Readthrough Activators and Nonsense-Mediated mRNA Decay Inhibitor Molecules: Real Potential in Many Genetic Diseases Harboring Premature Termination Codons
Abstract
1. Introduction
1.1. Origin of PTC
1.1.1. DNA Level
1.1.2. RNA Level
2. Natural Translation Mechanism
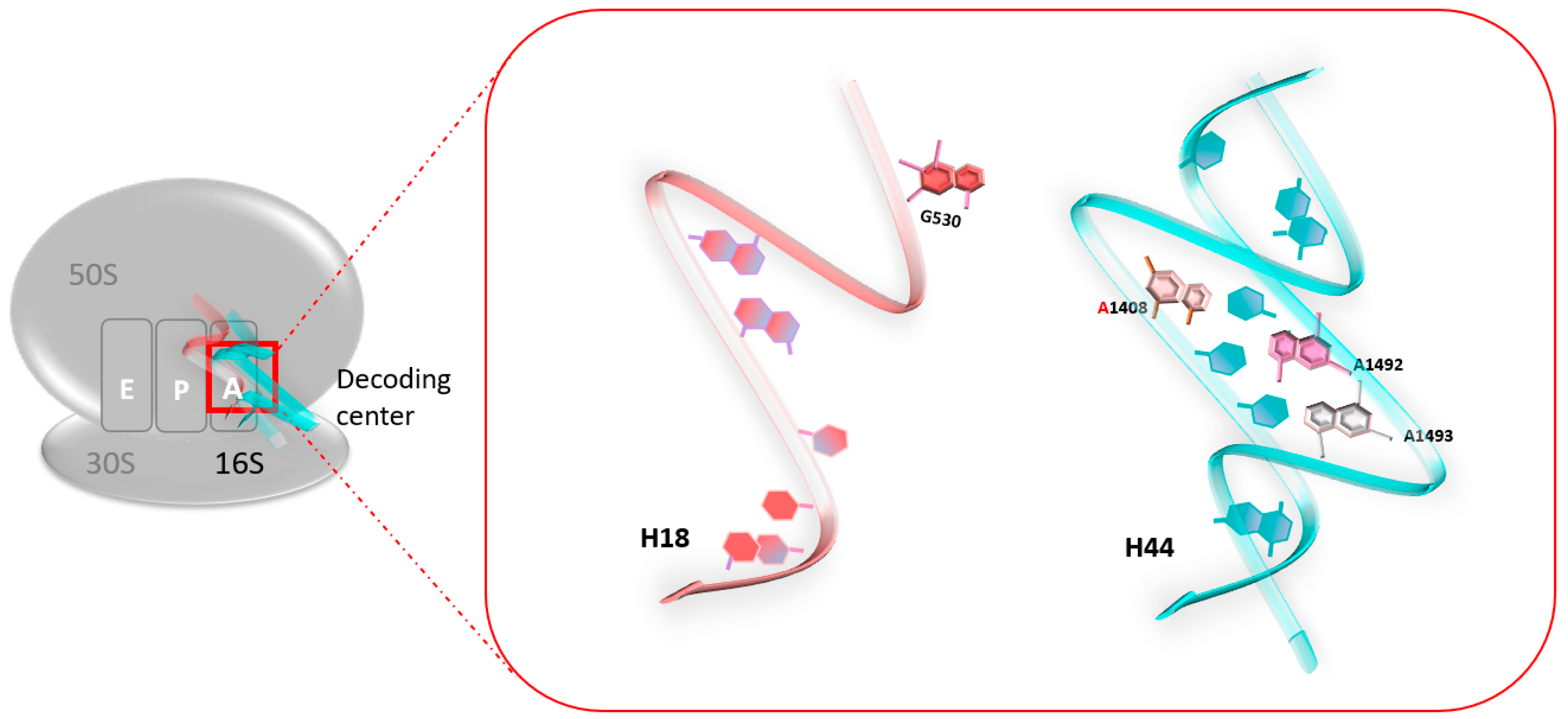
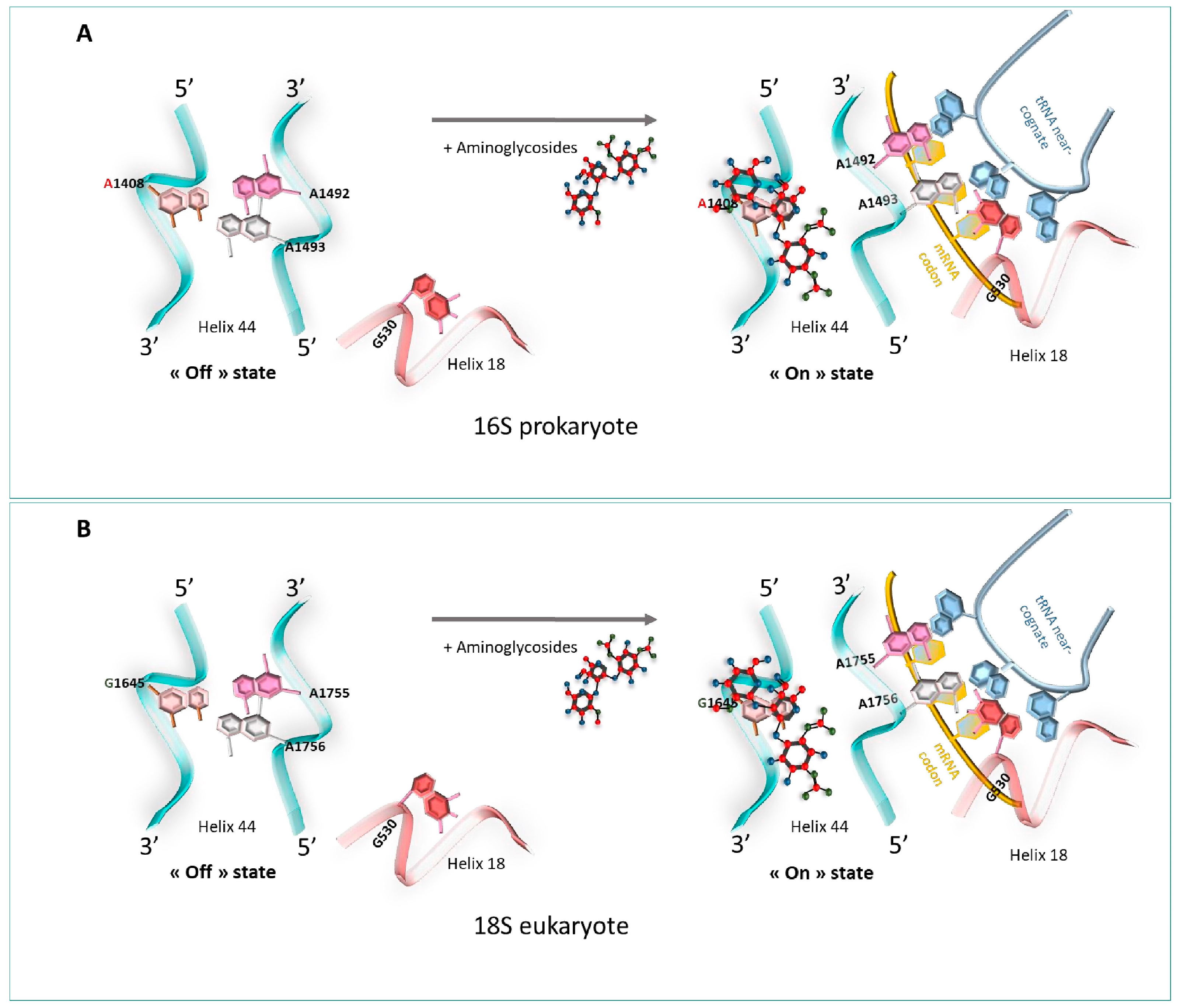
2.1. Ribosome Fidelity: The Role of the Decoding Center
2.2. Course of Natural Translation Termination Mechanism
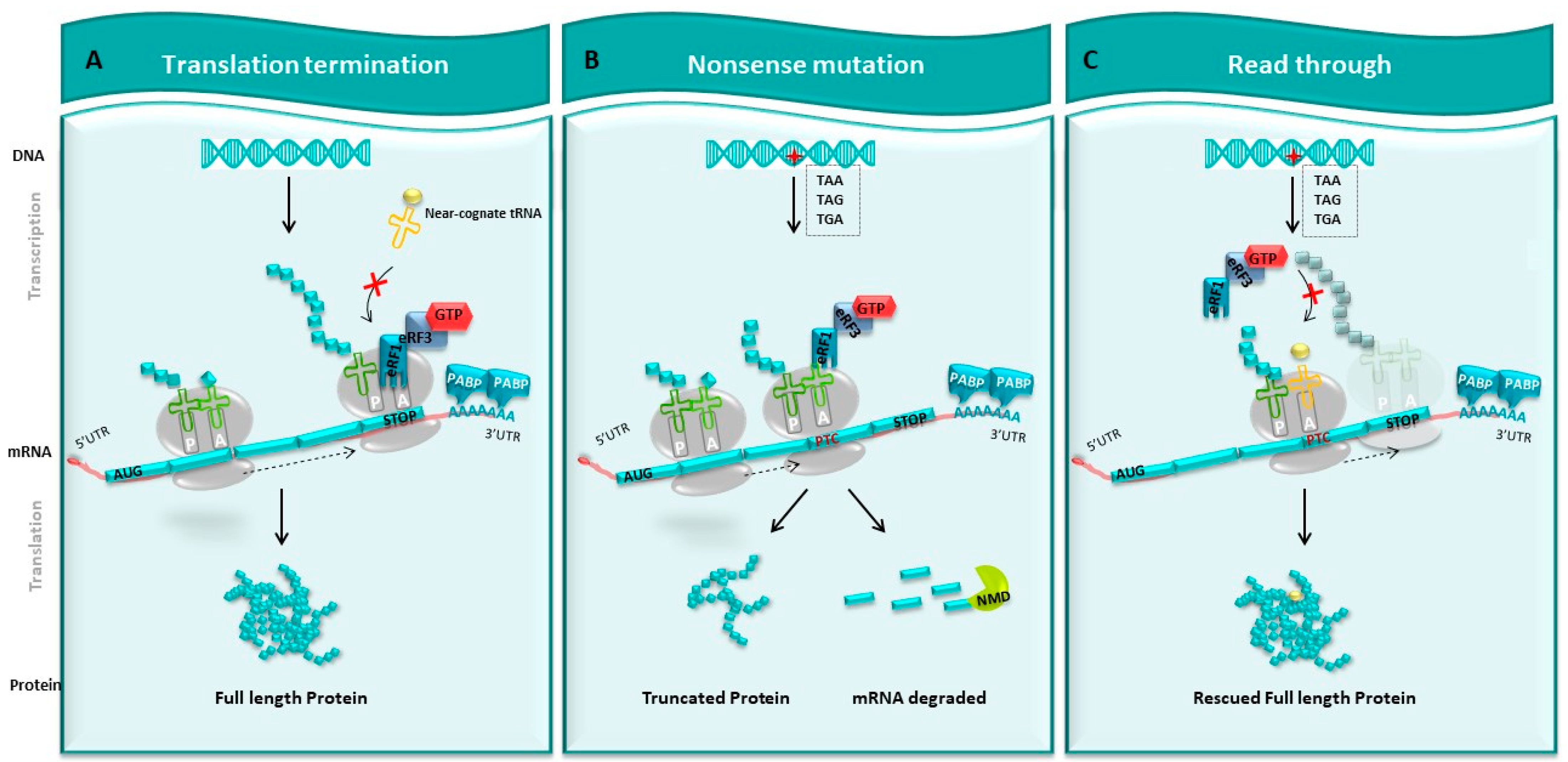
3. Disturbance of the Translation Mechanism: Consequences of PTCs within the mRNA
3.1. Truncated Protein: The Aftermath
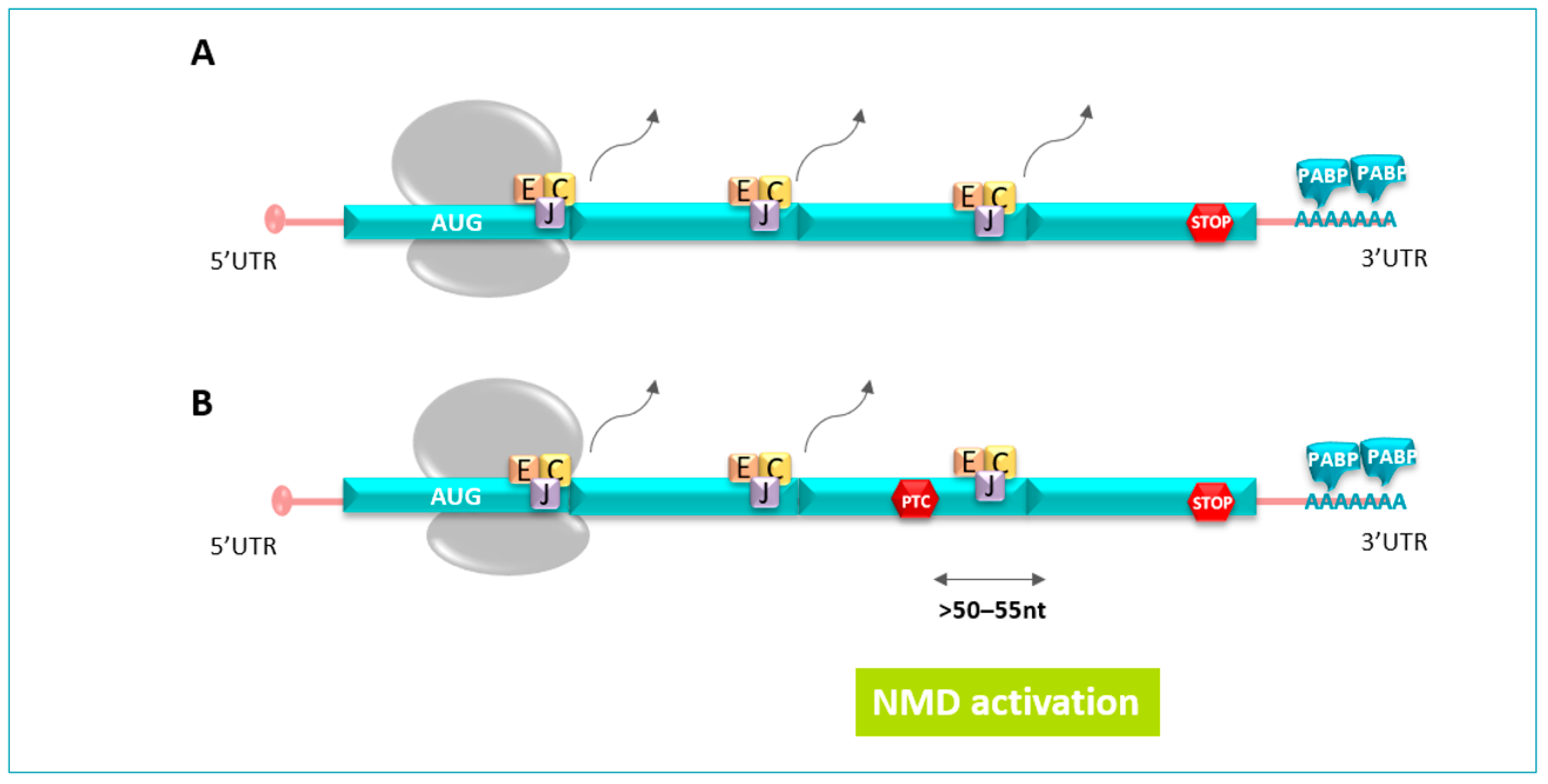
3.2. Degradation of Altered mRNA by the NMD System
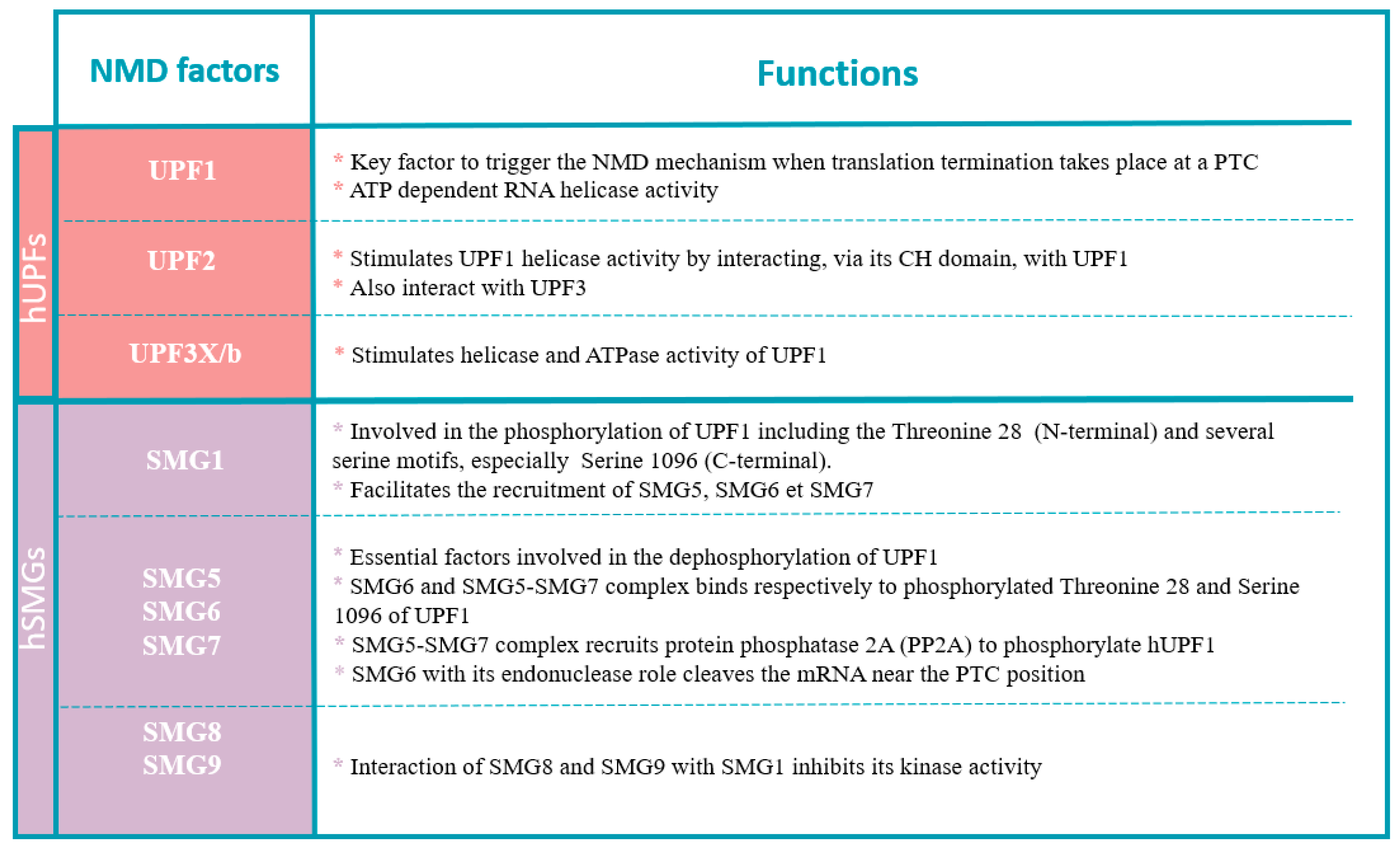
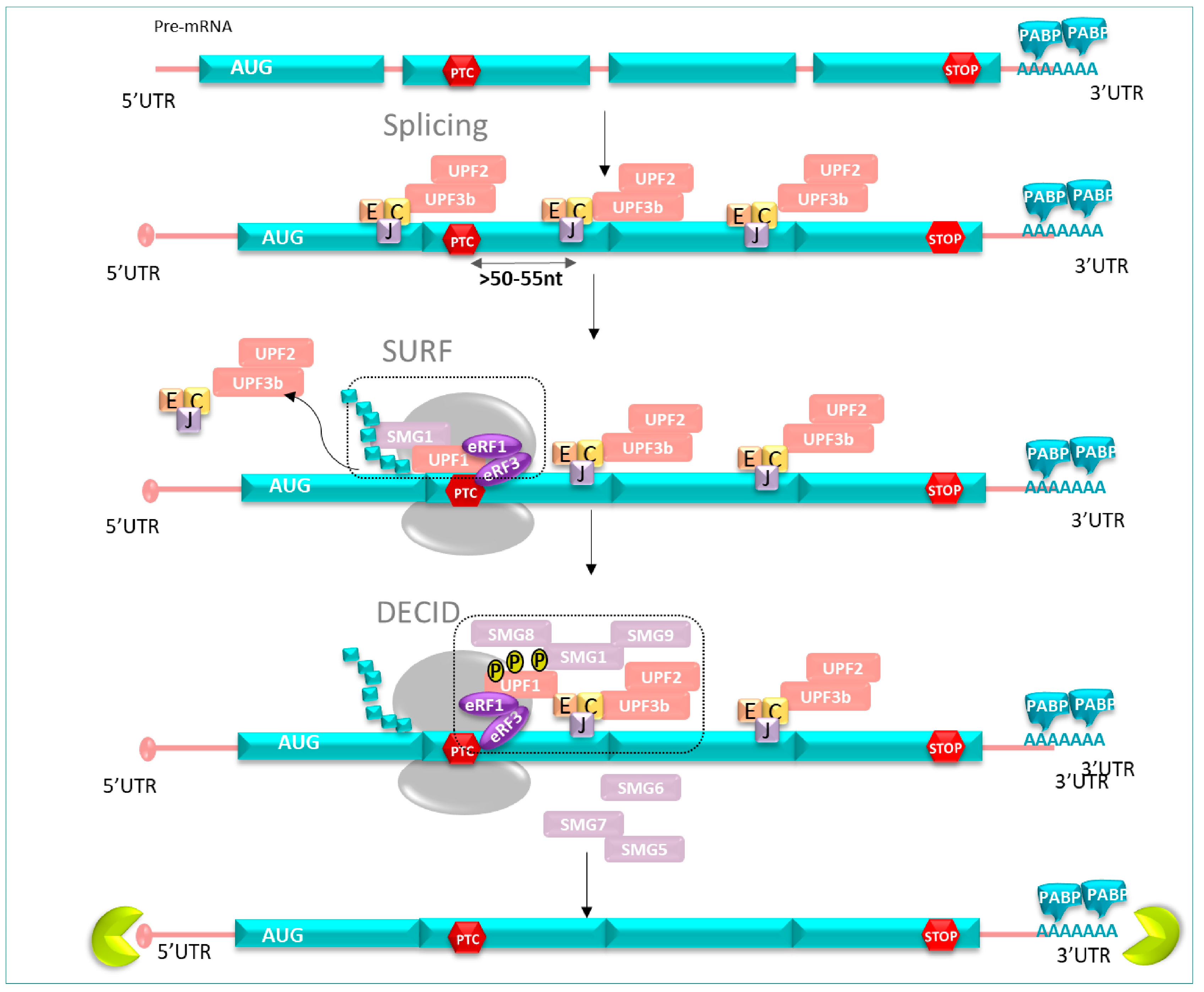
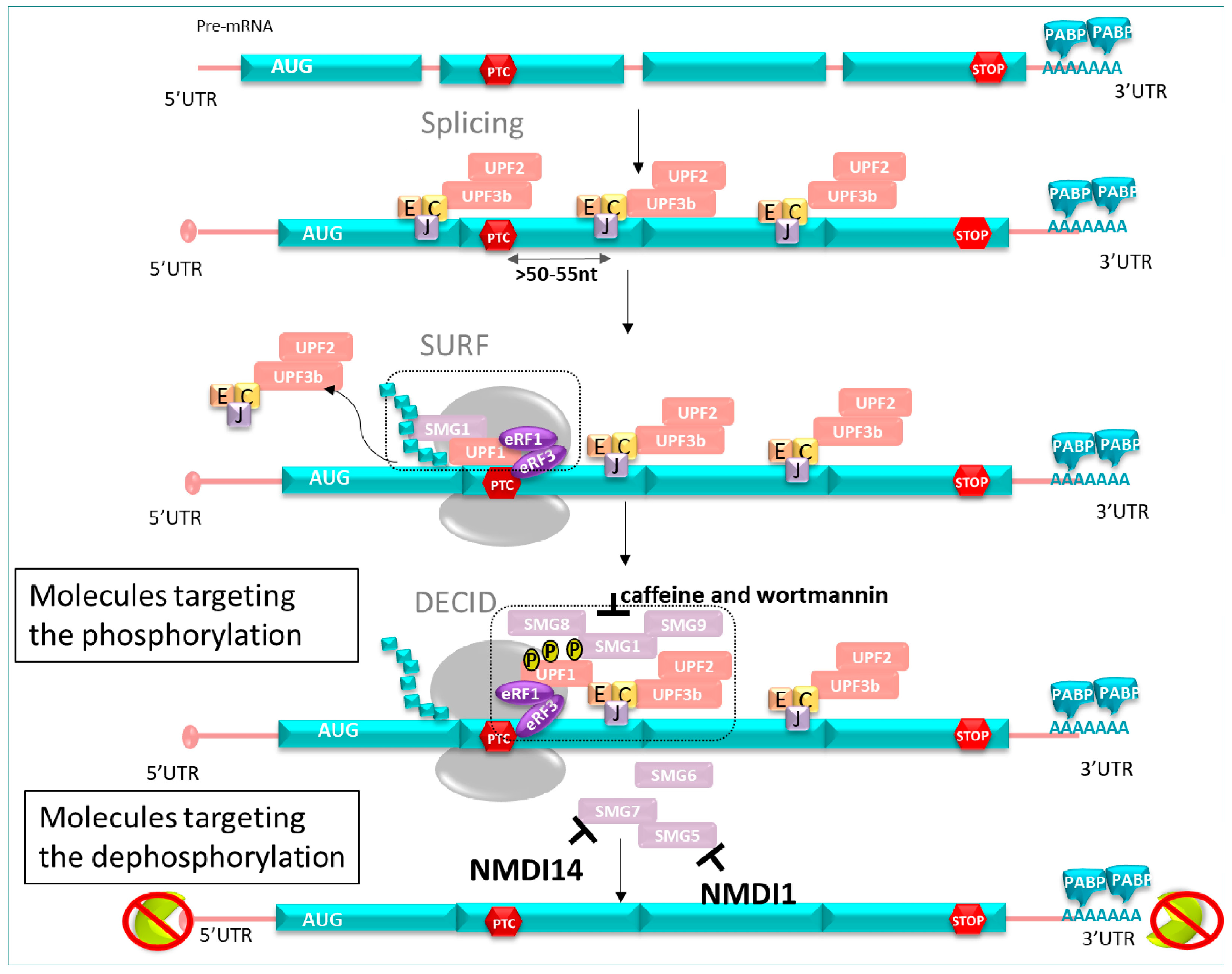
3.2.1. NMD Factors
3.2.2. NMD Mechanism Course
3.3. Impact of the PTC Position in the NMD System and Illustration in Peripheral Neuropathy
4. Readthrough Mechanism
4.1. The Translational Readthrough
4.2. Readthrough Therapeutic Approach
- Ribosome-targeting molecules;
- tRNA post-transcriptional inhibitors;
- eRF1-targeting molecules.
4.2.1. Ribosome-Targeting Molecules
- ➢
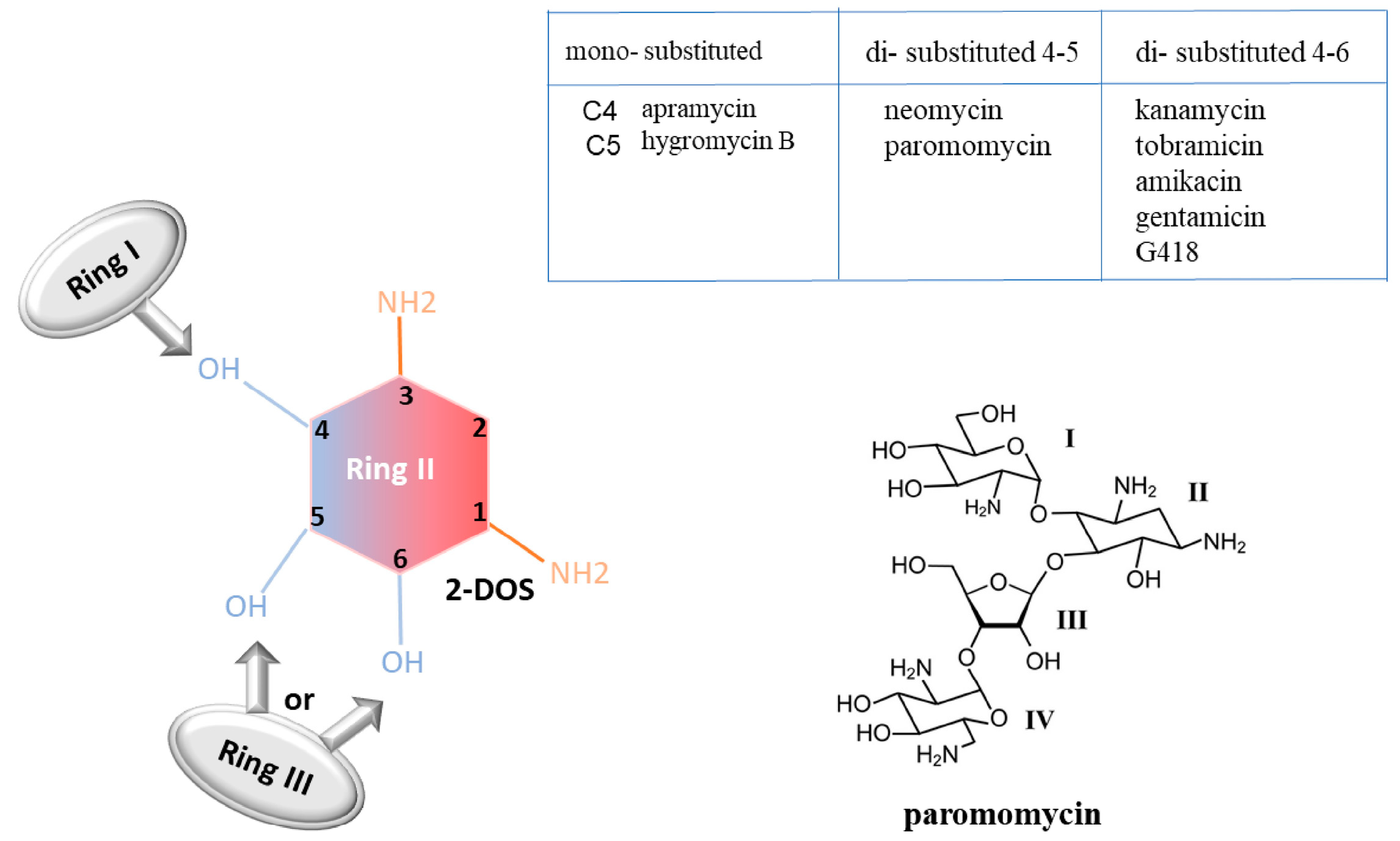
- Aminoglycoside
- ➢
- Ataluren (PTC124)
- ➢
- PTC414
- ➢
- RTC13 and RTC14
- ➢
- GJ071 and GJ072
- ➢
- TLN468
4.2.2. tRNA Post-Transcriptional Inhibitors
- ➢
- 2,6-diaminopurine
- ➢
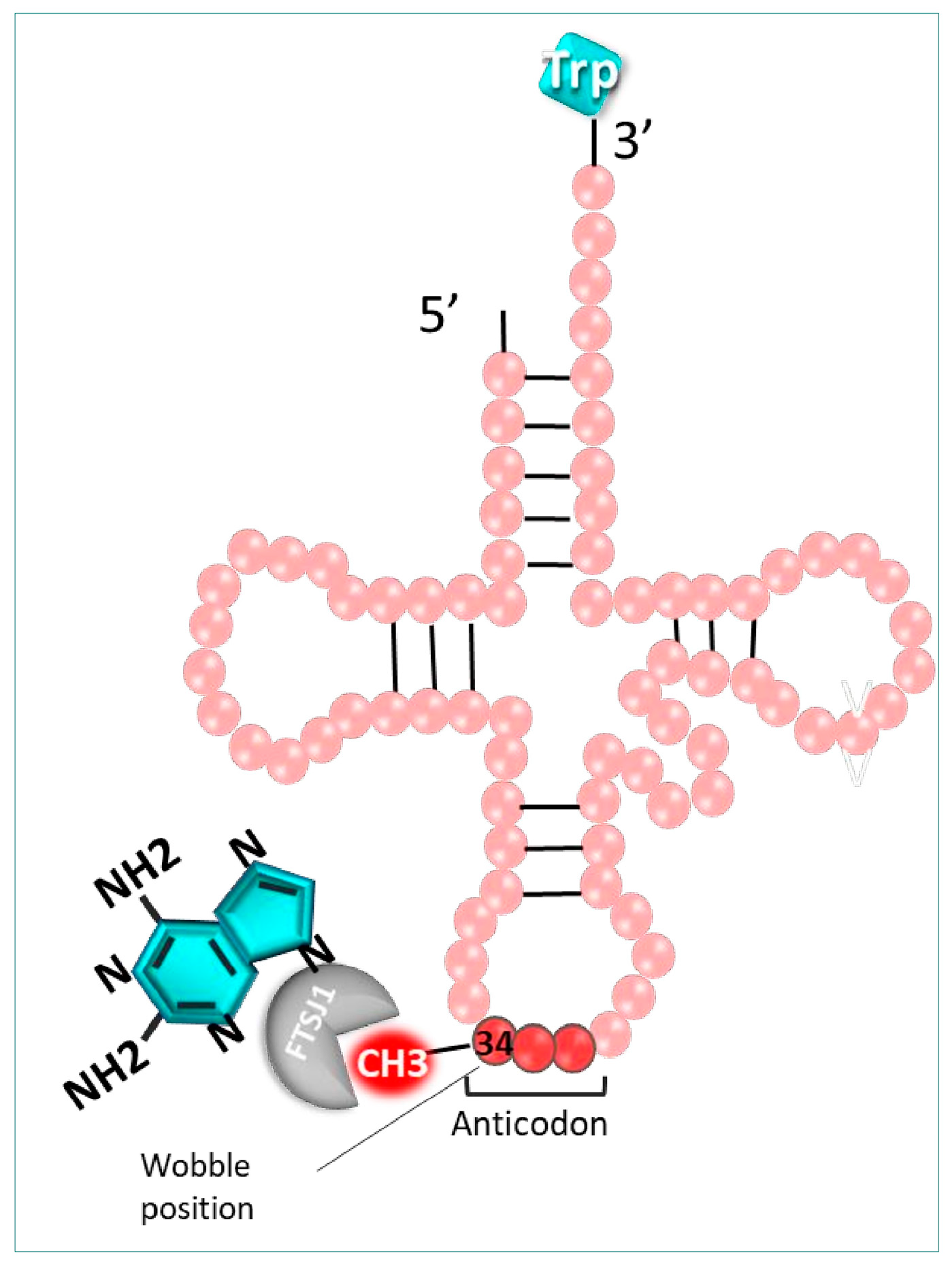
- NV derivatives
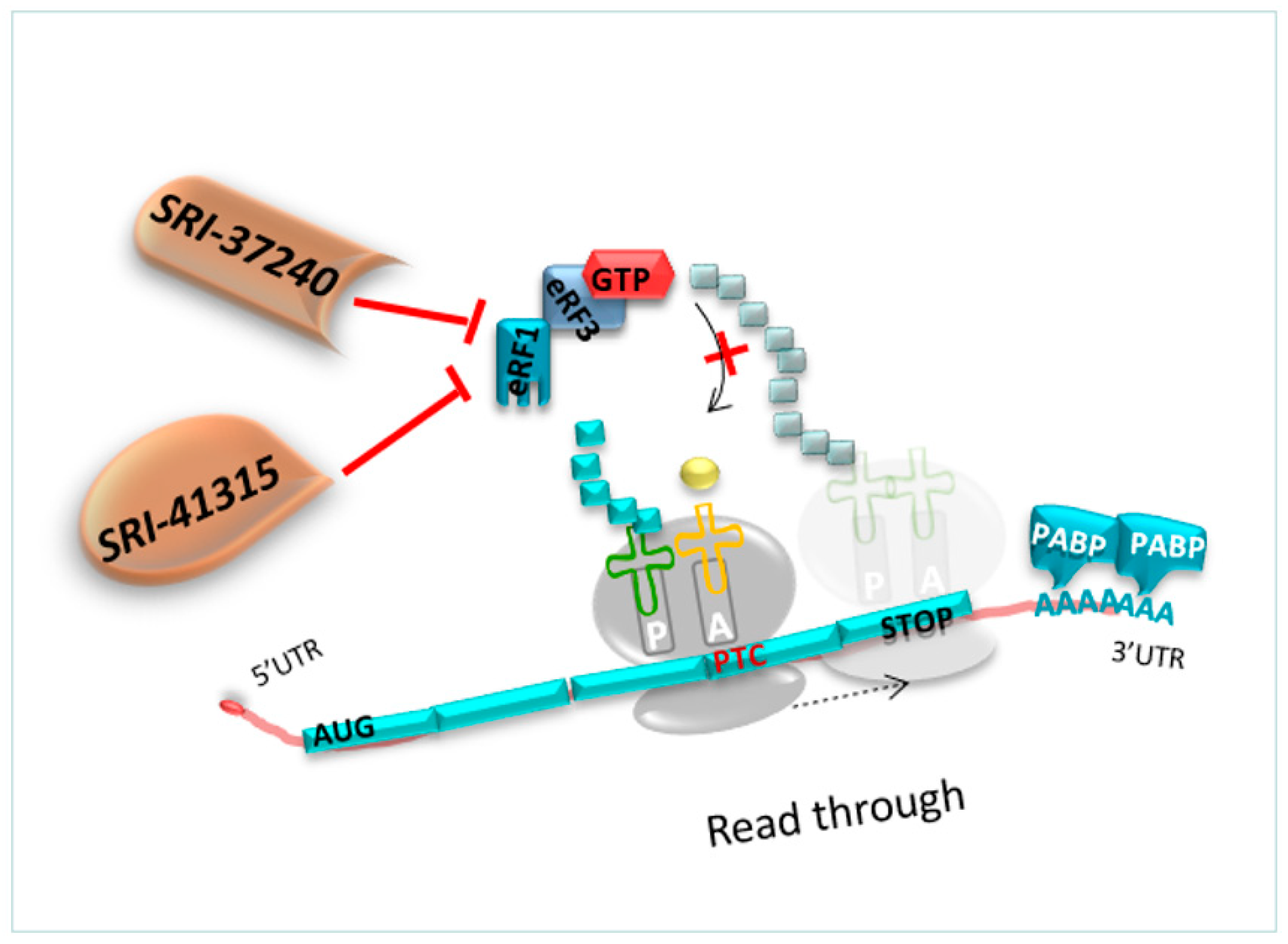
4.2.3. Molecules Targeting eRF1
- ➢
- SRI-37240 and SRI-41315
4.3. Readthrough Limits
- The stop codon nature
- The nucleotide context
- ➢
- 3′context
- ➢
- 5′context
5. NMD Inhibitors
5.1. Different Types of NMD Inhibitors (NMDIs)
5.2. Molecules Targeting the Phosphorylation Cycle of hUPF1
- ➢
- Caffeine and wortmannin
5.3. Molecules Targeting Dephosphorylation Cycle of hUPF1
- ➢
- NMDI 1
- ➢
- NMDI 14
- ➢
- Amlexanox
5.4. Limits of NMD Inhibition
6. By Itself, Readthrough Mechanism Is Insufficient
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frischmeyer, P.A.; Dietz, H.C. Nonsense-Mediated mRNA Decay in Health and Disease. Hum. Mol. Genet. 1999, 8, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Peltz, S.W.; Morsy, M.; Welch, E.M.; Jacobson, A. Ataluren as an Agent for Therapeutic Nonsense Suppression. Annu. Rev. Med. 2013, 64, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, S.; Testa, M.F.; Pinotti, M.; Branchini, A. Molecular Insights into Determinants of Translational Readthrough and Implications for Nonsense Suppression Approaches. Int. J. Mol. Sci. 2020, 21, 9449. [Google Scholar] [CrossRef] [PubMed]
- Miressi, F.; Benslimane, N.; Favreau, F.; Rassat, M.; Richard, L.; Bourthoumieu, S.; Laroche, C.; Magy, L.; Magdelaine, C.; Sturtz, F.; et al. GDAP1 Involvement in Mitochondrial Function and Oxidative Stress, Investigated in a Charcot-Marie-Tooth Model of hiPSCs-Derived Motor Neurons. Biomedicines 2021, 9, 945. [Google Scholar] [CrossRef] [PubMed]
- Pyromali, I.; Benslimane, N.; Favreau, F.; Goizet, C.; Lazaro, L.; Vitry, M.; Derouault, P.; Sturtz, F.; Magdelaine, C.; Lia, A.-S. From Negative to Positive Diagnosis: Structural Variation Could Be the Second Mutation You Are Looking for in a Recessive Autosomal Gene. J. Pers. Med. 2022, 12, 212. [Google Scholar] [CrossRef] [PubMed]
- DiVincenzo, C.; Elzinga, C.D.; Medeiros, A.C.; Karbassi, I.; Jones, J.R.; Evans, M.C.; Braastad, C.D.; Bishop, C.M.; Jaremko, M.; Wang, Z.; et al. The Allelic Spectrum of Charcot-Marie-Tooth Disease in over 17,000 Individuals with Neuropathy. Mol. Genet. Genom. Med. 2014, 2, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Bang, J.; Spina, S.; Miller, B.L. Non-Alzheimer’s Dementia 1. Lancet Lond. Engl. 2015, 386, 1672–1682. [Google Scholar] [CrossRef] [PubMed]
- Takada, L.T. The Genetics of Monogenic Frontotemporal Dementia. Dement. Neuropsychol. 2015, 9, 219–229. [Google Scholar] [CrossRef][Green Version]
- Gass, J.; Cannon, A.; Mackenzie, I.R.; Boeve, B.; Baker, M.; Adamson, J.; Crook, R.; Melquist, S.; Kuntz, K.; Petersen, R.; et al. Mutations in Progranulin Are a Major Cause of Ubiquitin-Positive Frontotemporal Lobar Degeneration. Hum. Mol. Genet. 2006, 15, 2988–3001. [Google Scholar] [CrossRef]
- Mühlemann, O.; Eberle, A.; Stalder, L.; Orozco, R. Recognition and Elimination of Nonsense mRNA. Biochim. Biophys. Acta 2008, 1779, 538–549. [Google Scholar] [CrossRef]
- Mort, M.; Ivanov, D.; Cooper, D.N.; Chuzhanova, N.A. A Meta-Analysis of Nonsense Mutations Causing Human Genetic Disease. Hum. Mutat. 2008, 29, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.; Martin, R. Mutations to Nonsense Codons in Human Genetic Disease: Implications for Gene Therapy by Nonsense Suppressor tRNAs. Nucleic Acids Res. 1994, 22, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Kewalramani, A. Messenger RNA Surveillance and the Evolutionary Proliferation of Introns. Mol. Biol. Evol. 2003, 20, 563–571. [Google Scholar] [CrossRef]
