Abstract
Background/Objectives: During cancer development, tumor cells accumulate somatic mutations, which could generate tumor-specific neoantigens. The aberrant protein can be recognized by the immune system as no-self, triggering an immune response against cells expressing this aberrant protein which could mediate tumor control or rejection. Since the expression of this mutated protein is exclusive to tumor cells, great efforts are being made to identify neoantigens of relevance in the development of new cancer treatment strategies. In comparison to adulthood tumors, pediatric malignancies present fewer mutations and thus fewer potential neoantigens. Acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy worldwide that can be benefited by the identification of neoantigens for immunotherapy approaches, the landscape of neoantigens in ALL is not well known, therefore the aim of our study was to identify potential neoantigens in ALL pediatric patients. Methods: To identify neoantigens in ALL, whole-exome sequencing of matched tumor-normal cells from pediatric cases was performed, with these data HLA-I alleles predicted and somatic mutations identified to propose potential neoantigens based on binding affinity of mutated peptide-HLA-I. Results: We found a strong correlation between tumor mutational burden (TMB) and neoantigen load (p < 0.001) but no correlation with prognosis. Furthermore, TMB and neoantigens were greater in ALL patients with at least one mutated DNA mismatch repair gene (p < 0.001). Also, differences between B- and T-cell ALL were found but statistical significance did not remain after permutation. Conclusions: The presence of neoantigens in pediatric cases with ALL makes the neoantigen-based immunotherapy a promising new strategy for the treatment of this malignancy, especially for patients with relapse.
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy worldwide, accounting for 30% of all pediatric malignancies in developed countries and up to 50% in developing ones [1,2]. It is also well documented that ALL displays a differential incidence rate among ethnic groups, being higher in Hispanics (~45 cases per million) than in Caucasians and Asians. ALL in the Mexican population not only shows one of the highest incidence rates (53 cases per million), but also has low frequency of good prognosis biomarkers [3,4]. For instance, around 10% of pediatric ALL cases from Mexico are positive to ETV6-RUNX1 but this biomarker accounts for ~25% of cases in other populations [5,6,7,8]. Furthermore, approximately 40% of Mexican pediatric cases die after an average follow-up of 3.9 years and more than a half of these deaths are related to relapse [9,10]. A plethora of evidence has highlighted the importance of acquired somatic mutations during cancer development, which is also considered a potential mechanism associated with relapse and overall survival (OS) rate in ALL [11]. Furthermore, acquired genetic alterations could result in the production of tumor-specific antigens or neoantigens, which might trigger an immune response against tumor cells expressing those mutated peptides and control tumor progression [12,13]. As the mismatch repair (MMR) system is responsible for recognizing and repairing incorrectly paired nucleotides that arise in DNA, its deficiency or inactivation may drive the rise in mutational burden. Mutations in DNA MMR genes have been previously related with risk and recurrence in ALL, which also impact neoantigen renewal [14,15,16,17]. Triggering the immune system through the use of neoantigens is constantly being highlighted as an important therapeutic target for cancer treatment [18,19,20,21]. As ALL emerges in lymphoid organs, where anti-tumor immune responses are typically initiated, its origin can cause immune sensing mechanisms failure or suppression of immune response, which might suggest that this hematological malignancy is poorly immunogenic. Nevertheless, the immune responsiveness in allogenic hematopoietic stem cell transplantation and in immunotherapy for relapsed or refractory ALL set the basis to hypothesize that leukemic cells could express new immunogenic specific peptides [22]. Even though it has been described that most hematologic malignancies have fewer protein coding mutations which generate fewer potential neoantigens than solid tumors, targeting a single high-quality neoantigen can be enough for disease control or even cure [18,23,24]. Supporting this hypothesis, a recent paper reported an increased neoantigen-specific CD8+ T-cell response in marrow-infiltrating lymphocytes from pediatric patients with ALL, including a neoantigen from the good prognosis biomarker ETV6-RUNX1, evidencing a responsiveness of immune system in ALL patients carrying this fusion gene [12]. Neoantigens are self-antigens produced in tumor cells that result from acquired somatic mutations during the cells malignant transformation, like chromosomal translocations, point mutations, and insertions and deletions (indels). Neoantigens can also derive from unique proteins or peptides produced by dysregulated RNA splicing, disordered post-translational modification and integrated viral open reading frames [13]. These neoantigens can be presented by human leukocyte antigen (HLA) molecules of malignant cells for T-cell recognition to initiate an anticancer immune response in patients [25,26,27,28]. Identification of candidate neoantigens with high-throughput genomics and bioinformatics technologies is generally based on the standard approach in which affinity of the putative peptides encoded by the identified mutations, for a given HLA I allele, can predict T-cell response [18,29,30,31]. Discovering unique tumor-specific neoantigens in ALL offers an enormous opportunity for more effective personalized treatment implementation. Considering that mutated gene products can act as tumor neoantigens, by exome sequencing of normal-tumor paired samples from pediatric patients we explored the landscape of coding somatic mutations in ALL to identify potential neoantigens associated with OS.
2. Results
2.1. Clinical Characteristics of Studied Population
Sixty-four children with de novo ALL were included. Saliva samples and bone marrow (BM) were collected at diagnosis; 35 of them were matched normal (saliva)-tumor (BM) tissues. Overall, 58% were male and 42% female, median age of the population was 104.2 months at diagnosis (range: 6.48–202.36 months) and 88% of patients were diagnosed with pre-B immunophenotype. Fourteen (22%) patients presented relapse and 12 (19%) died. The median of the follow-up of the patients was 72 months (range: 60–84 months) after diagnosis confirmation (Table 1).

Table 1.
Clinical characteristics of pediatric patients with acute lymphoblastic leukemia.
2.2. Distribution of HLA Class I Alleles
HLA genotype frequencies data were estimated for 64 ALL patients, of which 24 HLA–A*, 40 HLA–B* and 23 HLA–C* different alleles were identified. The most common HLA-A alleles in the ALL cohort were HLA-A*02:01 (15.6%) and HLA-A*24:02 (14.8%) present in 10 and 9 of the 64. Among HLA-B and -C alleles, HLA-B*39:05 (9.4%), HLA-B*07:02 (7%), HLA-C*07:01 (26.6%) and HLA-C*04:01 (16.4%) were the most frequent (Figure 1a–c). We evaluated the correlation among these six most frequent alleles with overall survival (OS) and event free survival (EFS), where event is defined as relapse or death, but no statistically significant associations were found (Table S1).
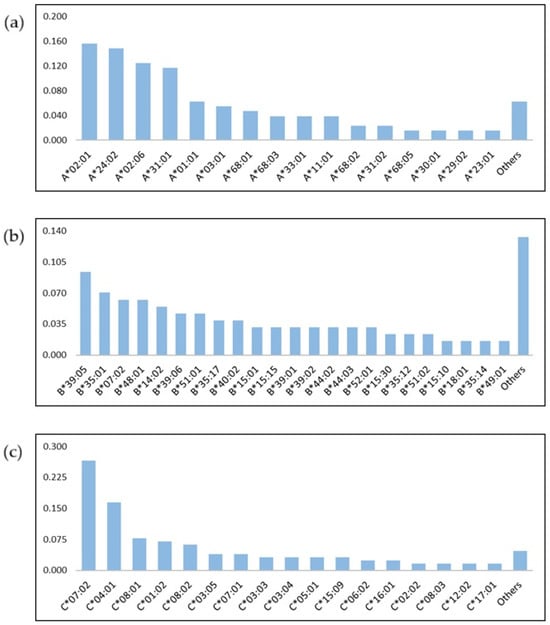
Figure 1.
