; De Luca Canto, G.; Acevedo, A.C.; Guerra, E.N. Salivary biomarkers in the diagnosis of breast cancer: A review. Crit. Rev. Oncol./Hematol. 2017, 110, 62–73. [Google Scholar] [CrossRef]
- Zhong, L.; Cheng, F.; Lu, X.; Duan, Y.; Wang, X. Untargeted saliva metabonomics study of breast cancer based on ultraperformance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Talanta 2016, 158, 351–360. [Google Scholar] [CrossRef]
- Takayama, T.; Tsutsui, H.; Shimizu, I.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Inoue, K.; Todoroki, K.; Min, J.Z.; Mizuno, H.; et al. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin. Chim. Acta 2016, 452, 18–26. [Google Scholar] [CrossRef]
- Jinno, H.; Murata, T.; Sunamura, M.; Sugimoto, M.; Kitagawa, Y. Breast cancer-specific signatures in saliva metabolites for early diagnosis. Poster Abstr. I/Breast 2015, 24 (Suppl. S1), S26–S86. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, Z.; Huang, Y.; Duan, Y.; Wang, X. Investigation of salivary free amino acid profile for early diagnosis of breast cancer with ultra-performance liquid chromatography-mass spectrometry. Clin. Chim. Acta 2015, 447, 23–31. [Google Scholar] [CrossRef]
- Liu, X.; Yu, H.; Qiao, Y.; Yang, J.; Shu, J.; Zhang, J.; Zhang, Z.; He, J.; Li, Z. Salivary Glycopatterns as Potential Biomarkers for Screening of Early-Stage Breast Cancer. eBioMedicine 2018, 28, 70–79. [Google Scholar] [CrossRef]
- Pereira, J.A.M.; Taware, R.; Porto-Figueira, P.; Rapole, S.; Câmara, J.S. Chapter 29—The salivary volatome in breast cancer. In Precision Medicine for Investigators, Practitioners and Providers; Faintuch, J., Faintuch, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 301–307. [Google Scholar]
- López-Jornet, P.; Aznar, C.; Ceron, J.J.; Tvarijonaviciute, A. Salivary biomarkers in breast cancer: A cross-sectional study. Support Care Cancer 2021, 29, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Assad, D.X.; Mascarenhas, E.C.P.; de Lima, C.L.; de Toledo, I.P.; Chardin, H.; Combes, A.; Acevedo, A.C.; Guerra, E.N.S. Salivary metabolites to detect patients with cancer: A systematic review. Int. J. Clin. Oncol. 2020, 25, 1016–1036. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, X.; Shu, J.; Hou, Y.; Chen, M.; Yu, H.; Ma, T.; Du, H.; Zhang, J.; Qiao, Y.; et al. Abnormal Galactosylated–Glycans recognized by Bandeiraea Simplicifolia Lectin I in saliva of patients with breast Cancer. Glycoconj. J. 2020, 37, 373–394. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillaryelectrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Bigler, L.R.; Streckfus, C.F.; Copeland, L.; Burns, R.; Dai, X.; Kuhn, M.; Martin, P.; Bigler, S.A. The potential use of saliva to detect recurrence of disease in women with breast carcinoma. J. Oral Pathol. Med. 2002, 31, 421–431. [Google Scholar] [CrossRef]