- Green, R.E.; Lewis, B.P.; Hillman, R.T.; Blanchette, M.; Lareau, L.F.; Garnett, A.T.; Rio, D.C.; Brenner, S.E. Widespread Predicted Nonsense-Mediated mRNA Decay of Alternatively-Spliced Transcripts of Human Normal and Disease Genes. Bioinforma. Oxf. Engl. 2003, 19 (Suppl. S1), i118–i121. [Google Scholar] [CrossRef] [PubMed]
- Morais, P.; Adachi, H.; Yu, Y.-T. Suppression of Nonsense Mutations by New Emerging Technologies. Int. J. Mol. Sci. 2020, 21, E4394. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-L.R.; Dougherty, J.P. Pharmaceutical Therapies to Recode Nonsense Mutations in Inherited Diseases. Pharmacol. Ther. 2012, 136, 227–266. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V. Ribosome Structure and the Mechanism of Translation. Cell 2002, 108, 557–572. [Google Scholar] [CrossRef]
- Schrode, P.; Huter, P.; Clementi, N.; Erlacher, M. Atomic Mutagenesis at the Ribosomal Decoding Site. RNA Biol. 2017, 14, 104–112. [Google Scholar] [CrossRef]
- Fan-Minogue, H.; Bedwell, D.M. Eukaryotic Ribosomal RNA Determinants of Aminoglycoside Resistance and Their Role in Translational Fidelity. RNA 2008, 14, 148–157. [Google Scholar] [CrossRef]
- Ivanov, P.V.; Gehring, N.H.; Kunz, J.B.; Hentze, M.W.; Kulozik, A.E. Interactions between UPF1, eRFs, PABP and the Exon Junction Complex Suggest an Integrated Model for Mammalian NMD Pathways. EMBO J. 2008, 27, 736–747. [Google Scholar] [CrossRef]
- Guissart, C.; Mouzat, K.; Kantar, J.; Louveau, B.; Vilquin, P.; Polge, A.; Raoul, C.; Lumbroso, S. Premature Termination Codons in SOD1 Causing Amyotrophic Lateral Sclerosis Are Predicted to Escape the Nonsense-Mediated mRNA Decay. Sci. Rep. 2020, 10, 20738. [Google Scholar] [CrossRef] [PubMed]
- Sylvain, V.; Lafarge, S.; Bignon, Y.-J. Dominant-Negative Activity of a Brca1 Truncation Mutant: Effects on Proliferation, Tumorigenicity in Vivo, and Chemosensitivity in a Mouse Ovarian Cancer Cell Line. Int. J. Oncol. 2002, 20, 845–853. [Google Scholar] [CrossRef]
- Thein, S.L.; Hesketh, C.; Taylor, P.; Temperley, I.J.; Hutchinson, R.M.; Old, J.M.; Wood, W.G.; Clegg, J.B.; Weatherall, D.J. Molecular Basis for Dominantly Inherited Inclusion Body Beta-Thalassemia. Proc. Natl. Acad. Sci. USA 1990, 87, 3924–3928. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Pouya, A.; Bitar, S.; Pfeiffer, A.; Bueno, D.; Rojas-Charry, L.; Arndt, S.; Gomez-Zepeda, D.; Tenzer, S.; Bello, F.D.; et al. GDAP1 Loss of Function Inhibits the Mitochondrial Pyruvate Dehydrogenase Complex by Altering the Actin Cytoskeleton. Commun. Biol. 2022, 5, 541. [Google Scholar] [CrossRef] [PubMed]
- Barneo-Muñoz, M.; Juárez, P.; Civera-Tregón, A.; Yndriago, L.; Pla-Martin, D.; Zenker, J.; Cuevas-Martín, C.; Estela, A.; Sánchez-Aragó, M.; Forteza-Vila, J.; et al. Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy. PLoS Genet. 2015, 11, e1005115. [Google Scholar] [CrossRef] [PubMed]
- Melero, R.; Buchwald, G.; Castaño, R.; Raabe, M.; Gil, D.; Lázaro, M.; Urlaub, H.; Conti, E.; Llorca, O. The Cryo-EM Structure of the UPF-EJC Complex Shows UPF1 Poised toward the RNA 3’ End. Nat. Struct. Mol. Biol. 2012, 19, 498–505. [Google Scholar] [CrossRef]
- Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. The Spliceosome Deposits Multiple Proteins 20-24 Nucleotides Upstream of mRNA Exon-Exon Junctions. EMBO J. 2000, 19, 6860–6869. [Google Scholar] [CrossRef]
- Lejeune, F.; Li, X.; Maquat, L.E. Nonsense-Mediated mRNA Decay in Mammalian Cells Involves Decapping, Deadenylating, and Exonucleolytic Activities. Mol. Cell 2003, 12, 675–687. [Google Scholar] [CrossRef]
- Lewis, B.P.; Green, R.E.; Brenner, S.E. Evidence for the Widespread Coupling of Alternative Splicing and Nonsense-Mediated mRNA Decay in Humans. Proc. Natl. Acad. Sci. USA 2003, 100, 189–192. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Czaplinski, K.; Trifillis, P.; He, F.; Jacobson, A.; Peltz, S.W. Characterization of the Biochemical Properties of the Human Upf1 Gene Product That Is Involved in Nonsense-Mediated mRNA Decay. RNA 2000, 6, 1226–1235. [Google Scholar] [CrossRef]
- Schweingruber, C.; Rufener, S.C.; Zünd, D.; Yamashita, A.; Mühlemann, O. Nonsense-Mediated mRNA Decay—Mechanisms of Substrate mRNA Recognition and Degradation in Mammalian Cells. Biochim. Biophys. Acta 2013, 1829, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Jayachandran, U.; Bonneau, F.; Fiorini, F.; Basquin, C.; Domcke, S.; Le Hir, H.; Conti, E. Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2. Mol. Cell 2011, 41, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Rebbapragada, I.; Lykke-Andersen, J. A Competition between Stimulators and Antagonists of Upf Complex Recruitment Governs Human Nonsense-Mediated mRNA Decay. PLoS Biol. 2008, 6, e111. [Google Scholar] [CrossRef] [PubMed]
