Mulberry Leaf Polyphenol Extract and Rutin Induces Autophagy Regulated by p53 in Human Hepatoma HepG2 Cells
Abstract
:1. Introduction
2. Results
2.1. HPLC Characterization of MLPE Polyphenols
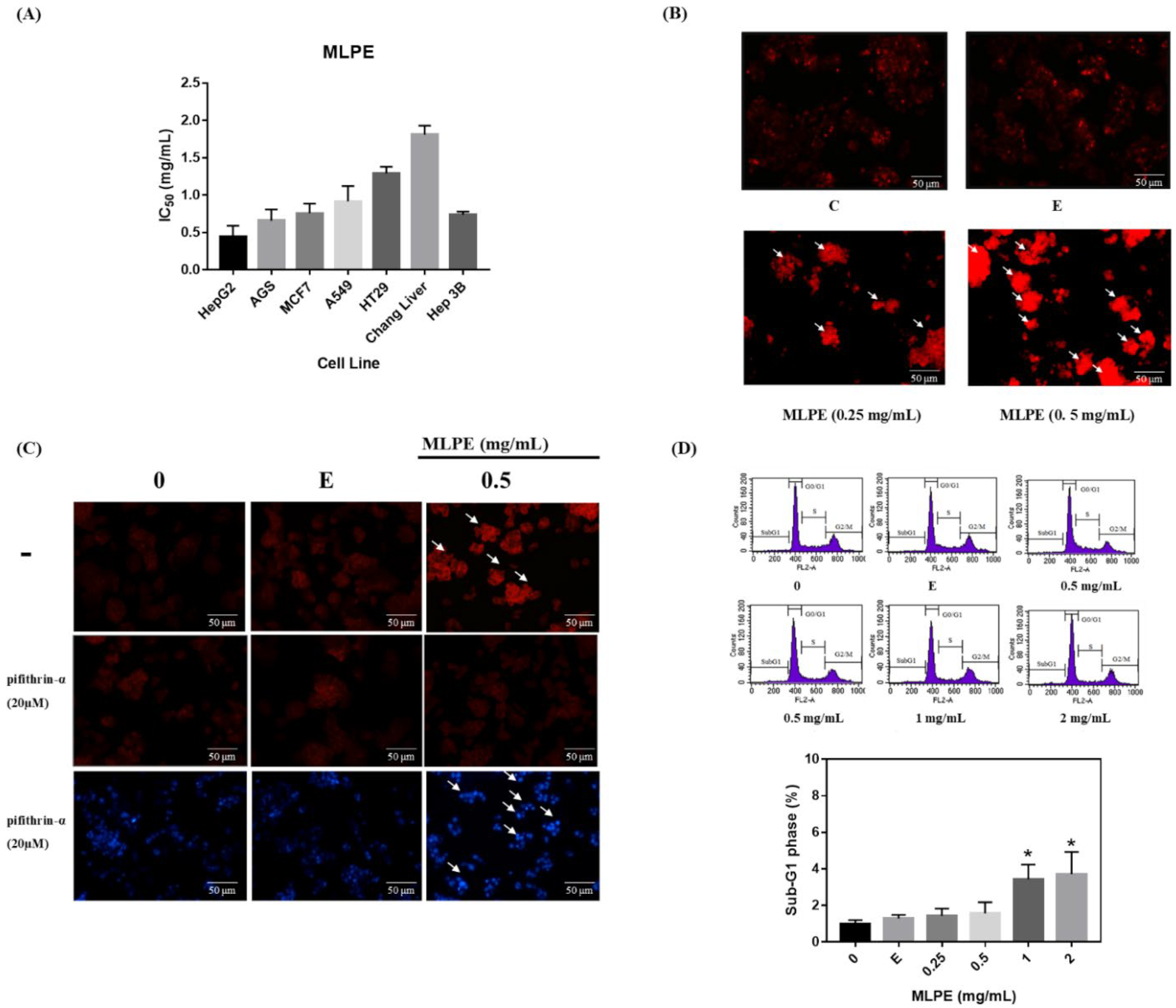
2.2. MLPE Cytotoxicity to Various Cell Lines
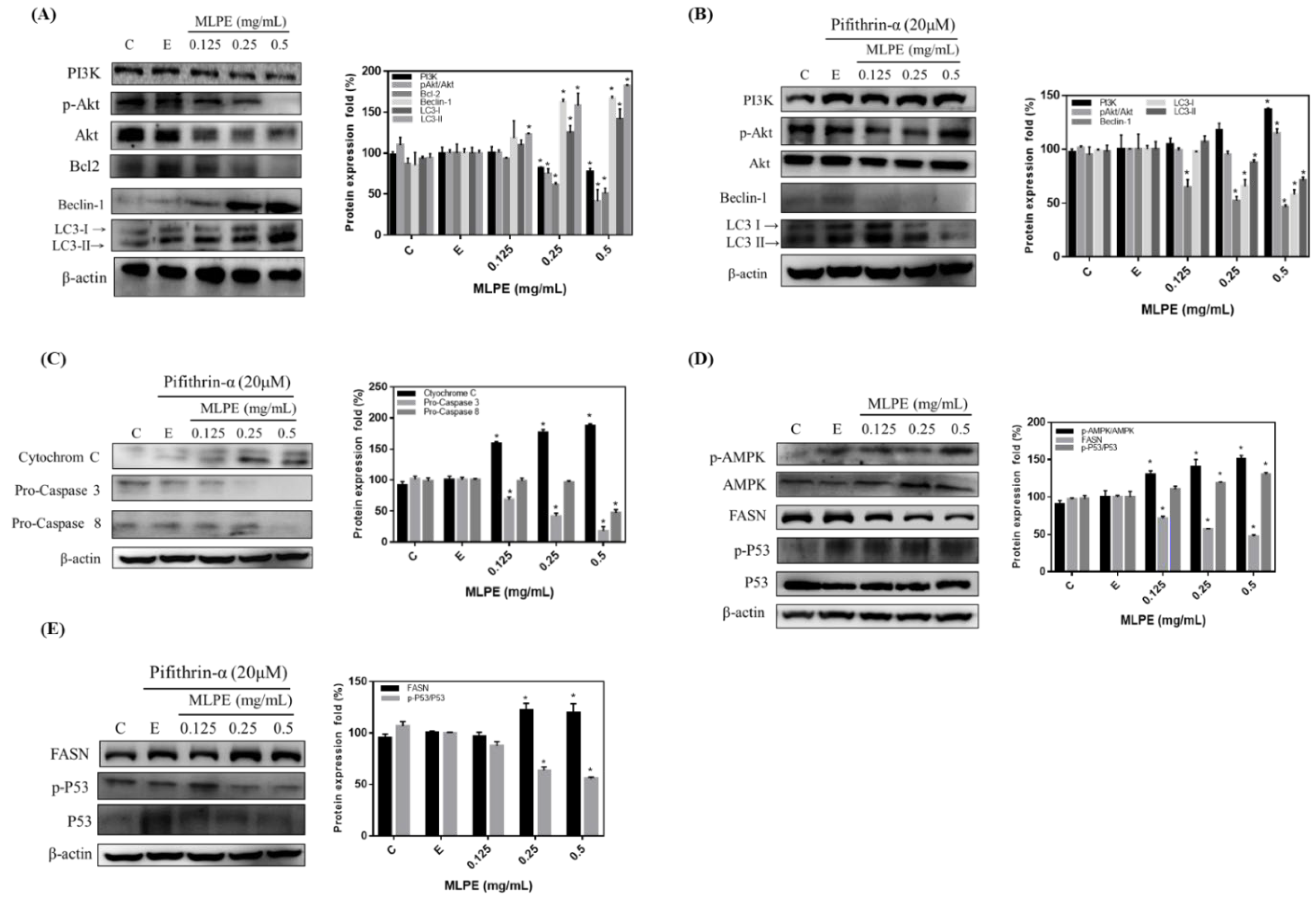
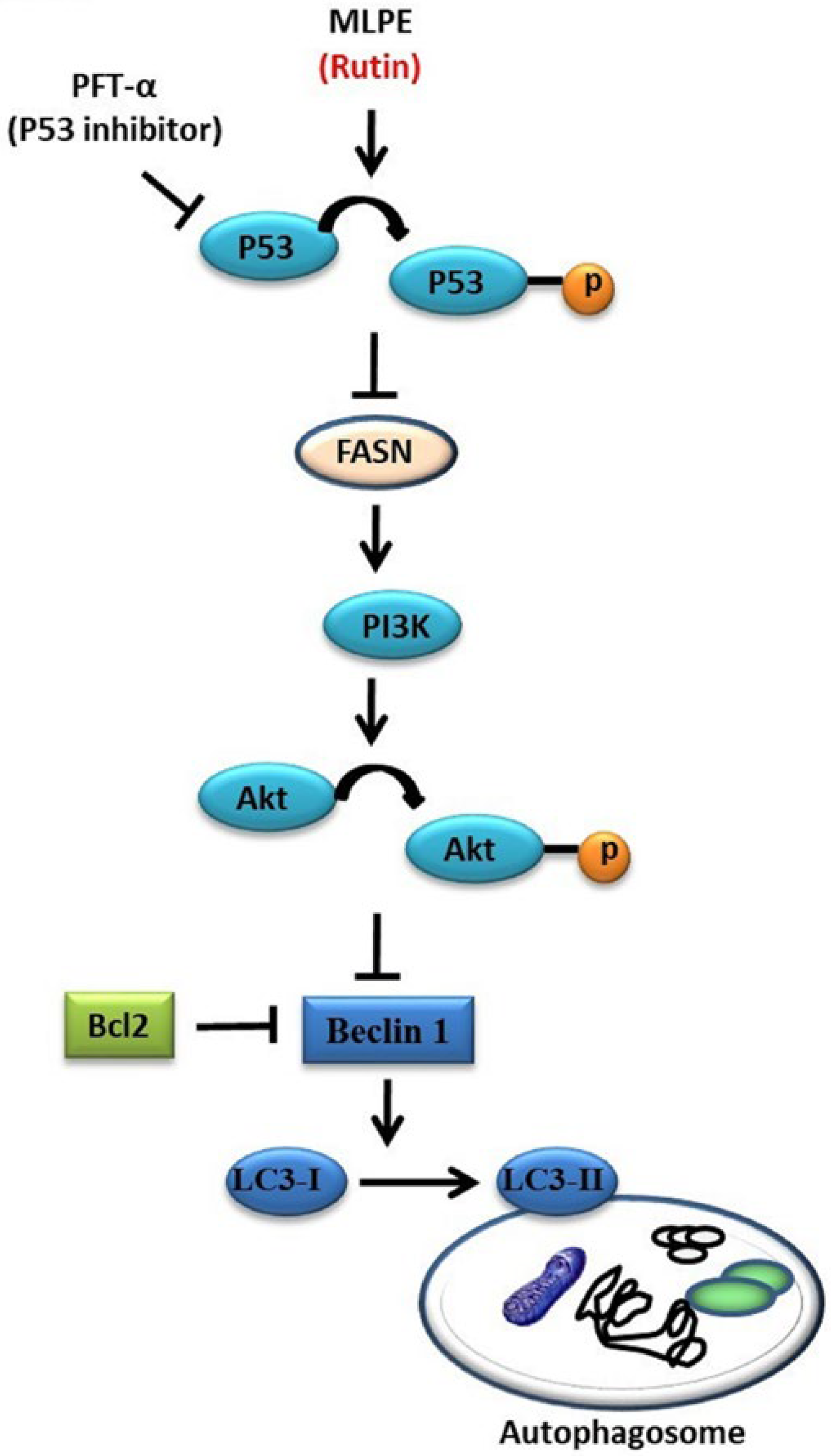
2.3. Regulation of MLPE-Induced Autophagy in HepG2 Cells by p53
2.4. Effect of MLPE on HepG2 Autophagy
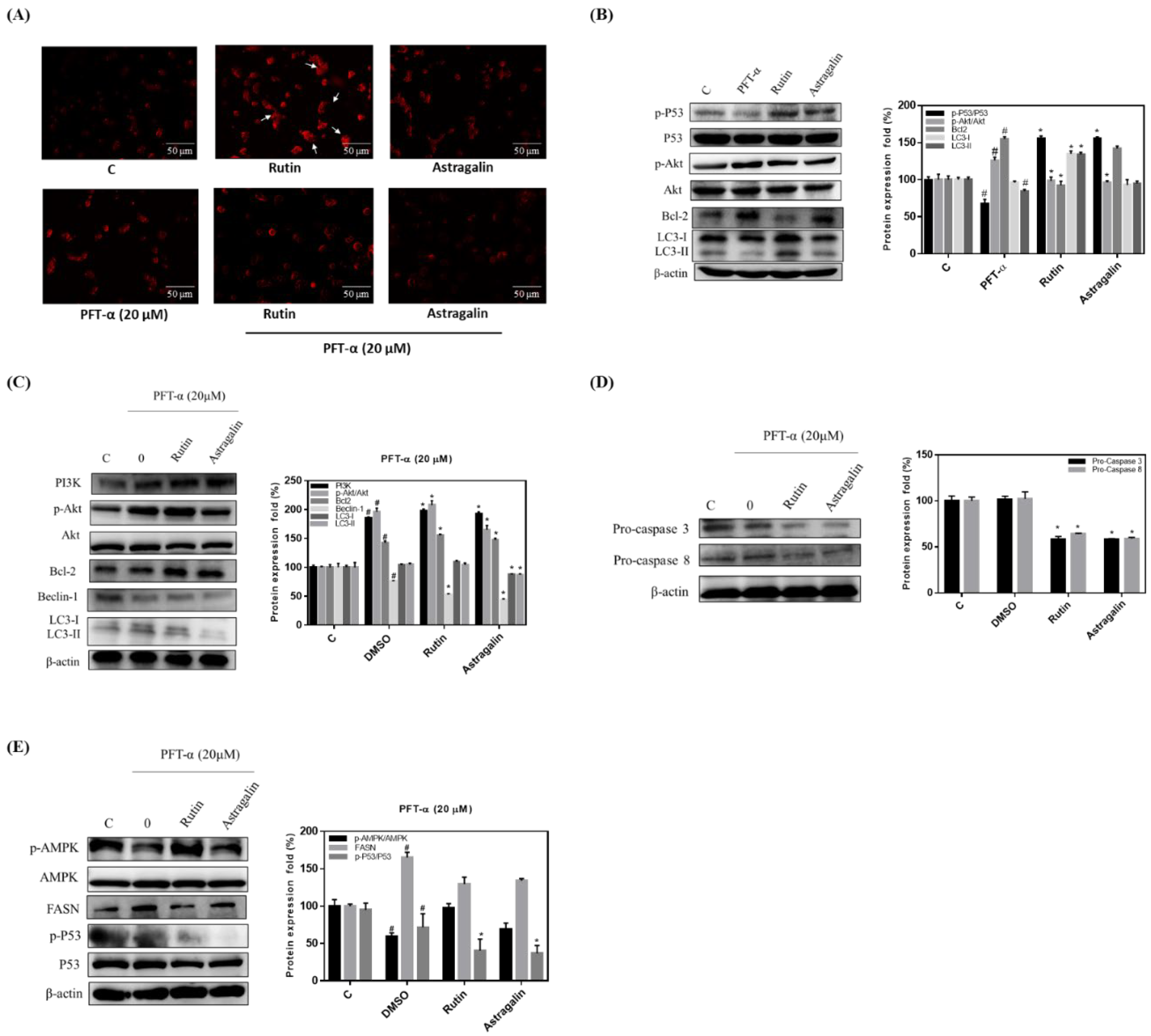
2.5. Rutin Mediation of Autophagy in MLPE-Treated HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. MLPE Preparation and High-Performance Liquid Chromatography
4.3. Cell Line Culture
4.4. Cell Viability Assay
4.5. DAPI and Acidic Vesicular Organelle Fluorescence Staining
4.6. Flow Cytometry for Cell Cycle Analysis
4.7. Protein Extraction and Western Blot
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hartke, J.; Johnson, M.; Ghabril, M. The diagnosis and treatment of hepatocellular carcinoma. Semin. Diagn. Pathol 2017, 34, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Sayiner, M.; Golabi, P.; Younossi, Z.M. Disease burden of hepatocellular carcinoma: A global