Frequency of HLA alleles in pediatric patients with acute lymphoblastic leukemia. HLA–A* (a), HLA–B* (b) and HLA–C* (c) alleles.
2.3. High Correlation of Missense Mutation and Neoantigen Load
The identification of potential neoantigens was carried out by including 35 paired normal-tumor samples. A total of 33,315 somatically acquired missense mutations were identified. A total of 9880 neoantigens were predicted by considering those peptides that could bind to at least one of the patients’ HLA-A, -B, or -C alleles with an affinity < 500 nM or %Rank < 2%. Even though neoantigens must ultimately be validated for their presentation and recognition by T lymphocytes, we will use the term “neoantigens” to describe “potential neoantigens” throughout the text. A single mutation can generate multiple neoantigens by binding to diverse HLA alleles or in distinct registers; nevertheless, predicted neoantigens were excluded if they displayed the same affinity binding to HLA as their wild-type counterparts. The number of neoantigens ranged from 0 to 4385, with a median of 47 per patient. Among these, the most common were EELRLDHPA, EHLDLDFSI, ERGLRWLVT, LAICLACSL, LRLDHPAMA, LYQGQLNKL, RALEIQPGL, SDDNSASLL, THKSDIYSF, TVVAVDRYM, YSKEHLAMM and YYRAHIAQL, which had frequencies ranging from 6% to 9%. ALL patients had low percentage of shared neoantigens (6–9%). We further assessed expression of SYNGAP1, CUL1, COX11, PORCN, CPA2, PEX5L, TRAF3IP1, IRAK4, RRH, CLK4, CLK1, CLTCL1 genes in which the most frequent neoantigens were identified. We compared ALL patients and healthy subjects, from data retrieved by the TARGET initiative, RNA-seq level 3 data from cBioPortal [32] and GTEx project. Except for PORCN, all genes were differentially expressed among cases and controls (Figure S1). We identified sixteen HLA-corresponding alleles for each shared neoantigen and most of them belonged to HLA-B (9 alleles, 56%) and HLA-C (6 alleles, 37%); HLA-B*39:06 and HLA-C*07:02 alleles were found to bind to four neoantigens each. Only one allele corresponded to HLA-A (*24:02) group (Table S2).
Notably, we found that the number of neoantigens was positively correlated with the number of missense mutations (R = 0.994, p < 0.001) in ALL pediatric patients (Figure 2).
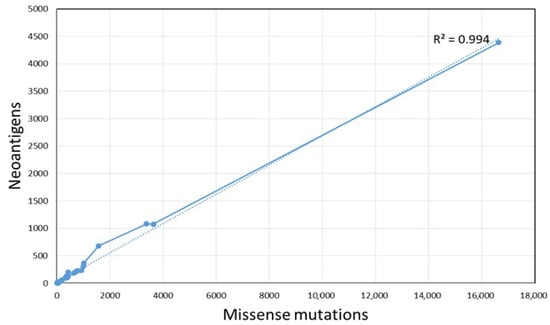
Figure 2.
Correlation of mutation burden and the number of potential neoantigens.
2.4. Association Among Clinical Features, Mutations and Neoantigens Numbers
When analyzing by gender, we found that male presented more mutations and neoantigens than female (X = 1301 vs. 687 and X= 360 vs. 217, respectively) but it did not reach statistical significance (p = 0.88 and 0.78; Table 2), whereas white blood cell (WBC), % of blasts in BM and risk stratification analyses showed no statistical significant differences, except for immunophenotype. Patients with T-cell ALL showed more missense mutations (X = 1991.7 vs. 872, p = 0.019) and neoantigens (X = 661.7 vs. 247.2, p = 0.025) (Table 2, Figure S2) but significance did not remain after permutation analysis.

Table 2.
Correlation mutations and neoantigens in clinical features groups.
2.5. Number of Mutations, Neoantigens and Neoantigen Frequency Survival Analysis
Patients were stratified in two groups to determine the association between the number of mutations (median = 174) and neoantigens (median = 47) compared to EFS and OS. We found no significant differences between them. As it has been previously described that low number of neoantigenic mutations could be used as an independent prognostic factor for clinical outcome in other cancers [33,34,35], we assessed the ratio of neoantigens per missense mutations and evaluated its association with EFS or OS. No statistically significant differences were found (Figure S3).
2.6. High Correlation Between Number of Mutations in DNA Mismatch Repair Genes and Neoantigens Load
As was previously documented, MMR-deficient cancers present a hypermutable state, which could generate a greater number of neoantigens that might be recognized by the immune system [26,36,37]. To test whether mutated DNA MMR genes could be implicated in an increase in number of mutations and consequently of neoantigens in ALL, we explored the mutations in MSH2, MSH6, MLH1 and PMS2. A total of 29 mutations were detected in 8 (23%) of the 35 patients; 2 (9%) of them had mutations in MSH2, 2 (6%) in MSH6, 8 (23%) in MLH1 and only 1 case (3%) in PMS2 (Table S3). Patients were stratified in two groups: (a) presented at least one mutation in one of these genes (MUT) and (b) presented no mutations (WT). The MUT group displayed higher mutational burden (Figure 3a) and neoantigen load (Figure 3b) than the WT group (p < 0.001) and statistical significance remains after the permutation test (p = 0.002 and p = 0.0008 for mutations and neoantigens, respectively). Notably, the patient carrying the highest mutation burden (n = 16,633) and neoantigen load (n = 4385) was the one who harbors mutations in most of the DNA mismatch repaired evaluated genes (12/29 in three genes) (Figure 3). No associations were found among number of mutations in these genes and EFS and OS (Figure S4).
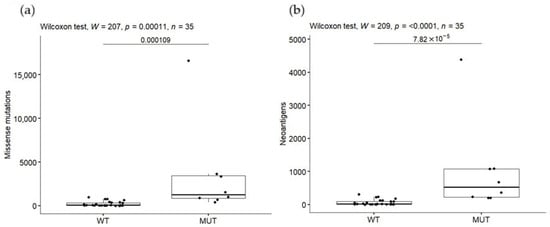
Figure 3.
Mutational burden (a) and tumor neoantigen load (b) in patients carrying missense mutations in DNA mismatch repair genes.
3. Discussion
The development of immunotherapy in recent years for ALL treatment has been focused on tumor-associated antigens which are overexpressed in leukemic cells but also expressed by normal cells in lower levels or in a specific stage or condition [38,39,40]. For instance, CD19 which is highly expressed during B-lineage neoplastic transformation, is a target for blinatumomab, a bispecific antibody, and for Tisagenlecleucel, a chimeric antigen receptor (CAR)-T cells therapy [41,42,43]. Nevertheless, this protein is also expressed during the differentiation of the hematopoietic stem cells, and its expression continues through B-cells lineage differentiation [44]. Additionally, CAR-T cells therapy still faces some challenges, such as reduced effectiveness in relapsed or chemoresistant patients, and high toxic side effects (cytokine release syndrome, B-cell aplasia, etc.). In fact, neurotoxicity for treatments based on a combination of bispecific antibodies and CAR-T has been observed [43,45,46]. Thus, discovering unique tumor-specific proteins in leukemic cells offers an enormous opportunity for more effective treatments. Even though it has been described that hematologic malignancies have fewer neoantigens than solid tumors [18,23,24], the immune response in allogenic hematopoietic stem cell transplantation and in immunotherapy for relapsed or refractory ALL [22] sets the basis to hypothesize that leukemic cells could express new immunogenic specific peptides derived from acquired coding mutations. Accordingly, we determined the missense mutational burden and neoantigen load in pediatric cases with ALL. Based on the germline exome data, we determined the frequency of HLA-A, -B and -C alleles in our ALL pediatric patients; further, we predicted the neoantigens with high binding affinity (affinity < 500 nM or %Rank < 2%) to patients’ HLA alleles.