- Raimondeau, E.; Bufton, J.C.; Schaffitzel, C. New Insights into the Interplay between the Translation Machinery and Nonsense-Mediated mRNA Decay Factors. Biochem. Soc. Trans. 2018, 46, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a Novel SMG-1-Upf1-eRF1-eRF3 Complex (SURF) to the Exon Junction Complex Triggers Upf1 Phosphorylation and Nonsense-Mediated mRNA Decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Hug, N.; Cáceres, J.F. The RNA Helicase DHX34 Activates NMD by Promoting a Transition from the Surveillance to the Decay-Inducing Complex. Cell Rep. 2014, 8, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Ohnishi, T.; Kashima, I.; Taya, Y.; Ohno, S. Human SMG-1, a Novel Phosphatidylinositol 3-Kinase-Related Protein Kinase, Associates with Components of the mRNA Surveillance Complex and Is Involved in the Regulation of Nonsense-Mediated mRNA Decay. Genes Dev. 2001, 15, 2215–2228. [Google Scholar] [CrossRef]
- Palma, M.; Leroy, C.; Salomé-Desnoulez, S.; Werkmeister, E.; Kong, R.; Mongy, M.; Le Hir, H.; Lejeune, F. A Role for AKT1 in Nonsense-Mediated mRNA Decay. Nucleic Acids Res. 2021, 49, 11022–11037. [Google Scholar] [CrossRef]
- Cho, H.; Abshire, E.T.; Popp, M.W.; Pröschel, C.; Schwartz, J.L.; Yeo, G.W.; Maquat, L.E. AKT Constitutes a Signal-Promoted Alternative Exon-Junction Complex That Regulates Nonsense-Mediated mRNA Decay. Mol. Cell 2022, 82, 2779–2796.e10. [Google Scholar] [CrossRef]
- Boehm, V.; Kueckelmann, S.; Gerbracht, J.V.; Kallabis, S.; Britto-Borges, T.; Altmüller, J.; Krüger, M.; Dieterich, C.; Gehring, N.H. SMG5-SMG7 Authorize Nonsense-Mediated mRNA Decay by Enabling SMG6 Endonucleolytic Activity. Nat. Commun. 2021, 12, 3965. [Google Scholar] [CrossRef]
- Inoue, K.; Khajavi, M.; Ohyama, T.; Hirabayashi, S.; Wilson, J.; Reggin, J.D.; Mancias, P.; Butler, I.J.; Wilkinson, M.F.; Wegner, M.; et al. Molecular Mechanism for Distinct Neurological Phenotypes Conveyed by Allelic Truncating Mutations. Nat. Genet. 2004, 36, 361–369. [Google Scholar] [CrossRef]
- Dabrowski, M.; Bukowy-Bieryllo, Z.; Zietkiewicz, E. Translational Readthrough Potential of Natural Termination Codons in Eucaryotes—The Impact of RNA Sequence. RNA Biol. 2015, 12, 950–958. [Google Scholar] [CrossRef]
- Floquet, C.; Hatin, I.; Rousset, J.-P.; Bidou, L. Statistical Analysis of Readthrough Levels for Nonsense Mutations in Mammalian Cells Reveals a Major Determinant of Response to Gentamicin. PLoS Genet. 2012, 8, e1002608. [Google Scholar] [CrossRef] [PubMed]
- Beier, H.; Barciszewska, M.; Krupp, G.; Mitnacht, R.; Gross, H.J. UAG Readthrough during TMV RNA Translation: Isolation and Sequence of Two tRNAsTyr with Suppressor Activity from Tobacco Plants. EMBO J. 1984, 3, 351–356. [Google Scholar] [CrossRef]
- Hofstetter, H.; Monstein, H.J.; Weissmann, C. The Readthrough Protein A1 Is Essential for the Formation of Viable Q Beta Particles. Biochim. Biophys. Acta 1974, 374, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Napthine, S.; Yek, C.; Powell, M.L.; Brown, T.D.K.; Brierley, I. Characterization of the Stop Codon Readthrough Signal of Colorado Tick Fever Virus Segment 9 RNA. RNA 2012, 18, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Rice, C.M. The Signal for Translational Readthrough of a UGA Codon in Sindbis Virus RNA Involves a Single Cytidine Residue Immediately Downstream of the Termination Codon. J. Virol. 1993, 67, 5062–5067. [Google Scholar] [CrossRef]
- Lin, M.F.; Carlson, J.W.; Crosby, M.A.; Matthews, B.B.; Yu, C.; Park, S.; Wan, K.H.; Schroeder, A.J.; Gramates, L.S.; Pierre, S.E.; et al. Revisiting the Protein-Coding Gene Catalog of Drosophila Melanogaster Using 12 Fly Genomes. Genome Res. 2007, 17, 1823–1836. [Google Scholar] [CrossRef]
- Schueren, F.; Thoms, S. Functional Translational Readthrough: A Systems Biology Perspective. PLoS Genet. 2016, 12, e1006196. [Google Scholar] [CrossRef]
- Eswarappa, S.M.; Potdar, A.A.; Koch, W.J.; Fan, Y.; Vasu, K.; Lindner, D.; Willard, B.; Graham, L.M.; DiCorleto, P.E.; Fox, P.L. Programmed Translational Readthrough Generates Anti-Angiogenic VEGF-Ax. Cell 2014, 157, 1605–1618. [Google Scholar] [CrossRef]
- Schatz, A.; Bugle, E.; Waksman, S.A. Streptomycin, a Substance Exhibiting Antibiotic Activity Against Gram-Positive and Gram-Negative Bacteria. Proc. Soc. Exp. Biol. Med. 1944, 55, 66–69. [Google Scholar] [CrossRef]