perspective. Dig. Dis. Sci 2019, 64, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Merchant, T.E.; Bendel, A.E.; Sabin, N.D.; Burger, P.C.; Shaw, D.W.; Chang, E.; Wu, S.; Zhou, T.; Eisenstat, D.D.; Foreman, N.K. Conformal radiation therapy for pediatric ependymoma, chemotherapy for incompletely resected ependymoma, and observation for completely resected, supratentorial ependymoma. J. Clin. Oncol. 2019, 37, 974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makary, M.S.; Khandpur, U.; Cloyd, J.M.; Mumtaz, K.; Dowell, J.D. Locoregional therapy approaches for hepatocellular carcinoma: Recent advances and management strategies. Cancers 2020, 12, 1914. [Google Scholar] [CrossRef]
- Lee, H.-J.; Lee, H.; Choi, Y.-I.; Lee, J.-J. Effect of lactic acid bacteria-fermented mulberry leaf extract on the improvement of intestinal function in rats. Korean J. Food Sci. Anim. Resour. 2017, 37, 561. [Google Scholar] [CrossRef] [Green Version]
- Ai, J.; Bao, B.; Battino, M.; Giampieri, F.; Chen, C.; You, L.; Cespedes, C.; Ognyanov, M.; Tian, L.; Bai, W. Recent advances on bioactive polysaccharides from mulberry. Food Funct. 2021, 12, 5219–5523. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, B.; Xia, Y.; Li, H.; Shi, X.; Wang, J.; Deng, Z. Bioaccessibility and transformation pathways of phenolic compounds in processed mulberry (Morus alba L.) leaves after in vitro gastrointestinal digestion and faecal fermentation. J. Funct. Foods 2019, 60, 103406. [Google Scholar] [CrossRef]
- Peng, C.-H.; Lin, H.-T.; Chung, D.-J.; Huang, C.-N.; Wang, C.-J. Mulberry leaf extracts prevent obesity-induced NAFLD with regulating adipocytokines, inflammation and oxidative stress. J. Food Drug Anal. 2018, 26, 778–787. [Google Scholar] [CrossRef]
- Li, Q.; Liu, F.; Liu, J.; Liao, S.; Zou, Y. Mulberry leaf polyphenols and fiber induce synergistic antiobesity and display a modulation effect on gut microbiota and metabolites. Nutrients 2019, 11, 1017. [Google Scholar] [CrossRef] [Green Version]
- Demir, S.; Turan, I.; Aliyazicioglu, Y.; Kilinc, K.; Yaman, S.O.; Ayazoglu Demir, E.; Arslan, A.; Mentese, A.; Deger, O. Morus rubra extract induces cell cycle arrest and apoptosis in human colon cancer cells through endoplasmic reticulum stress and telomerase. Nutr. Cancer 2017, 69, 74–83. [Google