In HLA-I alleles distribution, A*02:01, A*24:02, B*35:01, B*39:05, C*07:02 and C*04:01 were among the most frequent alleles as has been reported in Mexican ethnic groups like Tarasco, Tarahumara and Mestizo [47,48,49,50,51,52], as well as ALL cases [53,54]. Identifying the most common HLA alleles could be relevant for developing vaccines against human malignancies. For instance, the most frequent HLA-A alleles in our study, HLA-A*02:01 and -A*24:02, have been previously shown to be targeted for immunogenic approaches in cancer [55,56]. Both alleles have been associated with a variety of clinical outcomes in several diseases, but no association was found among them and clinical and demographic characteristics was identified in the present analysis [55,57,58,59,60].
A correlation between high missense mutational burden and predicted neoantigens load was found. Interestingly, our data differs from those published previously. First of all, a study of 100,000 human cancer genomes reported that pediatric malignancies, including pediatric ALL, had low abundance of somatic mutations (tumor mutational burden) [61]. Secondly, the coding mutations of this hematologic malignancy have been poorly explored as sources of neoantigens. To our knowledge, the only existing study was carried out by Zamora, whom by screening tumor-specific somatic missense mutations and gene fusions for their potential to generate neoantigens (strong putative neoantigens if IC50 < 150 nM, intermediate to weak < IC50 150–500 nM and as putative nonbinders > 500 nM) found few predicted neoantigens per patient (2–23) [12]. Discrepancies among our study and Zamora’s findings could be associated with the data source (RNASeq) and neoantigen filtering criteria. Our study not only includes neoepitopes with affinity (IC50 < 500 nM) values as Zamora’s, but also, we consider the percent rank (<2%) score of binding affinity and the difference in binding affinity of the mutant peptide to its wild-type counterpart (differential agretopic index: DAI). Rank score is less biased than binding affinity when comparing binding between multiple HLA alleles and candidate neoantigens; rank < 2% have higher probability to be presented by the MHC-I molecule than neopeptides with percent rank score > 2 [62]. High DAI score is related to the ability of neoantigens to protect against the tumor [31,63,64]. In addition, we selected nonamer peptides, while Zamora’s included wider peptides (14aa); also, most of the included cases by Zamora were ETV6-RUNX1 positive (7/9). Due to the presence of common coding mutations and/or fusions genes (ETV6-RUNX1, E2A-PBX1, BCR-ABL, etc.) within subgroups of patients, it has been reported that hematologic malignancies have more shared neoantigens than most solid tumors [24,65]. Contrary to what was expected, shared neoantigens in our cohort were lower than private or personal neoantigens. Even when data of fusion genes for our patients was not available, the frequency of the highest shared predicted neoantigen was around the same reported for children ETV6-RUNX1 positive (7.4%) with ALL from Mexico [66]. Even predicted neoantigens were heterogeneous across different patients; shared neoantigens such as EELRLDHPA, EHLDLDFSI, ERGLRWLVT, LAICLACSL, LRLDHPAMA, LYQGQLNKL, RALEIQPGL, SDDNSASLL, THKSDIYSF, TVVAVDRYM, YSKEHLAMM and YYRAHIAQ could be promising candidates for the development of immunotherapy regimens based on neoantigen vaccines. The aim of these vaccines is training the immune system against tumor cells in patients which are positive to tumor-specific neoantigens [67,68]. Neoantigen vaccines can be developed using peptides, mRNA, DNA and dendritic cells. Although these neoantigen vaccines could act as individualized drugs, combinations with chemotherapy, radiotherapy, immune checkpoint inhibitors, etc., might improve the efficacy of anticancer treatments based on tumor-specific neoantigens [67]. In addition to ALL and accounting that the identified neoantigens in this study were located in genes previously associated with other malignancies (breast cancer, melanoma, non-small cell lung cancer, etc.), we cannot discard their potential clinical application in the treatment of those tumors [69,70,71,72,73,74,75,76].
After in silico testing of whether the neoantigens can be presented by the MHC using computational prediction tools, additional steps are needed to know if the predicted neoantigens can be recognized by T-cells and induce an immune response: (a) assessing peptide transport from the cytoplasm to the endoplasmic reticulum mediated by transporters associated with antigen processing is a crucial process in the intracellular presentation of cytotoxic T-cell epitopes [77]; (b) determining the proteins’ or peptides’ expression abundance, either by evaluating the source of the proteins, the gene expression, or peptides’ expression by mass spectrometry analyses [78,79,80,81]; and (c) functional analysis to test whether the neoantigen can be recognized by a T-cell receptor and trigger T-cell activation. These tests could be made through assays as enzyme-linked immunosorbent spot (ELISPOT), which measure cytokine release, supported lipid bilayer (SLB)-based T-cell activation that allows monitoring interactions between the neoantigen-HLA and the T-cells and DNA barcode-based neoantigen–HLA tetramers and flow cytometry to identify the neoantigen–HLA–T-cells bounds [82,83,84,85]. Besides this, the expression levels of those genes having potential neoantigens were explored in ALL samples and normal tissue. RNA-seq data from cases and controls were retrieved from the TARGET initiative, RNA-seq phase 2 [32] and the GTEx project, respectively. Comparative analysis between cases and controls showed that except for PORCN, all genes were differentially expressed among ALL patients and controls, which increases the possibility of antigen expression abundance.
Another interesting finding was the relation among the higher mutational burden and neoantigen load with the presence of mutations in the DNA MMR genes MSH2, MSH6, MLH1 and PMS2. Patients negative for mutations in these genes have lower mutational burden and neoantigen load than the positive ones (X = 3532.25 vs. 1028 and 297.47 vs. 89.06, respectively; p = 0.0001 and 0.00007). These discoveries are similar not only to those previously documented in solid tumors such as colorectal cancer and lung adenocarcinoma but also to pediatric cancers [26]. Overall, MLH1 (41.5%) was the most commonly mutated DNA MMR gene, followed by MSH6 (34.5%) and MSH2 (21%). DNA MMR genes, particularly MLH1 and MSH2, have an important role in maintaining replication fidelity; thus, mutations in these genes lead to genome instability by not correcting base mismatches generated during DNA replication and cancer development [86]. This is particularly relevant since it has been shown that mutations in DNA MMR genes are sensitive to immune checkpoint blockade, even in healthy cells [36].
The association analysis among neoantigen frequency per mutation and clinical (WBC, immunophenotype, EFS and OS) variables showed no statistical differences among groups, which could be explained by the sample size. Since it has been reported that mutational burden and neoantigen load is correlated with prognosis in diverse cancer types (i.e., melanomas, non-small cell lung cancers and colorectal cancer, etc.) [87,88,89,90,91], predicted neoantigens identified in this study and preferentially those found in higher frequencies must be validated for their presentation and recognition by T lymphocytes to be named as neoantigens. To be proposed as neoantigens and potential therapeutic targets and considering their relevance in immunotherapy for ALL, predicted neoantigens must be tested in vitro.