- Obrecht, D.; Bernardini, F.; Dale, G.; Dembowsky, K. Chapter 15—Emerging New Therapeutics Against Key Gram-Negative Pathogens. In Annual Reports in Medicinal Chemistry; Macor, J.E., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 46, pp. 245–262. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Chittapragada, M.; Roberts, S.; Ham, Y.W. Aminoglycosides: Molecular Insights on the Recognition of RNA and Aminoglycoside Mimics. Perspect. Med. Chem. 2009, 3, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Inaoka, T.; Okamoto, S.; Ochi, K. A Novel Insertion Mutation in Streptomyces Coelicolor Ribosomal S12 Protein Results in Paromomycin Resistance and Antibiotic Overproduction. Antimicrob. Agents Chemother. 2009, 53, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Borovinskaya, M.A.; Pai, R.D.; Zhang, W.; Schuwirth, B.S.; Holton, J.M.; Hirokawa, G.; Kaji, H.; Kaji, A.; Cate, J.H.D. Structural Basis for Aminoglycoside Inhibition of Bacterial Ribosome Recycling. Nat. Struct. Mol. Biol. 2007, 14, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.R.; Gonzalez, R.L.; Puglisi, J.D. Comparison of X-Ray Crystal Structure of the 30S Subunit-Antibiotic Complex with NMR Structure of Decoding Site Oligonucleotide-Paromomycin Complex. Structure 2003, 11, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Fourmy, D.; Yoshizawa, S.; Puglisi, J.D. Paromomycin Binding Induces a Local Conformational Change in the A-Site of 16 s rRNA11Edited by I. Tinoco. J. Mol. Biol. 1998, 277, 333–345. [Google Scholar] [CrossRef]
- Vicens, Q.; Westhof, E. Crystal Structure of Paromomycin Docked into the Eubacterial Ribosomal Decoding A Site. Structure 2001, 9, 647–658. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional Insights from the Structure of the 30S Ribosomal Subunit and Its Interactions with Antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Yusupova, G.; Prokhorova, I.; Altman, R.; Djumagulov, M.; Shrestha, J.; Urzhumtsev, A.; Ferguson, A.; Chang, C.-W.T.; Yusupov, M.; Blanchard, S. Aminoglycoside Interactions and Impacts on the Eukaryotic Ribosome. Proc. Natl. Acad. Sci. USA 2017, 114, E10899–E10908. [Google Scholar] [CrossRef]
- Hermann, T. Aminoglycoside Antibiotics: Old Drugs and New Therapeutic Approaches. Cell Mol. Life Sci. CMLS 2007, 64, 1841–1852. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.; Mogg, A. Suppression of a Nonsense Mutation in Mammalian Cells in Vivo by the Aminoglycoside Anthiotics G–418 and Paromomycin. Nucleic Acids Res. 1985, 13, 6265–6272. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.-H.; Chun, H.H.; Nahas, S.A.; Mitui, M.; Gamo, K.M.; Du, L.; Gatti, R.A. Correction of ATM Gene Function by Aminoglycoside-Induced Read-through of Premature Termination Codons. Proc. Natl. Acad. Sci. USA 2004, 101, 15676–15681. [Google Scholar] [CrossRef] [PubMed]
- Barton-Davis, E.R.; Cordier, L.; Shoturma, D.I.; Leland, S.E.; Sweeney, H.L. Aminoglycoside Antibiotics Restore Dystrophin Function to Skeletal Muscles of Mdx Mice. J. Clin. Investig. 1999, 104, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Dunant, P.; Walter, M.C.; Karpati, G.; Lochmüller, H. Gentamicin Fails to Increase Dystrophin Expression in Dystrophin-Deficient Muscle. Muscle Nerve 2003, 27, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Jones, J.R.; Lanier, J.; Keeling, K.M.; Lindsey, J.R.; Tousson, A.; Bebök, Z.; Whitsett, J.A.; Dey, C.R.; Colledge, W.H.; et al. Aminoglycoside Suppression of a Premature Stop Mutation in a Cftr-/- Mouse Carrying a Human CFTR-G542X Transgene. J. Mol. Med. Berl. Ger. 2002, 80, 595–604. [Google Scholar] [CrossRef]
- Wilschanski, M.; Famini, C.; Blau, H.; Rivlin, J.; Augarten, A.; Avital, A.; Kerem, B.; Kerem, E. A Pilot Study of the Effect of Gentamicin on Nasal Potential Difference Measurements in Cystic Fibrosis Patients Carrying Stop Mutations. Am. J. Respir. Crit. Care Med. 2000, 161, 860–865. [Google Scholar] [CrossRef]
- Clancy, J.P.; Bebök, Z.; Ruiz, F.; King, C.; Jones, J.; Walker, L.; Greer, H.; Hong, J.; Wing, L.; Macaluso, M.; et al. Evidence That Systemic Gentamicin Suppresses Premature Stop Mutations in Patients with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2001, 163, 1683–1692. [Google Scholar] [CrossRef]
- Wilschanski, M.; Yahav, Y.; Yaacov, Y.; Blau, H.; Bentur, L.; Rivlin, J.; Aviram, M.; Bdolah-Abram, T.; Bebok, Z.; Shushi, L.; et al. Gentamicin-Induced Correction of CFTR Function in Patients with Cystic Fibrosis and CFTR Stop Mutations. N. Engl. J. Med. 2003, 349, 1433–1441. [Google Scholar] [CrossRef]
- Wagner, K.R.; Hamed, S.; Hadley, D.W.; Gropman, A.L.; Burstein, A.H.; Escolar, D.M.; Hoffman, E.P.; Fischbeck, K.H. Gentamicin Treatment of Duchenne and Becker Muscular Dystrophy Due to Nonsense Mutations. Ann. Neurol. 2001, 49, 706–711. [Google Scholar] [CrossRef]