Scholar] [CrossRef]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.; Iqbal, H.; Singh, K.P.; Joshi, S.K. A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: Current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, A.; Hasanain, M.; Kathuria, M.; Singh, A.; Datta, D.; Sarkar, J.; Mitra, K. Ormeloxifene-induced unfolded protein response contributes to autophagy-associated apoptosis via disruption of Akt/mTOR and activation of JNK. Sci. Rep. 2018, 8, 2303. [Google Scholar] [CrossRef] [Green Version]
- Pihán, P.; Carreras-Sureda, A.; Hetz, C. BCL-2 family: Integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017, 24, 1478–1487. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, M.S.; Zhang, H.-J.; Tsang, S.W. Perspectives of plant natural products in inhibition of cancer invasion and metastasis by regulating multiple signaling pathways. Curr. Med. Chem. 2018, 25, 5057–5087. [Google Scholar] [CrossRef]
- Graur, F.; Furcea, L.; Mois, E.; Biliuta, A.; Rozs, A.-T.; Negrean, V.; Al Hajjar, N. Analysis of p53 Protein Expression in Hepatocellular Carcinoma. J. Gastrointest. Liver Dis. 2016, 25, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.-P.; Lee, H.-J.; Ou, T.-T.; Chang, Y.-J.; Wang, C.