4. Materials and Methods
4.1. Patients, Samples and Data
Samples of tumor (BM) and normal (saliva) tissues of ALL pediatric patients which were collected at diagnosis and before clinical treatment were included. To identify somatic mutations, which were defined as variants present in tumor tissue but not in normal tissue of the patient, we obtain germline DNA through saliva samples. All patients were under 18 years old and recruited from 2014 to 2016, with a follow-up of >36 months after diagnosis confirmation. Cases were residents of the metropolitan area of Mexico City and recruited from public hospitals and health institutions from Mexico City by the Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia (MIGICCL). ALL diagnosis was confirmed by pediatric hematologists or oncologists according to morphology and immunophenotype of leukemic cells. Clinical data such as gender, age at diagnosis, WBC count, immunophenotype, risk of relapse, relapse and death were registered from the patients’ medical records. Risk stratification criteria from The National Cancer Institute (NCI) was employed. Standard risk: from 1 to 9.99 years of age or WBC count < 50 × 109/L, and high risk: ≤1 or ≥10 years of age and/or WBC ≥ 50 × 109/L. Relapse was defined as disease recurrence in patients who had previously achieved morphologic remission, characterized by <5% blasts in bone marrow and clearance of extramedullary disease. Cases with Down syndrome were excluded from the analysis. This study was approved by the National Ethics and Research Committees of the Instituto Mexicano del Seguro Social (IMSS) (R-2015-785-121). Written informed consent was obtained from the children’s parents and children assented when possible.
4.2. DNA Isolation and Exome Sequencing
Germline DNA from saliva was isolated using prepIT.L2P ORAGENE Purification Kit (DNA Genotek Inc., Kanata, ON, Canada). Tumor DNA was obtained from mononuclear cells, which were isolated from BM mononuclear by density gradient centrifugation and using the Gentra Puregene Blood Kit (Gentra Systems Inc., Minneapolis, MN, USA) according to the manufacturer’s instructions. DNA purity and concentration were determined by Nanodrop spectrophotometer ND1000 (Thermo Fisher Scientific, Waltham, MA, USA).
Paired-end whole-exome sequencing (WES) was performed with Illumina NEXSeq, kit NexteraXT as previously described [92] and a depth of 100X. The sequenced raw data were aligned against hg38 human genome using the Burrows–Wheeler Aligner (BWA) [93,94]. In addition, removal of duplicate reads, SNPs and InDels realignment, and Base Quality Score Recalibration were performed using the Genome Analysis Toolkit (GATK), as previously described [95,96]. Mutations were annotated with Variant Effect Predictor (VEP, https://www.ensembl.org/info/docs/tools/vep/index.html, accessed on 1 February 2022) and MuTect (https://gatk.broadinstitute.org, accessed on 1 January 2022) was used to identify variants on processed data.
4.3. Class I HLA Alleles Prediction and Peptides–HLA Binding Predictions
Optitype (https://github.com/FRED-2/OptiType, accessed on 5 April 2021) with default settings was used for prediction of class I HLA alleles with 97% accuracy [97].
Tumor-specific missense mutations were identified to generate an amino acid sequence of potential neoantigens. Peptides covering nine tiling nonamers overlapping each missense mutation were extracted through a program designed with the SeqIO module of Biopython [98]. We tested peptide binding affinity with each of the patient’s HLA class I alleles by using NetMHCcons v1.1 program [99]. Predictions were made in May 2023, using the latest version of the algorithm at the time. Peptides were defined as candidate epitopes based on the affinity definition as half maximal inhibitory concentration (IC50) of <500 nM or % Rank < 2% of prediction score to a set of 200,000 random natural 9mer peptides for any HLA. Predicted binders, whether strong or intermediate, were considered as potential neoantigens.
4.4. Gene Expression Analysis of the Most Frequent Neoantigens
To know whether SYNGAP1, CUL1, COX11, PORCN, CPA2, PEX5L, TRAF3IP1, IRAK4, RRH, CLK4, CLK1 and CLTCL1 genes are expressed in ALL, RNA-seq data from ALL cases and controls were retrieved from the TARGET (Therapeutically Applicable Research to Generate Effective Treatments) initiative, RNA-seq level 3 [32] and The GTEx (Genotype-Tissue Expression) project (https://gtexportal.org/home/, accessed on 19 August 2025), respectively. The TARGET cohort consists of 463 ALL cases (phs000464) and GTEx of 407 non-cancerous subjects. Comparative analysis between cases and controls was performed using the TNMplot platform [100].
4.5. Mutations in DNA Mismatch Repair Genes
Based on the knowledge that genes involved in DNA MMR are associated with the number of neoantigens and favorable prognosis in diverse types of cancer [17,101], we explored the landscape of MSH2, MSH6, MLH1 and PMS2 genes through exome sequencing data.
4.6. Statistical Analysis
Statistical analyses were performed in R-Studio (R version 4.3.2) [102]. The Wilcoxon–Mann–Whitney tests were calculated to identify the statistical significance of differences between comparative groups regarding distribution of clinical and molecular characteristics. A p-value < 0.05 was considered statistically significant. Permutation test was performed by using lmPerm package (version 2.1.4). Default settings were used, in which iteration number reached 5000, and/or the standard error of the estimate of the p-value goes below the threshold (default = 10% of p-value). Linear regression analysis was performed to evaluate correlation between number of mutations and number of potential neoantigens. The hazard ratio was determined using a Cox proportional model to describe the effect of neoantigen burden in patients’ outcome. Correlations between nonsynonymous mutation burden and neoantigen burden, neoantigen burden and overall response were calculated using Spearman correlation formula.
5. Conclusions
In this study, we observed a high correlation between mutational burden and antigens load in ALL pediatric cases. Our data favors immunotherapy based on the discovery of neoantigens as an opportunity for developing new ALL treatment protocols in pediatric patients, particularly for treating ALL relapse, which can substantially reduce sequelae derived from conventional chemotherapy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph18091405/s1, Table S1: Association of most frequent HLA-I alleles with relapse and death; Table S2: Neoantigens with higher frequency; Table S3: Neoantigens and mutations in ALL patients with mutated and non-mutated MMR genes; Figure S1: Expression boxplots of genes in which potential neoantigens are located. (A) SYNGAP1, (B) CUL1, (C) COX11, (D) PORCN, (E) CPA2, (F) PEX5L, (G) TRAF3IP1, (H) IRAK4, (I) RRH, (J) CLK4, (K) CLK1, (L) CLTCL1; Figure S2: Mutations and neoantigens in clinical features groups; Figure S3: Numbers of mutations, neoantigens and frequency of neoantigens are not correlated with prognosis in ALL patients. A, Kaplan–Meier event-free (left) and overall (right) survival curves in 35 ALL patients stratified according to the number of mutations. B, Kaplan–Meier event-free (left) and overall (right) survival curves in 35 ALL patients stratified according to the number of neoantigens. C, Kaplan–Meier progression-free (left) and overall (right) survival curves for ALL patients stratified according to the frequency of neoantigens per missense mutation; Figure S4: Numbers of mutations in MMR genes are not correlated with prognosis in ALL patients. Kaplan–Meier event-free (left) and overall (right) survival curves in 35 ALL patients stratified according to the number of mutations in MMR genes. MUT: at least one mutation in one, WT: no mutation.