- Streckfus, C.F. Salivary Biomarkers to Assess Breast Cancer Diagnosis and Progression: Are We There Yet? In Saliva and Salivary Diagnostics; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L. A Catalogue of Altered Salivary Proteins Secondary to Invasive Ductal Carcinoma: A Novel In Vivo Paradigm to Assess Breast Cancer Progression. Sci. Rep. 2016, 6, 30800. [Google Scholar] [CrossRef]
- Ragusa, A.; Romano, P.; Lenucci, M.S.; Civino, E.; Vergara, D.; Pitotti, E.; Neglia, C.; Distante, A.; Romano, G.D.; Di Renzo, N.; et al. Differential Glycosylation Levels in Saliva from Patients with Lung or Breast Cancer: A Preliminary Assessment for Early Diagnostic Purposes. Metabolites 2021, 11, 566. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Martínez-Reglero, C.; Salgado-Barreira, Á.; Takkouche, B.; López-López, R.; Suárez-Cunqueiro, M.M.; Muinelo-Romay, L. Salivary biomarkers for cancer diagnosis: A meta-analysis. Ann. Med. 2020, 52, 131–144. [Google Scholar] [CrossRef]
- Murata, T.; Yanagisawa, T.; Kurihara, T.; Kaneko, M.; Ota, S.; Enomoto, A.; Tomita, M.; Sugimoto, M.; Sunamura, M.; Hayashida, T.; et al. Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res. Treat. 2019, 177, 591–601. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Solomatin, D.V.; Kosenok, V.K. Metabolic Features of Saliva in Breast Cancer Patients. Metabolites 2022, 12, 166. [Google Scholar] [CrossRef]
- Ilić, I.R.; Stojanović, N.M.; Radulović, N.S.; Živković, V.V.; Randjelović, P.J.; Petrović, A.S.; Božić, M.; Ilić, R.S. The Quantitative ER Immunohistochemical Analysis in Breast Cancer: Detecting the 3 + 0, 4 + 0, and 5 + 0 Allred Score Cases. Medicina 2019, 55, 461. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.; Dowsett, M.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V.; Kosenok, V.K.; Sarf, E.A. Chronophysiological features of the normal mineral composition of human saliva. Arch. Oral Biol. 2017, 82, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V.; Kosenok, V.K.; Massard, G. Endogenous Intoxication and Saliva Lipid Peroxidation in Patients with Lung Cancer. Diagnostics 2016, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Smolyakova, R.M.; Prokhorova, V.I.; Zharkov, V.V.; Lappo, S.V. Assessment of the binding capacity and transport function of serum albumin in patients with lung cancer. Nov. Khirurgii 2005, 13, 78–84. [Google Scholar]
- Chesnokova, N.P.; Barsukov, V.Y.; Ponukalina, E.V.; Agabekov, A.I. Regularities of changes in free radical destabilization processes of biological membranes in cases of colon ascendens adenocarcinoma and the role of such regularities in neoplastic proliferation development. Fundam. Res. 2015, 1, 164–168. [Google Scholar]
- Pankova, O.V.; Perelmuter, V.M.; Savenkova, O.V. Characteristics of proliferation marker expression and apoptosis regulation depending on the character of disregenerator changes in bronchial epithelium of patients with squamous cell lung cancer. Sib. Oncol. J. 2010, 41, 36–41. [Google Scholar]