-J. Mulberry leaf polyphenol extract induced apoptosis involving regulation of adenosine monophosphate-activated protein kinase/fatty acid synthase in a p53-negative hepatocellular carcinoma cell. J. Agric. Food Chem. 2012, 60, 6891–6898. [Google Scholar] [CrossRef]
- Dehelean, C.A.; Marcovici, I.; Soica, C.; Mioc, M.; Coricovac, D.; Iurciuc, S.; Cretu, O.M.; Pinzaru, I. Plant-Derived Anticancer Compounds as New Perspectives in Drug Discovery and Alternative Therapy. Molecules 2021, 26, 1109. [Google Scholar] [CrossRef]
- Zhu, J.; Singh, M.; Selivanova, G.; Peuget, S. Pifithrin-α alters p53 post-translational modifications pattern and differentially inhibits p53 target genes. Sci. Rep. 2020, 10, 1049. [Google Scholar] [CrossRef] [Green Version]
- Han, X.-X.; Jiang, Y.-P.; Liu, N.; Wu, J.; Yang, J.-M.; Li, Y.-X.; Sun, M.; Sun, T.; Zheng, P.; Yu, J.-Q. Protective effects of astragalin on spermatogenesis in streptozotocin-induced diabetes in male mice by improving antioxidant activity and inhibiting inflammation. Biomed. Pharmacother. 2019, 110, 561–570. [Google Scholar] [CrossRef]
- Linder, B.; Kögel, D. Autophagy in cancer cell death. Biology 2019, 8, 82. [Google Scholar] [CrossRef] [Green Version]