- Malik, V.; Rodino-Klapac, L.R.; Viollet, L.; Wall, C.; King, W.; Al-Dahhak, R.; Lewis, S.; Shilling, C.J.; Kota, J.; Serrano-Munuera, C.; et al. Gentamicin-Induced Readthrough of Stop Codons in Duchenne Muscular Dystrophy. Ann. Neurol. 2010, 67, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Dukovski, D.; Shumate, J.; Scampavia, L.; Miller, J.P.; Spicer, T.P. Identification of Compounds That Promote Readthrough of Premature Termination Codons in the CFTR. SLAS Discov. 2021, 26, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Woodley, D.T.; Cogan, J.; Hou, Y.; Lyu, C.; Marinkovich, M.P.; Keene, D.; Chen, M. Gentamicin Induces Functional Type VII Collagen in Recessive Dystrophic Epidermolysis Bullosa Patients. J. Clin. Investig. 2017, 127, 3028–3038. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Santamaría, L.; Maseda, R.; del Carmen de Arriba, M.; Membrilla, J.A.; Sigüenza, A.I.; Mascías, J.; García, M.; Quintana, L.; Esteban-Rodríguez, I.; Hernández-Fernández, C.P.; et al. Evaluation of Systemic Gentamicin as Translational Readthrough Therapy for a Patient With Epidermolysis Bullosa Simplex With Muscular Dystrophy Owing to PLEC1 Pathogenic Nonsense Variants. JAMA Dermatol. 2022, 158, 439. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, O.W. Aminoglycoside Induced Ototoxicity. Toxicology 2008, 249, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Wargo, K.A.; Edwards, J.D. Aminoglycoside-Induced Nephrotoxicity. J. Pharm. Pract. 2014, 27, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wan, P.; Li, P.; Wang, J.; Guo, S.; Zhang, Y.; An, Y.; Ye, C.; Liu, Z.; Gao, J.; et al. Mechanism and Prevention of Ototoxicity Induced by Aminoglycosides. Front. Cell Neurosci. 2021, 15, 692762. [Google Scholar] [CrossRef]
- Laurent, G.; Rollman, B. Mechanism of aminoglycoside-induced lysosomal phospholipidosis: In vitro and in vivo studies with Gentamicin and Amikacin. Biochem. Pharmacol. 1982, 31, 3861–3870. [Google Scholar] [CrossRef]
- Sha, S.H.; Schacht, J. Stimulation of Free Radical Formation by Aminoglycoside Antibiotics. Hear. Res. 1999, 128, 112–118. [Google Scholar] [CrossRef]
- Vicens, Q.; Westhof, E. RNA as a Drug Target: The Case of Aminoglycosides. Chembiochem. Eur. J. Chem. Biol. 2003, 4, 1018–1023. [Google Scholar] [CrossRef]
- Baradaran-Heravi, A.; Balgi, A.D.; Zimmerman, C.; Choi, K.; Shidmoossavee, F.S.; Tan, J.S.; Bergeaud, C.; Krause, A.; Flibotte, S.; Shimizu, Y.; et al. Novel Small Molecules Potentiate Premature Termination Codon Readthrough by Aminoglycosides. Nucleic Acids Res. 2016, 44, 6583–6598. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Farahabadi, S.; Baradaran-Heravi, A.; Zimmerman, C.; Choi, K.; Flibotte, S.; Roberge, M. Small Molecule Y-320 Stimulates Ribosome Biogenesis, Protein Synthesis, and Aminoglycoside-Induced Premature Termination Codon Readthrough. PLoS Biol. 2021, 19, e3001221. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, I.; Glikin, D.; Smolkin, B.; Hainrichson, M.; Belakhov, V.; Baasov, T. Repairing Faulty Genes by Aminoglycosides: Development of New Derivatives of Geneticin (G418) with Enhanced Suppression of Diseases-Causing Nonsense Mutations. Bioorg. Med. Chem. 2010, 18, 3735–3746. [Google Scholar] [CrossRef]
- Bidou, L.; Bugaud, O.; Belakhov, V.; Baasov, T.; Namy, O. Characterization of New-Generation Aminoglycoside Promoting Premature Termination Codon Readthrough in Cancer Cells. RNA Biol. 2017, 14, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, J.; Atia-Glikin, D.; Shulman, E.; Shapira, K.; Shavit, M.; Belakhov, V.; Baasov, T. Increased Selectivity toward Cytoplasmic versus Mitochondrial Ribosome Confers Improved Efficiency of Synthetic Aminoglycosides in Fixing Damaged Genes: A Strategy for Treatment of Genetic Diseases Caused by Nonsense Mutations. J. Med. Chem. 2012, 55, 10630–10643. [Google Scholar] [CrossRef] [PubMed]
- Mattis, V.B.; Ebert, A.D.; Fosso, M.Y.; Chang, C.-W.; Lorson, C.L. Delivery of a Read-through Inducing Compound, TC007, Lessens the Severity of a Spinal Muscular Atrophy Animal Model. Hum. Mol. Genet. 2009, 18, 3906–3913. [Google Scholar] [CrossRef] [PubMed]
- Nudelman, I.; Rebibo-Sabbah, A.; Shallom-Shezifi, D.; Hainrichson, M.; Stahl, I.; Ben-Yosef, T.; Baasov, T. Redesign of Aminoglycosides for Treatment of Human Genetic Diseases Caused by Premature Stop Mutations. Bioorg. Med. Chem. Lett. 2006, 16, 6310–6315. [Google Scholar] [CrossRef]
- Sabbavarapu, N.M.; Shavit, M.; Degani, Y.; Smolkin, B.; Belakhov, V.; Baasov, T. Design of Novel Aminoglycoside Derivatives with Enhanced Suppression of Diseases-Causing Nonsense Mutations. ACS Med. Chem. Lett. 2016, 7, 418–423. [Google Scholar] [CrossRef]