- Sato, E.F.; Choudhury, T.; Nishikawa, T.; Inoue, M. Dynamic aspect of reactive oxygen and nitric oxide in oral cavity. J. Clin. Biochem. Nutr. 2008, 42, 8–13. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K.; Gundyrev, I.A. Biochemical Markers of Saliva in Lung Cancer: Diagnostic and Prognostic Perspectives. Diagnostics 2020, 10, 186. [Google Scholar] [CrossRef]
- Smirmova, O.V.; Borisov, V.I.; Guens, G.P. Immediate and long-term outcomes of drug treatment in patients with metastatic triple negative breast cancer. Malig. Tumours 2018, 8, 68–77. [Google Scholar] [CrossRef][Green Version]
- Knaś, M.; Maciejczyk, M.; Waszkiel, D.; Zalewska, A. Oxidative stress and salivary antioxidants. Dent. Med. Probl. 2013, 50, 461–466. [Google Scholar]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Indicators of L-arginine metabolism in saliva: A focus on breast cancer. J. Oral Biosci. 2021, 63, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Zhou, M.; Liu, H.; Ding, Y.; Khong, H.T.; Yu, D.; Fodstad, O.; Tan, M. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene 2009, 28, 3689–3701. [Google Scholar] [CrossRef] [PubMed]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Age and gender characteristics of the biochemical composition of saliva: Correlations with the composition of blood plasma. J. Oral Biol. Craniofacial Res. 2020, 10, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Almasri, N.M.; Al Hamad, M. Immunohistochemical evaluation of human epidermal growth factor receptor 2 and estrogen and progesterone receptors in breast carcinoma. Breast Cancer Res. 2005, 7, 598–604. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L.; Dellinger, T.; Dai, X.; Cox, W.J.; McArthur, A.; Kingman, A.; Thigpen, J.T. Reliability assessment of soluble c-erb B-2 concentrations in the saliva of healthy women and men. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2001, 91, 174–179. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L.; Tucci, M.; Thigpen, J.T. A preliminary study of CA15-3, c-erbB-2, epidermal growth factor receptor, cathepsin-D, and p53 in saliva among women with breast carcinoma. Cancer Investig. 2000, 18, 101–109. [Google Scholar] [CrossRef]
- Streckfus, C.; Bigler, L. The Use of Soluble, Salivary c-erb B-2 for the Detection and Post-operative Follow-up of Breast Cancer in Women: The Results of a Five-year Translational Research Study. Adv. Dent. Res. 2005, 18, 17–24. [Google Scholar] [CrossRef]

| Feature | Breast Cancer, n = 487 | Control Group, n = 298 |
|---|---|---|
| Age, years | 54.5 [47.0; 56.0] | 49.3 [43.8; 56.1] |
| Histological type | ||
| Ductal | 227 (46.6%) | - |
| Lobular | 86 (17.7%) | - |
| Mixed (Ductal + Lobular) | 12 (2.5%) | - |
| Rare forms | 58 (11.9%) | - |