Author Contributions
Conceptualization, D.K.M.-S. and S.J.-M.; Data curation, D.K.M.-S., L.G.-R., C.J.P.-A. and D.I.M.-H.; Formal analysis, D.K.M.-S. and S.J.-M.; Funding acquisition, S.J.-M.; Investigation, D.K.M.-S. and S.J.-M.; Methodology, D.K.M.-S. and S.J.-M.; Project administration, S.J.-M.; Resources, J.C.N.-E., J.F.-L., E.J.-H., A.M.-S., V.C.B.-M., M.M.-R., M.L.P.-S., D.A.D.-R., J.R.T.-N., J.G.P.-G., L.V.F.-V., R.A.-S., M.M.V.-A., J.A.M.-T., L.E.M.-P., K.A.S.-L., R.M.E.-E., C.J.P.-A., O.A.S.-R., H.R.-V., J.M.M.-A. and S.J.-M.; Software, D.K.M.-S. and L.G.-R.; Supervision, J.M.M.-A. and S.J.-M.; Validation, D.I.M.-H.; Writing—original draft, D.K.M.-S. and S.J.-M.; Writing—review and editing, D.K.M.-S., L.G.-R., J.C.N.-E., J.F.-L., E.J.-H., A.M.-S., V.C.B.-M., M.M.-R., M.L.P.-S., D.A.D.-R., J.R.T.-N., J.G.P.-G., L.V.F.-V., R.A.-S., M.M.V.-A., J.A.M.-T., L.E.M.-P., K.A.S.-L., R.M.E.-E., C.J.P.-A., D.I.M.-H., O.A.S.-R., H.R.-V., J.M.M.-A. and S.J.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Instituto Nacional de Medicina Genómica (FALTA), Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONACYT, IFC 2016-2119), National Institutes of Health 3R01CA262263-01A1S1 sub-award number SCON-00004408 and 2U24ES028524 sub-award number 00011320; Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCYT, CF-2023-G 1399 and Dirección General de Políticas de Investigación en Salud (DGPIS), Financiamiento de Proyectos de Investigación para la Salud (FPIS 2024) FPIS2024-INMEGEN-7593. Diana Karen Mendiola Soto, a student of the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) has granted with the scholarhip CONAHCY-706961. The funding body did not have a role in the design of the study, collection, analysis, and interpretation of the data or manuscript preparation.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Instituto Mexicano del Seguro Social and Instituto Nacional de Medicina Genómica (protocol codes R-2013-785-068 and 01/2018/I, respectively).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data are not publicly available because they are protected by the Instituto Nacional de Medicina Genómica; however, data are available on reasonable request to the corresponding author.
Acknowledgments
The authors acknowledges the patients and their parents for their participation in the present study. Additionally, we thank Haydee Miranda Ortíz and Julio César Canseco Méndez from the Sequencing Unit of the National Insitute of Genomic Medicine for their technical support.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ALL | Acute Lymphoblastic Leukemia |
| TMB | Tumor Mutational Burden |
| OS | Overall Survival |
| MMR | Mismatch Repair |
| HLA | Human Leukocyte Antigen |
| EFS | Event Free Survival |
| WBC | White Blood Cells |
| BM | Bone Marrow |
| WT | Wild-Type, Non-Mutated |
| MUT | Mutated |
| CAR | Chimeric Antigen Receptor |
| MIGICCL | Mexican Interinstitutional Group for the Identification of the Causes of Childhood Leukemia |
| NCI | The National Cancer Institute |
References
- Graiqevci-Uka, V.; Behluli, E.; Spahiu, L.; Liehr, T.; Temaj, G. Targeted Treatment and Immunotherapy in High-risk and Relapsed/Refractory Pediatric Acute Lymphoblastic Leukemia. Curr. Pediatr. Rev. 2023, 19, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Barcenas-Lopez, D.A.; Nunez-Enriquez, J.C.; Hidalgo-Miranda, A.; Beltran-Anaya, F.O.; May-Hau, D.I.; Jimenez-Hernandez, E.; Bekker-Mendez, V.C.; Flores-Lujano, J.; Medina-Sanson, A.; Tamez-Gomez, E.L.; et al. Transcriptome Analysis Identifies LINC00152 as a Biomarker of Early Relapse and Mortality in Acute Lymphoblastic Leukemia. Genes 2020, 11, 302. [Google Scholar] [CrossRef]
- Burke, W.; Thummel, K. Precision medicine and health disparities: The case of pediatric acute lymphoblastic leukemia. Nurs. Outlook 2019, 67, 331–336. [Google Scholar] [CrossRef]
- Flores-Lujano, J.; Duarte-Rodriguez, D.A.; Jimenez-Hernandez, E.; Martin-Trejo, J.A.; Allende-Lopez, A.; Penaloza-Gonzalez, J.G.; Perez-Saldivar, M.L.; Medina-Sanson, A.; Torres-Nava, J.R.; Solis-Labastida, K.A.; et al. Persistently high incidence rates of childhood acute leukemias from 2010 to 2017 in Mexico City: A population study from the MIGICCL. Front. Public Health 2022, 10, 918921. [Google Scholar] [CrossRef]
- Jimenez-Morales, S.; Miranda-Peralta, E.; Saldana-Alvarez, Y.; Perez-Vera, P.; Paredes-Aguilera, R.; Rivera-Luna, R.; Velazquez-Cruz, R.; Ramirez-Bello, J.; Carnevale, A.; Orozco, L. BCR-ABL, ETV6-RUNX1 and E2A-PBX1: Prevalence of the most common acute lymphoblastic leukemia fusion genes in Mexican patients. Leuk. Res. 2008, 32, 1518–1522. [Google Scholar] [CrossRef]
- Rahnemoon, A.R.; Zaker, F.; Izadyar, M.; Ansari, S.; Poopak, B.; Tadavosyan, Y. Prevalence of ETV6/RUNX1 Fusion Gene in Pediatric Patients with Acute Lymphoblastic Leukemia in Iran. Iran. J. Pediatr. 2013, 23, 681–686. [Google Scholar] [PubMed]
- Mata-Rocha, M.; Rangel-Lopez, A.; Jimenez-Hernandez, E.; Nunez-Enriquez, J.C.; Morales-Castillo, B.A.; Sanchez-Escobar, N.; Sepulveda-Robles, O.A.; Bravata-Alcantara, J.C.; Najera-Cortes, A.S.; Perez-Saldivar, M.L.; et al. Low Prevalence of ETV6::RUNX1 Fusion Gene in a Hispanic Population. Front. Pediatr. 2022, 10, 837656. [Google Scholar] [CrossRef]
- Romana, S.P.; Poirel, H.; Leconiat, M.; Flexor, M.A.; Mauchauffe, M.; Jonveaux, P.; Macintyre, E.A.; Berger, R.; Bernard, O.A. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood 1995, 86, 4263–4269. [Google Scholar] [CrossRef]
- Jimenez-Hernandez, E.; Jaimes-Reyes, E.Z.; Arellano-Galindo, J.; Garcia-Jimenez, X.; Tiznado-Garcia, H.M.; Duenas-Gonzalez, M.T.; Martinez Villegas, O.; Sanchez-Jara, B.; Bekker-Mendez, V.C.; Ortiz-Torres, M.G.; et al. Survival of Mexican Children with Acute Lymphoblastic Leukaemia under Treatment with the Protocol from the Dana-Farber Cancer Institute 00-01. BioMed Res. Int. 2015, 2015, 576950. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Perez, J.C.; Lopez-Razo, O.N.; Garcia-Arellano, G.; Pinzon-Uresti, M.A.; Jimenez-Castillo, R.A.; Gonzalez-Llano, O.; Gomez-Almaguer, D. Results of Treating Childhood Acute Lymphoblastic Leukemia in a Low-middle Income Country: 10 Year Experience in Northeast Mexico. Arch. Med. Res. 2016, 47, 668–676. [Google Scholar] [CrossRef]