- Goldmann, T.; Rebibo-Sabbah, A.; Overlack, N.; Nudelman, I.; Belakhov, V.; Baasov, T.; Ben-Yosef, T.; Wolfrum, U.; Nagel-Wolfrum, K. Beneficial Read-Through of a USH1C Nonsense Mutation by Designed Aminoglycoside NB30 in the Retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6671–6680. [Google Scholar] [CrossRef]
- Nudelman, I.; Rebibo-Sabbah, A.; Cherniavsky, M.; Belakhov, V.; Hainrichson, M.; Chen, F.; Schacht, J.; Pilch, D.S.; Ben-Yosef, T.; Baasov, T. Development of Novel Aminoglycoside (NB54) with Reduced Toxicity and Enhanced Suppression of Disease-Causing Premature Stop Mutations. J. Med. Chem. 2009, 52, 2836–2845. [Google Scholar] [CrossRef]
- Xue, X.; Mutyam, V.; Tang, L.; Biswas, S.; Du, M.; Jackson, L.A.; Dai, Y.; Belakhov, V.; Shalev, M.; Chen, F.; et al. Synthetic Aminoglycosides Efficiently Suppress Cystic Fibrosis Transmembrane Conductance Regulator Nonsense Mutations and Are Enhanced by Ivacaftor. Am. J. Respir. Cell Mol. Biol. 2014, 50, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, T.; Overlack, N.; Möller, F.; Belakhov, V.; van Wyk, M.; Baasov, T.; Wolfrum, U.; Nagel-Wolfrum, K. A Comparative Evaluation of NB30, NB54 and PTC124 in Translational Read-through Efficacy for Treatment of an USH1C Nonsense Mutation. EMBO Mol. Med. 2012, 4, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Kerem, E. ELX-02: An Investigational Read-through Agent for the Treatment of Nonsense Mutation-Related Genetic Disease. Expert Opin. Investig. Drugs 2020, 29, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.K.; Alroy, I.; Sharpe, N.; Goddeeris, M.M.; Williams, G. ELX-02 Generates Protein via Premature Stop Codon Read-Through without Inducing Native Stop Codon Read-Through Proteins. J. Pharmacol. Exp. Ther. 2020, 374, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Crawford, D.K.; Mullenders, J.; Pott, J.; Boj, S.F.; Landskroner-Eiger, S.; Goddeeris, M.M. Targeting G542X CFTR Nonsense Alleles with ELX-02 Restores CFTR Function in Human-Derived Intestinal Organoids. J. Cyst. Fibros. 2021, 20, 436–442. [Google Scholar] [CrossRef] [PubMed]
- De Poel, E.; Spelier, S.; Suen, S.W.F.; Kruisselbrink, E.; Graeber, S.Y.; Mall, M.A.; Weersink, E.J.M.; van der Eerden, M.M.; Koppelman, G.H.; van der Ent, C.K.; et al. Functional Restoration of CFTR Nonsense Mutations in Intestinal Organoids. J. Cyst. Fibros. 2022, 21, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 Targets Genetic Disorders Caused by Nonsense Mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef]
- Huang, S.; Bhattacharya, A.; Ghelfi, M.D.; Li, H.; Fritsch, C.; Chenoweth, D.M.; Goldman, Y.E.; Cooperman, B.S. Ataluren Binds to Multiple Protein Synthesis Apparatus Sites and Competitively Inhibits Release Factor-Dependent Termination. Nat. Commun. 2022, 13, 2413. [Google Scholar] [CrossRef]
- Beznosková, P.; Cuchalová, L.; Wagner, S.; Shoemaker, C.J.; Gunišová, S.; von der Haar, T.; Valášek, L.S. Translation Initiation Factors eIF3 and HCR1 Control Translation Termination and Stop Codon Read-Through in Yeast Cells. PLoS Genet. 2013, 9, e1003962. [Google Scholar] [CrossRef]
- Samanta, A.; Stingl, K.; Kohl, S.; Ries, J.; Linnert, J.; Nagel-Wolfrum, K. Ataluren for the Treatment of Usher Syndrome 2A Caused by Nonsense Mutations. Int. J. Mol. Sci. 2019, 20, 6274. [Google Scholar] [CrossRef]
- Berger, J.; Li, M.; Berger, S.; Meilak, M.; Rientjes, J.; Currie, P.D. Effect of Ataluren on Dystrophin Mutations. J. Cell Mol. Med. 2020, 24, 6680. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Liu, X.; Welch, E.M.; Hirawat, S.; Peltz, S.W.; Bedwell, D.M. PTC124 Is an Orally Bioavailable Compound That Promotes Suppression of the Human CFTR-G542X Nonsense Allele in a CF Mouse Model. Proc. Natl. Acad. Sci. USA 2008, 105, 2064–2069. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.N.; Kovács, A.D.; Pearce, D.A. The Novel Cln1R151X Mouse Model of Infantile Neuronal Ceroid Lipofuscinosis (INCL) for Testing Nonsense Suppression Therapy. Hum. Mol. Genet. 2015, 24, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Guglieri, M.; Bushby, K. Molecular Treatments in Duchenne Muscular Dystrophy. Curr. Opin. Pharmacol. 2010, 10, 331–337. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Campbell, C.; Torricelli, R.E.; Finkel, R.S.; Flanigan, K.M.; Goemans, N.; Heydemann, P.; Kaminska, A.; Kirschner, J.; Muntoni, F.; et al. Ataluren in Patients with Nonsense Mutation Duchenne Muscular Dystrophy (ACT DMD): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Kerem, E.; Konstan, M.W.; De Boeck, K.; Accurso, F.J.; Sermet-Gaudelus, I.; Wilschanski, M.; Elborn, J.S.; Melotti, P.; Bronsveld, I.; Fajac, I.; et al. Ataluren for the Treatment of Nonsense-Mutation Cystic Fibrosis: A Randomised, Double-Blind, Placebo-Controlled Phase 3 Trial. Lancet Respir. Med. 2014, 2, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Flume, P.; McNamara, J.; Solomon, M.; Chilvers, M.; Chmiel, J.; Harris, R.S.; Haseltine, E.; Stiles, D.; Li, C.; et al. A Phase 3 Study of Tezacaftor in Combination with Ivacaftor in Children Aged 6 through 11 years with Cystic Fibrosis. J. Cyst. Fibros. Off. J. Eur. Cyst. Fibros. Soc. 2019, 18, 708–713. [Google Scholar] [CrossRef]