| Unknown | 104 (21.3%) | - |
| Clinical Stage | ||
| Stage I | 119 (24.4%) | - |
| Stage IIa | 123 (25.3%) | - |
| Stage IIb | 88 (18.1%) | - |
| Stage IIIa | 55 (11.3%) | - |
| Stage IIIb | 47 (9.6%) | - |
| Stage IV | 55 (11.3%) | - |
| Subtype | ||
| Luminal A-like | 64 (13.1%) | - |
| Luminal B-like (HER2+) | 230 (47.4%) | - |
| Luminal B-like (HER2−) | 63 (12.9%) | - |
| Non-Luminal (HER2+) | 38 (7.8%) | - |
| Basal-like (Triple-negative) | 28 (5.7%) | - |
| Unknown | 64 (13.1%) | - |
| HER2-status | ||
| HER2-negative HER2 (−) | 156 (36.1%) | - |
| HER2-positive | 276 (63.9%) | - |
| HER2 (+) | 124 (44.9%) | - |
| HER2 (++) | 83 (30.1%) | - |
| HER2 (+++) | 69 (25.0%) | - |
| ER-status | ||
| ER-negative ER (−) | 77 (17.7%) | - |
| ER-positive | 359 (82.3%) | - |
| ER (+) | 60 (16.7%) | - |
| ER (++) | 77 (21.4%) | - |
| ER (+++) | 222 (61.9%) | - |
| PR-status | ||
| PR-negative PR (−) | 125 (28.7%) | - |
| PR-positive | 310 (71.3%) | - |
| PR (+) | 64 (20.6%) | - |
| PR (++) | 79 (25.5%) | - |
| PR (+++) | 167 (53.9%) | - |
| No. | Indicators | Lum A | Lum B (+) | Lum B (−) | BL | Non-Lum | Kruskal–Wallis Test (H, p) |
|---|---|---|---|---|---|---|---|
| 1 | pH | 0.6 | −0.1 | −0.2 | 1.1 | −1.3 | 4.125; 0.5316 |
| 2 | Calcium, mmol/L | 8.7 | −4.2 | −1.2 | −11.5 | −9.7 | 6.424; 0.2671 |
| 3 | Phosphorus, mmol/L | 0.7 | 7.5 | 8.7 | 9.7 | −5.4 | 7.439; 0.1900 |
| 4 | Ca/P-ratio, c.u. | 0.5 | −8.4 | −12.4 | −20.2 | −6.0 | 8.226; 0.1442 |
| 5 | Sodium, mmol/L | −21.4 | −12.5 | 7.9 | 5.0 | −15.2 | 6.519; 0.2589 |
| 6 | Potassium, mmol/L | −1.3 | 2.1 | 21.6 | 23.0 | 5.3 | 7.879; 0.1631 |
| 7 | Na/K-ratio, c.u. | −15.5 | −14.0 | −13.4 | −17.0 | 3.3 | 5.517; 0.3560 |
| 8 | Chlorides, mmol/L | −3.9 | −2.0 | 6.6 | 12.8 | −2.1 | 7.662; 0.1759 |
| 9 | Magnesium, mmol/L | −6.5 | −0.7 | 0.1 | −7.4 | −1.4 | 1.992; 0.8503 |
| 10 | NO, μmol/L | 22.4 | 21.6 | 40.7 | 19.0 | 49.3 | 16.02; 0.0068 * |
| 11 | Protein, mg/mL | −24.5 | −21.4 | −12.8 | −5.7 | −10.7 | 70.13; 0.0000 * |
| 12 | Urea, mmol/L | 33.0 | 42.6 | 46.5 | 41.0 | 36.0 | 66.45; 0.0000 * |
| 13 | Uric acid, μmol/L | −28.8 | −34.0 | −21.9 | 2.6 | −9.2 | 7.819; 0.1665 |
| 14 | Lactic acid, mmol/L | 10.4 | −3.0 | −6.4 | −1.5 | −4.9 | 4.204; 0.5205 |
| 15 | Pyruvic acid, μmol/L | −1.8 | 1.8 | 10.7 | 8.9 | 5.4 | 4.440; 0.4879 |
| 16 | Albumin, mg/mL | 14.7 | 8.8 | 40.1 | 0.5 | 29.5 | 6.786; 0.2371 |
| 17 | α-Aminoacids, mmol/L | 3.8 | 4.8 | 9.1 | 6.5 | 5.2 | 29.84; 0.0000 * |
| 18 | Imidazole compounds, mmol/L | −7.9 | −3.9 | 13.2 | 3.9 | −2.6 | 6.158; 0.2912 |
| 19 | Sialic acids, mmol/L | 0.0 | 13.8 | 13.8 | 37.9 | −5.2 | 7.038; 0.2179 |
| 20 | Seromucoids, c.u. | 8.8 | 6.6 | 8.2 | 8.8 | 2.7 | 6.123; 0.2944 |
| 21 | ALT, U/L | 2.0 | 10.0 | 6.0 | 10.0 | 16.0 | 0.9695; 0.9650 |
| 22 | AST, U/L | 16.4 | 10.4 | 16.4 | 26.9 | 10.4 | 9.703; 0.0841 |
| 23 | AST/ALT-ratio, c.u. | 16.7 | 1.4 | 19.0 | 26.0 | 11.8 | 8.854; 0.1150 |