- Jimenez-Morales, S.; Aranda-Uribe, I.S.; Perez-Amado, C.J.; Ramirez-Bello, J.; Hidalgo-Miranda, A. Mechanisms of Immunosuppressive Tumor Evasion: Focus on Acute Lymphoblastic Leukemia. Front. Immunol. 2021, 12, 737340. [Google Scholar] [CrossRef]
- Zamora, A.E.; Crawford, J.C.; Allen, E.K.; Guo, X.J.; Bakke, J.; Carter, R.A.; Abdelsamed, H.A.; Moustaki, A.; Li, Y.; Chang, T.C.; et al. Pediatric patients with acute lymphoblastic leukemia generate abundant and functional neoantigen-specific CD8+ T cell responses. Sci. Transl. Med. 2019, 11, eaat8549. [Google Scholar] [CrossRef]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Gerard, B.; Chambon-Pautas, C.; Cave, H.; Duval, M.; Vilmer, E.; Grandchamp, B. Microsatellite instability and frameshift mutations in BAX and transforming growth factor-beta RII genes are very uncommon in acute lymphoblastic leukemia in vivo but not in cell lines. Blood 1998, 92, 230–233. [Google Scholar] [CrossRef]
- Mathonnet, G.; Krajinovic, M.; Labuda, D.; Sinnett, D. Role of DNA mismatch repair genetic polymorphisms in the risk of childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2003, 123, 45–48. [Google Scholar] [CrossRef]
- Best, A.; Matheson, E.; Minto, L.; Hall, A.G.; Irving, J.A. Mismatch repair and the downstream target genes, PAX5 and Ikaros, in childhood acute lymphoblastic leukemia. Leuk. Res. 2010, 34, 1098–1102. [Google Scholar] [CrossRef]
- Germano, G.; Lamba, S.; Rospo, G.; Barault, L.; Magri, A.; Maione, F.; Russo, M.; Crisafulli, G.; Bartolini, A.; Lerda, G.; et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature 2017, 552, 116–120. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, A.; Larkin, J. Checkpoint inhibitors in advanced melanoma: Effect on the field of immunotherapy. Expert Rev. Anticancer Ther. 2017, 17, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Lauss, M.; Donia, M.; Harbst, K.; Andersen, R.; Mitra, S.; Rosengren, F.; Salim, M.; Vallon-Christersson, J.; Torngren, T.; Kvist, A.; et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat. Commun. 2017, 8, 1738. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhao, W.; Wu, J.; Lu, J.; Ding, Y.; Wu, S.; Wang, H.; Ding, D.; Mo, F.; Zhou, Z.; et al. Neoantigens Derived from Recurrently Mutated Genes as Potential Immunotherapy Targets for Gastric Cancer. BioMed Res. Int. 2019, 2019, 8103142. [Google Scholar] [CrossRef] [PubMed]
- Bachireddy, P.; Burkhardt, U.E.; Rajasagi, M.; Wu, C.J. Haematological malignancies: At the forefront of immunotherapeutic innovation. Nat. Rev. Cancer 2015, 15, 201–215. [Google Scholar] [CrossRef]
- Zacharakis, N.; Chinnasamy, H.; Black, M.; Xu, H.; Lu, Y.C.; Zheng, Z.; Pasetto, A.; Langhan, M.; Shelton, T.; Prickett, T.; et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 2018, 24, 724–730. [Google Scholar] [CrossRef]
- Biernacki, M.A.; Bleakley, M. Neoantigens in Hematologic Malignancies. Front. Immunol. 2020, 11, 121. [Google Scholar] [CrossRef] [PubMed]
- Hackl, H.; Charoentong, P.; Finotello, F.; Trajanoski, Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet. 2016, 17, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Carter, R.A.; Li, Y.; Li, Y.; Wang, H.; Edmonson, M.N.; Chen, X.; Arnold, P.; Geiger, T.L.; Wu, G.; et al. The neoepitope landscape in pediatric cancers. Genome Med. 2017, 9, 78. [Google Scholar] [CrossRef]
- Brennick, C.A.; George, M.M.; Corwin, W.L.; Srivastava, P.K.; Ebrahimi-Nik, H. Neoepitopes as cancer immunotherapy targets: Key challenges and opportunities. Immunotherapy 2017, 9, 361–371. [Google Scholar] [CrossRef]
- Ho, S.Y.; Chang, C.M.; Liao, H.N.; Chou, W.H.; Guo, C.L.; Yen, Y.; Nakamura, Y.; Chang, W.C. Current Trends in Neoantigen-Based Cancer Vaccines. Pharmaceuticals 2023, 16, 392. [Google Scholar] [CrossRef]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.T.; Lupardus, P.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, J.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef]
- Duan, F.; Duitama, J.; Al Seesi, S.; Ayres, C.M.; Corcelli, S.A.; Pawashe, A.P.; Blanchard, T.; McMahon, D.; Sidney, J.; Sette, A.; et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 2014, 211, 2231–2248. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, H.; Hasegawa, K.; Oda, K.; Yamamoto, S.; Nishijima, A.; Imai, Y.; Asada, K.; Ikeda, Y.; Karasaki, T.; Fujiwara, K.; et al. The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 2017, 6, e1338996. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Chae, Y.K.; Anker, J.F.; Bais, P.; Namburi, S.; Giles, F.J.; Chuang, J.H. Mutations in DNA repair genes are associated with increased neo-antigen load and activated T cell infiltration in lung adenocarcinoma. Oncotarget 2018, 9, 7949–7960. [Google Scholar] [CrossRef]
- Karnell, J.L.; Dimasi, N.; Karnell, F.G., 3rd; Fleming, R.; Kuta, E.; Wilson, M.; Wu, H.; Gao, C.; Herbst, R.; Ettinger, R. CD19 and CD32b differentially regulate human B cell responsiveness. J. Immunol. 2014, 192, 1480–1490. [Google Scholar] [CrossRef]
- Poe, J.C.; Hasegawa, M.; Tedder, T.F. CD19, CD21, and CD22: Multifaceted response regulators of B lymphocyte signal transduction. Int. Rev. Immunol. 2001, 20, 739–762. [Google Scholar] [CrossRef]
- Walker, K.; Mistry, A.; Watson, C.M.; Nadat, F.; O’Callaghan, E.; Care, M.; Crinnion, L.A.; Arumugakani, G.; Bonthron, D.T.; Carter, C.; et al. Inherited CD19 Deficiency Does Not Impair Plasma Cell Formation or Response to CXCL12. J. Clin. Immunol. 2023, 43, 1543–1556. [Google Scholar] [CrossRef]
- von Stackelberg, A.; Locatelli, F.; Zugmaier, G.; Handgretinger, R.; Trippett, T.M.; Rizzari, C.; Bader, P.; O’Brien, M.M.; Brethon, B.; Bhojwani, D.; et al. Phase I/Phase II Study of Blinatumomab in Pediatric Patients with Relapsed/Refractory Acute Lymphoblastic Leukemia. J. Clin. Oncol. 2016, 34, 4381–4389. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Panuciak, K.; Margas, M.; Makowska, K.; Lejman, M. Insights into Modern Therapeutic Approaches in Pediatric Acute Leukemias. Cells 2022, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.D.; Birch, J.; Accogli, T.; Criado, I.; Khabirova, E.; Parks, C.; Wood, Y.; Young, M.D.; Porter, T.; Richardson, R.; et al. Transcriptional signatures associated with persisting CD19 CAR-T cells in children with leukemia. Nat. Med. 2023, 29, 1700–1709. [Google Scholar] [CrossRef]