- Ryan, N.J. Ataluren: First Global Approval. Drugs 2014, 74, 1709–1714. [Google Scholar] [CrossRef]
- Auld, D.S.; Thorne, N.; Maguire, W.F.; Inglese, J. Mechanism of PTC124 Activity in Cell-Based Luciferase Assays of Nonsense Codon Suppression. Proc. Natl. Acad. Sci. USA 2009, 106, 3585–3590. [Google Scholar] [CrossRef]
- Moosajee, M.; Tracey-White, D.; Smart, M.; Weetall, M.; Torriano, S.; Kalatzis, V.; da Cruz, L.; Coffey, P.; Webster, A.R.; Welch, E. Functional Rescue of REP1 Following Treatment with PTC124 and Novel Derivative PTC-414 in Human Choroideremia Fibroblasts and the Nonsense-Mediated Zebrafish Model. Hum. Mol. Genet. 2016, 25, 3416–3431. [Google Scholar] [CrossRef]
- Du, L.; Damoiseaux, R.; Nahas, S.; Gao, K.; Hu, H.; Pollard, J.M.; Goldstine, J.; Jung, M.E.; Henning, S.M.; Bertoni, C.; et al. Nonaminoglycoside Compounds Induce Readthrough of Nonsense Mutations. J. Exp. Med. 2009, 206, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Kayali, R.; Ku, J.-M.; Khitrov, G.; Jung, M.E.; Prikhodko, O.; Bertoni, C. Read-through Compound 13 Restores Dystrophin Expression and Improves Muscle Function in the Mdx Mouse Model for Duchenne Muscular Dystrophy. Hum. Mol. Genet. 2012, 21, 4007–4020. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Jung, M.E.; Damoiseaux, R.; Completo, G.; Fike, F.; Ku, J.-M.; Nahas, S.; Piao, C.; Hu, H.; Gatti, R.A. A New Series of Small Molecular Weight Compounds Induce Read through of All Three Types of Nonsense Mutations in the ATM Gene. Mol. Ther. J. Am. Soc. Gene Ther. 2013, 21, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.Y.; Li, H.; Ghelfi, M.D.; Goldman, Y.E.; Cooperman, B.S. Ataluren and Aminoglycosides Stimulate Read-through of Nonsense Codons by Orthogonal Mechanisms. Proc. Natl. Acad. Sci. USA 2021, 118, e2020599118. [Google Scholar] [CrossRef] [PubMed]
- Bidou, L.; Bugaud, O.; Merer, G.; Coupet, M.; Hatin, I.; Chirkin, E.; Karri, S.; Demais, S.; François, P.; Cintrat, J.-C.; et al. 2-Guanidino-Quinazoline Promotes the Readthrough of Nonsense Mutations Underlying Human Genetic Diseases. Proc. Natl. Acad. Sci. USA 2022, 119, e2122004119. [Google Scholar] [CrossRef] [PubMed]
- Komarova (Andreyanova), E.S.; Osterman, I.A.; Pletnev, P.I.; Ivanenkov, Y.A.; Majouga, A.G.; Bogdanov, A.A.; Sergiev, P.V. 2-Guanidino-Quinazolines as a Novel Class of Translation Inhibitors. Biochimie 2017, 133, 45–55. [Google Scholar] [CrossRef]
- Karri, S.; Bidou, L.; Cornu, D.; Serot, C.; Hinzpeter, A.; Sermet-Gaudelus, I.; Namy, O. TLN468 Changes the Pattern of tRNA Used to Readthrough Premature Termination Codons in CFTR. Mol. Biol. 2023. preprint. [Google Scholar] [CrossRef]
- Benhabiles, H.; Gonzalez-Hilarion, S.; Amand, S.; Bailly, C.; Prévotat, A.; Reix, P.; Hubert, D.; Adriaenssens, E.; Rebuffat, S.; Tulasne, D.; et al. Optimized Approach for the Identification of Highly Efficient Correctors of Nonsense Mutations in Human Diseases. PLoS ONE 2017, 12, e0187930. [Google Scholar] [CrossRef]
- Trzaska, C.; Amand, S.; Bailly, C.; Leroy, C.; Marchand, V.; Duvernois-Berthet, E.; Saliou, J.-M.; Benhabiles, H.; Werkmeister, E.; Chassat, T.; et al. 2,6-Diaminopurine as a Highly Potent Corrector of UGA Nonsense Mutations. Nat. Commun. 2020, 11, 1509. [Google Scholar] [CrossRef]
- Leroy, C.; Spelier, S.; Essonghe, N.C.; Poix, V.; Kong, R.; Gizzi, P.; Bourban, C.; Amand, S.; Bailly, C.; Guilbert, R.; et al. Use of 2,6-Diaminopurine as a Potent Suppressor of UGA Premature Stop Codons in Cystic Fibrosis. Mol. Ther. 2023, 31, 970–985. [Google Scholar] [CrossRef]
- Carollo, P.S.; Tutone, M.; Culletta, G.; Fiduccia, I.; Corrao, F.; Pibiri, I.; Di Leonardo, A.; Zizzo, M.G.; Melfi, R.; Pace, A.; et al. Investigating the Inhibition of FTSJ1, a Tryptophan tRNA-Specific 2′-O-Methyltransferase by NV TRIDs, as a Mechanism of Readthrough in Nonsense Mutated CFTR. Int. J. Mol. Sci. 2023, 24, 9609. [Google Scholar] [CrossRef]
- Pibiri, I.; Melfi, R.; Tutone, M.; Di Leonardo, A.; Pace, A.; Lentini, L. Targeting Nonsense: Optimization of 1,2,4-Oxadiazole TRIDs to Rescue CFTR Expression and Functionality in Cystic Fibrosis Cell Model Systems. Int. J. Mol. Sci. 2020, 21, 6420. [Google Scholar] [CrossRef] [PubMed]
- Pibiri, I.; Pace, A.; Tutone, M.; Lentini, L.; Melfi, R.; Leonardo, A.D. Oxadiazole Derivatives for the Treatment of Genetic Diseases Due to Nonsense Mutations. US11203578B2. 2021. Available online: https://patents.google.com/patent/US11203578B2/en (accessed on 3 November 2023).