| 24 | LDH, U/L | 43.1 | 31.8 | 4.6 | 21.3 | 65.3 | 18.78; 0.0021 * |
| 25 | ALP, U/L | 13.8 | 6.9 | 44.8 | 34.7 | 24.1 | 21.68; 0.0006 * |
| 26 | GGT, U/L | 8.7 | 12.3 | 17.3 | 13.2 | 2.4 | 40.03; 0.0000 * |
| 27 | Catalase, nkat/mL | −14.9 | −22.5 | 7.7 | -18.2 | −10.8 | 15.24; 0.0094 * |
| 28 | Superoxide dismutase, c.u. | 2.3 | 31.8 | 45.5 | 27.3 | 15.9 | 10.79; 0.0557 |
| 29 | α-Amylase, U/L | 60.7 | 65.1 | 141.0 | 60.5 | 20.6 | 17.41; 0.0038 * |
| 30 | Antioxidant activity, mmol/L | −14.1 | −0.8 | 4.9 | 2.4 | −11.0 | 2.688; 0.7479 |
| 31 | Peroxidase, c.u. | −9.6 | 9.6 | 16.4 | 20.5 | 86.3 | 7.194; 0.2066 |
| 32 | SOD/Catalase-ratio, c.u. | 3.2 | 53.6 | 53.3 | 80.3 | 36.9 | 12.12; 0.0332 * |
| 33 | SOD/Peroxidase-ratio, c.u. | −9.6 | 12.4 | 88.9 | 51.8 | −35.0 | 13.97; 0.0158 * |
| 34 | Diene conjugates, c.u. | −3.1 | −0.3 | −2.8 | 1.7 | 0.7 | 8.617; 0.1254 |
| 35 | Triene conjugates, c.u. | 6.6 | −0.7 | −0.9 | 1.1 | 2.0 | 3.988; 0.5511 |
| 36 | Schiff bases, c.u. | 10.2 | 0.6 | −3.2 | −3.3 | 3.1 | 7.175; 0.2079 |
| 37 | MDA, μmol/L | 1.3 | 6.4 | 9.6 | 35.9 | 4.5 | 22.95; 0.0003 * |
| 38 | SB/(DC+TC)-ratio, c.u. | 5.9 | −0.4 | −2.2 | −0.7 | −0.2 | 9.043; 0.1074 |
| 39 | SB/TC-ratio, c.u. | 7.2 | 1.6 | −1.1 | 3.0 | 7.2 | 10.05; 0.0739 |
| 40 | MM 254, c.u. | −18.8 | −5.4 | 5.4 | 24.1 | 1.7 | 7.962; 0.1583 |
| 41 | MM 280, c.u. | −16.7 | −8.0 | −10.3 | 22.0 | −5.9 | 6.170; 0.2901 |
| 42 | MM 280/254 | 2.8 | 0.9 | 3.1 | 4.5 | 1.0 | 4.729; 0.4498 |
| No. | Indicators | Kruskal–Wallis Test (H, p) + Control Group | ||
|---|---|---|---|---|
| HER2 | ER | PR | ||
| 1 | pH | 2.858; 0.5818 | 3.467; 0.4829 | 0.7895; 0.9399 |
| 2 | Calcium, mmol/L | 5.365; 0.2518 | 8.055; 0.0896 ** | 7.971; 0.0926 ** |
| 3 | Phosphorus, mmol/L | 6.013; 0.1982 | 9.168; 0.0570 ** | 3.688; 0.4498 |
| 4 | Ca/P-ratio, c.u. | 6.397; 0.1714 | 5.822; 0.2128 | 6.498; 0.1649 |
| 5 | Sodium, mmol/L | 3.038; 0.5515 | 4.154; 0.3856 | 3.286; 0.5112 |
| 6 | Potassium, mmol/L | 5.475; 0.2419 | 1.370; 0.8494 | 1.119; 0.8913 |
| 7 | Na/K-ratio, c.u. | 5.651; 0.2268 | 3.911; 0.4182 | 6.548; 0.1618 |
| 8 | Chlorides, mmol/L | 4.632; 0.3272 | 5.517; 0.2382 | 5.454; 0.2483 |
| 9 | Magnesium, mmol/L | 0.6162; 0.9612 | 1.312; 0.8593 | 0.6850; 0.9532 |
| 10 | NO, μmol/L | 16.04; 0.0030 * | 17.23; 0.0017 * | 20.47; 0.0004 * |
| 11 | Protein, mg/mL | 75.09; 0.0000 * | 89.55; 0.0000 * | 79.20; 0.0000 * |
| 12 | Urea, mmol/L | 65.41; 0.0000 * | 66.16; 0.0000 * | 65.78; 0.0000 * |
| 13 | Uric acid, μmol/L | 8.990; 0.0614 ** | 11.65; 0.0202 * | 9.063; 0.0596 ** |
| 14 | Lactic acid, mmol/L | 4.819; 0.3064 | 2.271; 0.6860 | 4.215; 0.3776 |
| 15 | Pyruvic acid, μmol/L | 5.060; 0.2812 | 2.375; 0.6672 | 4.344; 0.3614 |
| 16 | Albumin, mg/mL | 8.648; 0.0705 ** | 7.980; 0.0923 ** | 12.68; 0.0130 * |
| 17 | α-Aminoacids, mmol/L | 24.58; 0.0001 * | 25.68; 0.0000 * | 26.72; 0.0000 * |