- Giavridis, T.; van der Stegen, S.J.C.; Eyquem, J.; Hamieh, M.; Piersigilli, A.; Sadelain, M. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 2018, 24, 731–738. [Google Scholar] [CrossRef]
- Brudno, J.N.; Kochenderfer, J.N. Recent advances in CAR T-cell toxicity: Mechanisms, manifestations and management. Blood Rev. 2019, 34, 45–55. [Google Scholar] [CrossRef]
- Loeza, F.; Vargas-Alarcon, G.; Andrade, F.; Vergara, Y.; Rodriguez-Perez, J.M.; Ruiz-Morales, J.A.; Alarcon-Segovia, D.; Granados, J. Distribution of class I and class III MHC antigens in the Tarasco Amerindians. Hum. Immunol. 2002, 63, 143–148. [Google Scholar] [CrossRef]
- Garcia-Ortiz, J.E.; Sandoval-Ramirez, L.; Rangel-Villalobos, H.; Maldonado-Torres, H.; Cox, S.; Garcia-Sepulveda, C.A.; Figuera, L.E.; Marsh, S.G.; Little, A.M.; Madrigal, J.A.; et al. High-resolution molecular characterization of the HLA class I and class II in the Tarahumara Amerindian population. Tissue Antigens 2006, 68, 135–146. [Google Scholar] [CrossRef]
- Barquera, R.; Hernandez-Zaragoza, D.I.; Bravo-Acevedo, A.; Arrieta-Bolanos, E.; Clayton, S.; Acuna-Alonzo, V.; Martinez-Alvarez, J.C.; Lopez-Gil, C.; Adalid-Sainz, C.; Vega-Martinez, M.D.R.; et al. The immunogenetic diversity of the HLA system in Mexico correlates with underlying population genetic structure. Hum. Immunol. 2020, 81, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Barquera, R.; Zuniga, J.; Hernandez-Diaz, R.; Acuna-Alonzo, V.; Montoya-Gama, K.; Moscoso, J.; Torres-Garcia, D.; Garcia-Salas, C.; Silva, B.; Cruz-Robles, D.; et al. HLA class I and class II haplotypes in admixed families from several regions of Mexico. Mol. Immunol. 2008, 45, 1171–1178. [Google Scholar] [CrossRef]
- Del Angel-Pablo, A.D.; Juarez-Martin, A.I.; Perez-Rubio, G.; Ambrocio-Ortiz, E.; Lopez-Flores, L.A.; Camarena, A.E.; Falfan-Valencia, R. HLA Allele and Haplotype Frequencies in Three Urban Mexican Populations: Genetic Diversity for the Approach of Genomic Medicine. Diagnostics 2020, 10, 47. [Google Scholar] [CrossRef]
- Gonzalez-Quezada, B.A.; Creary, L.E.; Munguia-Saldana, A.J.; Flores-Aguilar, H.; Fernandez-Vina, M.A.; Gorodezky, C. Exploring the ancestry and admixture of Mexican Oaxaca Mestizos from Southeast Mexico using next-generation sequencing of 11 HLA loci. Hum. Immunol. 2019, 80, 157–162. [Google Scholar] [CrossRef]
- Fernandez-Torres, J.; Flores-Jimenez, D.; Arroyo-Perez, A.; Granados, J.; Lopez-Reyes, A. HLA-B*40 allele plays a role in the development of acute leukemia in Mexican population: A case-control study. BioMed Res. Int. 2013, 2013, 705862. [Google Scholar] [CrossRef]
- Bello-Lopez, J.M.; Cisneros, C.B.; Martinez-Albarran, A. HLA analysis of Mexican candidates for bone marrow transplantation and probability of finding compatible related donors. Transfus. Apher. Sci. 2018, 57, 82–87. [Google Scholar] [CrossRef]
- Tosch, C.; Bastien, B.; Barraud, L.; Grellier, B.; Nourtier, V.; Gantzer, M.; Limacher, J.M.; Quemeneur, E.; Bendjama, K.; Preville, X. Viral based vaccine TG4010 induces broadening of specific immune response and improves outcome in advanced NSCLC. J. Immunother. Cancer 2017, 5, 70. [Google Scholar] [CrossRef]
- Obara, W.; Eto, M.; Mimata, H.; Kohri, K.; Mitsuhata, N.; Miura, I.; Shuin, T.; Miki, T.; Koie, T.; Fujimoto, H.; et al. A phase I/II study of cancer peptide vaccine S-288310 in patients with advanced urothelial carcinoma of the bladder. Ann. Oncol. 2017, 28, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Stokidis, S.; Baxevanis, C.N.; Fortis, S.P. The Prognostic Significance of Selected HLA Alleles on Prostate Cancer Outcome. Int. J. Mol. Sci. 2023, 24, 14454. [Google Scholar] [CrossRef] [PubMed]
- Van Son, C.; Loan, N.T.H.; Trang, T.H.; Thinh, L.X.; Khanh, N.B.; Nhung, L.T.H.; Van Hung, N.; Que, T.N.; Van Lieu, N.; Tung, P.D.; et al. Predominant HLA Alleles and Haplotypes in Mild Adverse Drug Reactions Caused by Allopurinol in Vietnamese Patients with Gout. Diagnostics 2021, 11, 1611. [Google Scholar] [CrossRef]
- Teck, A.T.; Urban, S.; Quass, P.; Nelde, A.; Schuster, H.; Letsch, A.; Busse, A.; Walz, J.S.; Keilholz, U.; Ochsenreither, S. Cancer testis antigen Cyclin A1 harbors several HLA-A*02:01-restricted T cell epitopes, which are presented and recognized in vivo. Cancer Immunol. Immunother. 2020, 69, 1217–1227. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wei, Z.; Zhang, Z.; Zhang, B.; Zhu, C.; Chen, K.; Chuai, G.; Qu, S.; Xie, L.; Gao, Y.; et al. pTuneos: Prioritizing tumor neoantigens from next-generation sequencing data. Genome Med. 2019, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Rech, A.J.; Balli, D.; Mantero, A.; Ishwaran, H.; Nathanson, K.L.; Stanger, B.Z.; Vonderheide, R.H. Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer. Cancer Immunol. Res. 2018, 6, 276–287. [Google Scholar] [CrossRef]
- Ghorani, E.; Rosenthal, R.; McGranahan, N.; Reading, J.L.; Lynch, M.; Peggs, K.S.; Swanton, C.; Quezada, S.A. Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann. Oncol. 2018, 29, 271–279. [Google Scholar] [CrossRef]
- Cimen Bozkus, C.; Roudko, V.; Finnigan, J.P.; Mascarenhas, J.; Hoffman, R.; Iancu-Rubin, C.; Bhardwaj, N. Immune Checkpoint Blockade Enhances Shared Neoantigen-Induced T-cell Immunity Directed against Mutated Calreticulin in Myeloproliferative Neoplasms. Cancer Discov. 2019, 9, 1192–1207. [Google Scholar] [CrossRef]
- Bekker-Mendez, V.C.; Miranda-Peralta, E.; Nunez-Enriquez, J.C.; Olarte-Carrillo, I.; Guerra-Castillo, F.X.; Pompa-Mera, E.N.; Ocana-Mondragon, A.; Rangel-Lopez, A.; Bernaldez-Rios, R.; Medina-Sanson, A.; et al. Prevalence of gene rearrangements in Mexican children with acute lymphoblastic leukemia: A population study-report from the Mexican Interinstitutional Group for the identification of the causes of childhood leukemia. BioMed Res. Int. 2014, 2014, 210560. [Google Scholar] [CrossRef]
- Li, X.; You, J.; Hong, L.; Liu, W.; Guo, P.; Hao, X. Neoantigen cancer vaccines: A new star on the horizon. Cancer Biol. Med. 2023, 21, 274–311. [Google Scholar] [CrossRef]
- Blass, E.; Ott, P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 18, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yong, H.M.; Chen, F.F.; Mei, P.J.; Liu, H.; Li, C.; Pan, Z.Q.; Wu, Y.P.; Zheng, J.N. Cullin1 is a novel marker of poor prognosis and a potential therapeutic target in human breast cancer. Ann. Oncol. 2013, 24, 2016–2022. [Google Scholar] [CrossRef]