- Corrao, F.; Zizzo, M.G.; Tutone, M.; Melfi, R.; Fiduccia, I.; Carollo, P.S.; Leonardo, A.D.; Caldara, G.; Perriera, R.; Pace, A.; et al. Nonsense Codons Suppression. An Acute Toxicity Study of Three Optimized TRIDs in Murine Model, Safety and Tolerability Evaluation. Biomed. Pharmacother. 2022, 156, 113886. [Google Scholar] [CrossRef] [PubMed]
- England, C.G.; Ehlerding, E.B.; Cai, W. NanoLuc: A Small Luciferase Is Brightening up the Field of Bioluminescence. Bioconjug. Chem. 2016, 27, 1175–1187. [Google Scholar] [CrossRef]
- Sharma, J.; Du, M.; Wong, E.; Mutyam, V.; Li, Y.; Chen, J.; Wangen, J.; Thrasher, K.; Fu, L.; Peng, N.; et al. A Small Molecule That Induces Translational Readthrough of CFTR Nonsense Mutations by eRF1 Depletion. Nat. Commun. 2021, 12, 4358. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Aghajan, M.; Quesenberry, T.; Low, A.; Murray, S.F.; Monia, B.P.; Guo, S. Targeting Translation Termination Machinery with Antisense Oligonucleotides for Diseases Caused by Nonsense Mutations. Nucleic Acid Ther. 2019, 29, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Blanchet, S.; Cornu, D.; Hatin, I.; Grosjean, H.; Bertin, P.; Namy, O. Deciphering the Reading of the Genetic Code by Near-Cognate tRNA. Proc. Natl. Acad. Sci. USA 2018, 115, 3018–3023. [Google Scholar] [CrossRef]
- Roy, B.; Friesen, W.J.; Tomizawa, Y.; Leszyk, J.D.; Zhuo, J.; Johnson, B.; Dakka, J.; Trotta, C.R.; Xue, X.; Mutyam, V.; et al. Ataluren Stimulates Ribosomal Selection of Near-Cognate tRNAs to Promote Nonsense Suppression. Proc. Natl. Acad. Sci. USA 2016, 113, 12508–12513. [Google Scholar] [CrossRef]
- Simon, A.J.; Lev, A.; Wolach, B.; Gavrieli, R.; Amariglio, N.; Rosenthal, E.; Gazit, E.; Eyal, E.; Rechavi, G.; Somech, R. The Effect of Gentamicin-Induced Readthrough on a Novel Premature Termination Codon of CD18 Leukocyte Adhesion Deficiency Patients. PLoS ONE 2010, 5, e13659. [Google Scholar] [CrossRef]
- Bidou, L.; Hatin, I.; Perez, N.; Allamand, V.; Panthier, J.-J.; Rousset, J.-P. Premature Stop Codons Involved in Muscular Dystrophies Show a Broad Spectrum of Readthrough Efficiencies in Response to Gentamicin Treatment. Gene Ther. 2004, 11, 619–627. [Google Scholar] [CrossRef]
- Tate, W.P.; Poole, E.S.; Mannering, S.A. Hidden Infidelities of the Translational Stop Signal. Prog. Nucleic Acid Res. Mol. Biol. 1996, 52, 293–335. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.S.; Brown, C.M.; Tate, W.P. The Identity of the Base Following the Stop Codon Determines the Efficiency of in Vivo Translational Termination in Escherichia Coli. EMBO J. 1995, 14, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Cassan, M.; Rousset, J.-P. UAG Readthrough in Mammalian Cells: Effect of Upstream and Downstream Stop Codon Contexts Reveal Different Signals. BMC Mol. Biol. 2001, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Pelham, H.R. Leaky UAG Termination Codon in Tobacco Mosaic Virus RNA. Nature 1978, 272, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Tork, S.; Hatin, I.; Rousset, J.-P.; Fabret, C. The Major 5’ Determinant in Stop Codon Read-through Involves Two Adjacent Adenines. Nucleic Acids Res. 2004, 32, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Mikhailova, T.; Eliseev, B.; Yeramala, L.; Sokolova, E.; Susorov, D.; Shuvalov, A.; Schaffitzel, C.; Alkalaeva, E. PABP Enhances Release Factor Recruitment and Stop Codon Recognition during Translation Termination. Nucleic Acids Res. 2016, 44, 7766–7776. [Google Scholar] [CrossRef] [PubMed]
- Keeling, K.M.; Du, M.; Bedwell, D.M. Therapies of Nonsense-Associated Diseases; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Stephenson, L.S.; Maquat, L.E. Cytoplasmic mRNA for Human Triosephosphate Isomerase Is Immune to Nonsense-Mediated Decay despite Forming Polysomes. Biochimie 1996, 78, 1043–1047. [Google Scholar] [CrossRef]
- Torella, A.; Zanobio, M.; Zeuli, R.; Del Vecchio Blanco, F.; Savarese, M.; Giugliano, T.; Garofalo, A.; Piluso, G.; Politano, L.; Nigro, V. The Position of Nonsense Mutations Can Predict the Phenotype Severity: A Survey on the DMD Gene. PLoS ONE 2020, 15, e0237803. [Google Scholar] [CrossRef]
- Jia, J.; Werkmeister, E.; Gonzalez-Hilarion, S.; Leroy, C.; Gruenert, D.C.; Lafont, F.; Tulasne, D.; Lejeune, F. Premature Termination Codon Readthrough in Human Cells Occurs in Novel Cytoplasmic Foci and Requires UPF Proteins. J. Cell Sci. 2017, 130, 3009–3022. [Google Scholar] [CrossRef]
- Usuki, F.; Yamashita, A.; Higuchi, I.; Ohnishi, T.; Shiraishi, T.; Osame, M.; Ohno, S. Inhibition of Nonsense-Mediated mRNA Decay Rescues the Phenotype in Ullrich’s Disease. Ann. Neurol. 2004, 55, 740–744. [Google Scholar] [CrossRef]
- Kuzmiak, H.A.; Maquat, L.E. Applying Nonsense-Mediated mRNA Decay Research to the Clinic: Progress and Challenges. Trends Mol. Med. 2006, 12, 306–316. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Cougot, N.; Mahuteau-Betzer, F.; Nguyen, C.-H.; Grierson, D.S.; Bertrand, E.; Tazi, J.; Lejeune, F. Inhibition of Nonsense-Mediated mRNA Decay (NMD) by a New Chemical Molecule Reveals the Dynamic of NMD Factors in P-Bodies. J. Cell Biol. 2007, 178, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Grigoryan, A.; Wang, D.; Wang, J.; Breda, L.; Rivella, S.; Cardozo, T.; Gardner, L.B. Identification and Characterization of Small Molecules That Inhibit Nonsense-Mediated RNA Decay and Suppress Nonsense P53 Mutations. Cancer Res. 2014, 74, 3104–3113. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Hilarion, S.; Beghyn, T.; Jia, J.; Debreuck, N.; Berte, G.; Mamchaoui, K.; Mouly, V.; Gruenert, D.C.; Déprez, B.; Lejeune, F. Rescue of Nonsense Mutations by Amlexanox in Human Cells. Orphanet J. Rare Dis. 2012, 7, 58. [Google Scholar] [CrossRef]
- Atanasova, V.S.; Jiang, Q.; Prisco, M.; Gruber, C.; Piñón Hofbauer, J.; Chen, M.; Has, C.; Bruckner-Tuderman, L.; McGrath, J.A.; Uitto, J.; et al. Amlexanox Enhances Premature Termination Codon Read-Through in COL7A1 and Expression of Full Length Type VII Collagen: Potential Therapy for Recessive Dystrophic Epidermolysis Bullosa. J. Investig. Dermatol. 2017, 137, 1842–1849. [Google Scholar] [CrossRef] [PubMed]
- Benslimane, N.; Miressi, F.; Loret, C.; Richard, L.; Nizou, A.; Pyromali, I.; Faye, P.-A.; Favreau, F.; Lejeune, F.; Lia, A.-S. Amlexanox: Readthrough Induction and Nonsense-Mediated mRNA Decay Inhibition in a Charcot-Marie-Tooth Model of hiPSCs-Derived Neuronal Cells Harboring a Nonsense Mutation in GDAP1 Gene. Pharmaceuticals 2023, 16, 1034. [Google Scholar] [CrossRef]
- Ohguchi, Y.; Nomura, T.; Suzuki, S.; Takeda, M.; Miyauchi, T.; Mizuno, O.; Shinkuma, S.; Fujita, Y.; Nemoto, O.; Ono, K.; et al. Gentamicin-Induced Readthrough and Nonsense-Mediated mRNA Decay of SERPINB7 Nonsense Mutant Transcripts. J. Investig. Dermatol. 2018, 138, 836–843. [Google Scholar] [CrossRef]
- Floquet, C.; Deforges, J.; Rousset, J.-P.; Bidou, L. Rescue of Non-Sense Mutated P53 Tumor Suppressor Gene by Aminoglycosides. Nucleic Acids Res. 2011, 39, 3350–3362. [Google Scholar] [CrossRef]
- Pawlicka, K.; Kalathiya, U.; Alfaro, J. Nonsense-Mediated mRNA Decay: Pathologies and the Potential for Novel Therapeutics. Cancers 2020, 12, 765. [Google Scholar] [CrossRef]
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense Surveillance Regulates Expression of Diverse Classes of Mammalian Transcripts and Mutes Genomic Noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef]
- Wittmann, J.; Hol, E.M.; Jäck, H.-M. hUPF2 Silencing Identifies Physiologic Substrates of Mammalian Nonsense-Mediated mRNA Decay. Mol. Cell Biol. 2006, 26, 1272–1287. [Google Scholar] [CrossRef] [PubMed]
- Viegas, M.H.; Gehring, N.H.; Breit, S.; Hentze, M.W.; Kulozik, A.E. The Abundance of RNPS1, a Protein Component of the Exon Junction Complex, Can Determine the Variability in Efficiency of the Nonsense Mediated Decay Pathway. Nucleic Acids Res. 2007, 35, 4542–4551. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benslimane, N.; Loret, C.; Chazelas, P.; Favreau, F.; Faye, P.-A.; Lejeune, F.; Lia, A.-S. Readthrough Activators and Nonsense-Mediated mRNA Decay Inhibitor Molecules: Real Potential in Many Genetic Diseases Harboring Premature Termination Codons. Pharmaceuticals 2024, 17, 314. https://doi.org/10.3390/ph17030314
Benslimane N, Loret C, Chazelas P, Favreau F, Faye P-A, Lejeune F, Lia A-S. Readthrough Activators and Nonsense-Mediated mRNA Decay Inhibitor Molecules: Real Potential in Many Genetic Diseases Harboring Premature Termination Codons. Pharmaceuticals. 2024; 17(3):314. https://doi.org/10.3390/ph17030314
Chicago/Turabian StyleBenslimane, Nesrine, Camille Loret, Pauline Chazelas, Frédéric Favreau, Pierre-Antoine Faye, Fabrice Lejeune, and Anne-Sophie Lia. 2024. "Readthrough Activators and Nonsense-Mediated mRNA Decay Inhibitor Molecules: Real Potential in Many Genetic Diseases Harboring Premature Termination Codons" Pharmaceuticals 17, no. 3: 314. https://doi.org/10.3390/ph17030314
APA StyleBenslimane, N., Loret, C., Chazelas, P., Favreau, F., Faye, P.-A., Lejeune, F., & Lia, A.-S. (2024). Readthrough Activators and Nonsense-Mediated mRNA Decay Inhibitor Molecules: Real Potential in Many Genetic Diseases Harboring Premature Termination Codons. Pharmaceuticals, 17(3), 314. https://doi.org/10.3390/ph17030314