| 18 | Imidazole compounds, mmol/L | 4.478; 0.3452 | 4.757; 0.3132 | 4.123; 0.3896 |
| 19 | Sialic acids, mmol/L | 0.7944; 0.9392 | 2.169; 0.7047 | 1.640; 0.8017 |
| 20 | Seromucoids, c.u. | 6.399; 0.1713 | 7.597; 0.1075 | 10.70; 0.0302 * |
| 21 | ALT, U/L | 3.192; 0.5263 | 0.7748; 0.9418 | 3.865; 0.4245 |
| 22 | AST, U/L | 9.008; 0.0609 ** | 3.668; 0.4528 | 5.101; 0.2771 |
| 23 | AST/ALT-ratio, c.u. | 9.652; 0.0467 * | 3.293; 0.5101 | 9.622; 0.0473 * |
| 24 | LDH, U/L | 13.83; 0.0078 * | 14.50; 0.0059 * | 12.35; 0.0149 * |
| 25 | ALP, U/L | 14.46; 0.0060 * | 26.72; 0.0000 * | 18.83; 0.0008 * |
| 26 | GGT, U/L | 39.01; 0.0000 * | 36.20; 0.0000 * | 42.78; 0.0000 * |
| 27 | α-Amylase, U/L | 17.76; 0.0014 * | 16.05; 0.0030 * | 19.30; 0.0007 * |
| 28 | Catalase, nkat/mL | 14.51; 0.0058 * | 16.04; 0.0030 * | 19.41; 0.0007 * |
| 29 | Superoxide dismutase, c.u. | 6.113; 0.1908 | 5.349; 0.2533 | 6.311; 0.1771 |
| 30 | Antioxidant activity, mmol/L | 1.095; 0.8950 | 1.206; 0.8772 | 2.973; 0.5623 |
| 31 | Peroxidase, c.u. | 3.604; 0.4622 | 5.628; 0.2287 | 6.264; 0.1803 |
| 32 | SOD/Catalase-ratio, c.u. | 7.923; 0.0944 ** | 5.605; 0.2307 | 10.07; 0.0393 * |
| 33 | SOD/Peroxidase-ratio, c.u. | 5.470; 0.2424 | 6.314; 0.1769 | 11.47; 0.0217 * |
| 34 | Diene conjugates, c.u. | 7.459; 0.1135 | 9.019; 0.0606 ** | 2.224; 0.6946 |
| 35 | Triene conjugates, c.u. | 2.874; 0.5791 | 5.884; 0.2080 | 2.649; 0.6181 |
| 36 | Schiff bases, c.u. | 2.215; 0.6962 | 3.841; 0.4280 | 4.934; 0.2942 |
| 37 | MDA, μmol/L | 16.40; 0.0025 * | 19.98; 0.0005 * | 19.17; 0.0007 * |
| 38 | SB/(DC+TC)-ratio, c.u. | 2.115; 0.7146 | 8.384; 0.0785 ** | 4.159; 0.3849 |
| 39 | SB/TC-ratio, c.u. | 2.369; 0.6682 | 5.409; 0.2478 | 3.029; 0.5529 |
| 40 | MM 254, c.u. | 5.346; 0.2536 | 8.585; 0.0724 ** | 6.688; 0.1533 |
| 41 | MM 280, c.u. | 4.588; 0.3323 | 6.409; 0.1706 | 5.865; 0.2095 |
| 42 | MM 280/254 | 4.545; 0.3372 | 5.853; 0.2104 | 4.260; 0.3720 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bel’skaya, L.V.; Sarf, E.A. «Salivaomics» of Different Molecular Biological Subtypes of Breast Cancer. Curr. Issues Mol. Biol. 2022, 44, 3053-3074. https://doi.org/10.3390/cimb44070211
Bel’skaya LV, Sarf EA. «Salivaomics» of Different Molecular Biological Subtypes of Breast Cancer. Current Issues in Molecular Biology. 2022; 44(7):3053-3074. https://doi.org/10.3390/cimb44070211
Chicago/Turabian StyleBel’skaya, Lyudmila V., and Elena A. Sarf. 2022. "«Salivaomics» of Different Molecular Biological Subtypes of Breast Cancer" Current Issues in Molecular Biology 44, no. 7: 3053-3074. https://doi.org/10.3390/cimb44070211
APA StyleBel’skaya, L. V., & Sarf, E. A. (2022). «Salivaomics» of Different Molecular Biological Subtypes of Breast Cancer. Current Issues in Molecular Biology, 44(7), 3053-3074. https://doi.org/10.3390/cimb44070211