- Chen, G.; Cheng, Y.; Martinka, M.; Li, G. Cul1 expression is increased in early stages of human melanoma. Pigment. Cell Melanoma Res. 2010, 23, 572–574. [Google Scholar] [CrossRef]
- Esteves, F.; Xavier, J.M.; Ford, A.M.; Rocha, C.; Pharoah, P.D.P.; Caldas, C.; Chin, S.F.; Maia, A.T. Germline allelic expression of genes at 17q22 locus associates with risk of breast cancer. Eur. J. Cancer 2022, 172, 146–157. [Google Scholar] [CrossRef]
- Tang, L.; Xu, J.; Wei, F.; Wang, L.; Nie, W.W.; Chen, L.B.; Guan, X.X. Association of STXBP4/COX11 rs6504950 (G>A) polymorphism with breast cancer risk: Evidence from 17,960 cases and 22,713 controls. Arch. Med. Res. 2012, 43, 383–388. [Google Scholar] [CrossRef]
- Sens-Abuazar, C.; Napolitano, E.F.E.; Osorio, C.A.; Krepischi, A.C.; Ricca, T.I.; Castro, N.P.; da Cunha, I.W.; Maciel Mdo, S.; Rosenberg, C.; Brentani, M.M.; et al. Down-regulation of ANAPC13 and CLTCL1: Early Events in the Progression of Preinvasive Ductal Carcinoma of the Breast. Transl. Oncol. 2012, 5, 113–123. [Google Scholar] [CrossRef]
- Liu, J.; Pan, S.; Hsieh, M.H.; Ng, N.; Sun, F.; Wang, T.; Kasibhatla, S.; Schuller, A.G.; Li, A.G.; Cheng, D.; et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. USA 2013, 110, 20224–20229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qi, X.; Donnelly, L.; Elghobashi-Meinhardt, N.; Long, T.; Zhou, R.W.; Sun, Y.; Wang, B.; Li, X. Mechanisms and inhibition of Porcupine-mediated Wnt acylation. Nature 2022, 607, 816–822. [Google Scholar] [CrossRef]
- Chen, A.S.; Liu, H.; Wu, Y.; Luo, S.; Patz, E.F., Jr.; Glass, C.; Su, L.; Du, M.; Christiani, D.C.; Wei, Q. Genetic variants in DDO and PEX5L in peroxisome-related pathways predict non-small cell lung cancer survival. Mol. Carcinog. 2022, 61, 619–628. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Baeza, J.; Gu, K.; Zheng, Y.; Chen, S.; Zhou, Z. DeepTAP: An RNN-based method of TAP-binding peptide prediction in the selection of tumor neoantigens. Comput. Biol. Med. 2023, 164, 107247. [Google Scholar] [CrossRef]
- Wan, Y.R.; Kosaloglu-Yalcin, Z.; Peters, B.; Nielsen, M. A large-scale study of peptide features defining immunogenicity of cancer neo-epitopes. NAR Cancer 2024, 6, zcae002. [Google Scholar] [CrossRef]
- Hilf, N.; Kuttruff-Coqui, S.; Frenzel, K.; Bukur, V.; Stevanovic, S.; Gouttefangeas, C.; Platten, M.; Tabatabai, G.; Dutoit, V.; van der Burg, S.H.; et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 2019, 565, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Creech, A.L.; Ting, Y.S.; Goulding, S.P.; Sauld, J.F.K.; Barthelme, D.; Rooney, M.S.; Addona, T.A.; Abelin, J.G. The Role of Mass Spectrometry and Proteogenomics in the Advancement of HLA Epitope Prediction. Proteomics 2018, 18, e1700259. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.; Busby, J.; Palmer, C.D.; Davis, M.J.; Murphy, T.; Clark, A.; Busby, M.; Duke, F.; Yang, A.; Young, L.; et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 2018, 37, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Forsthuber, T.G. ELISPOT Techniques. Methods Mol. Biol. 2016, 1304, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhu, L.; Xu, Q.; Zhang, X.; Li, B.; Lee, L.J. The co-stimulation of anti-CD28 and IL-2 enhances the sensitivity of ELISPOT assays for detection of neoantigen-specific T cells in PBMC. J. Immunol. Methods 2020, 484–485, 112831. [Google Scholar] [CrossRef]
- Lin, J.J.Y.; Low-Nam, S.T.; Alfieri, K.N.; McAffee, D.B.; Fay, N.C.; Groves, J.T. Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 2019, 12, eaat8715. [Google Scholar] [CrossRef]
- Bentzen, A.K.; Marquard, A.M.; Lyngaa, R.; Saini, S.K.; Ramskov, S.; Donia, M.; Such, L.; Furness, A.J.; McGranahan, N.; Rosenthal, R.; et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 2016, 34, 1037–1045. [Google Scholar] [CrossRef]
- Guan, J.; Lu, C.; Jin, Q.; Lu, H.; Chen, X.; Tian, L.; Zhang, Y.; Ortega, J.; Zhang, J.; Siteni, S.; et al. MLH1 Deficiency-Triggered DNA Hyperexcision by Exonuclease 1 Activates the cGAS-STING Pathway. Cancer Cell 2021, 39, 109–121 e105. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef]
- Gong, L.; He, R.; Xu, Y.; Luo, T.; Jin, K.; Yuan, W.; Zheng, Z.; Liu, L.; Liang, Z.; Li, A.; et al. Neoantigen load as a prognostic and predictive marker for stage II/III non-small cell lung cancer in Chinese patients. Thorac. Cancer 2021, 12, 2170–2181. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, B.; Zhou, N.; Sun, Z.; Li, J.; Chen, Q.; Wu, X.; Zhou, Y.; Shi, Y.; Lu, X.; et al. Identification of WDFY3 Neoantigens as Prognostic Markers in Longterm Survivors of Extrahepatic Cholangiocarcinoma. Curr. Cancer Drug Targets 2020, 20, 875–886. [Google Scholar] [CrossRef]
- Miller, A.; Asmann, Y.; Cattaneo, L.; Braggio, E.; Keats, J.; Auclair, D.; Lonial, S.; Network, M.C.; Russell, S.J.; Stewart, A.K. High somatic mutation and neoantigen burden are correlated with decreased progression-free survival in multiple myeloma. Blood Cancer J. 2017, 7, e612. [Google Scholar] [CrossRef]
- Chen, R.; Im, H.; Snyder, M. Whole-Exome Enrichment with the Illumina TruSeq Exome Enrichment Platform. Cold Spring Harb. Protoc. 2015, 2015, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Szolek, A.; Schubert, B.; Mohr, C.; Sturm, M.; Feldhahn, M.; Kohlbacher, O. OptiType: Precision HLA typing from next-generation sequencing data. Bioinformatics 2014, 30, 3310–3316. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Karosiene, E.; Lundegaard, C.; Lund, O.; Nielsen, M. NetMHCcons: A consensus method for the major histocompatibility complex class I predictions. Immunogenetics 2012, 64, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Bartha, A.; Gyorffy, B. TNMplot.com: A Web Tool for the Comparison of Gene Expression in Normal, Tumor and Metastatic Tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- He, Y.; Zhang, L.; Zhou, R.; Wang, Y.; Chen, H. The role of DNA mismatch repair in immunotherapy of human cancer. Int. J. Biol. Sci. 2022, 18, 2821–2832. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 15 August 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).