- Turk, M.; Tatli, O.; Alkan, H.F.; Ozfiliz Kilbas, P.; Alkurt, G.; Dinler Doganay, G. Co-Chaperone Bag-1 Plays a Role in the Autophagy-Dependent Cell Survival through Beclin 1 Interaction. Molecules 2021, 26, 854. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Bonam, S.R.; Bayry, J.; Tschan, M.P.; Muller, S. Progress and challenges in the use of MAP1LC3 as a legitimate marker for measuring dynamic autophagy in vivo. Cells 2020, 9, 1321. [Google Scholar] [CrossRef]
- Kim, S.M.; Ha, S.E.; Lee, H.J.; Rampogu, S.; Vetrivel, P.; Kim, H.H.; Venkatarame Gowda Saralamma, V.; Lee, K.W.; Kim, G.S. Sinensetin induces autophagic cell death through p53-related AMPK/mTOR signaling in hepatocellular carcinoma HepG2 cells. Nutrients 2020, 12, 2462. [Google Scholar] [CrossRef] [PubMed]
- Fitzwalter, B.E.; Towers, C.G.; Sullivan, K.D.; Andrysik, Z.; Hoh, M.; Ludwig, M.; O’Prey, J.; Ryan, K.M.; Espinosa, J.M.; Morgan, M.J. Autophagy inhibition mediates apoptosis sensitization in cancer therapy by relieving FOXO3a turnover. Dev. Cell 2018, 44, 555–565.e553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, Y.-L.; Min, M.; Shen, W.; Liu, Y. Genistein induced anticancer effects on pancreatic cancer cell lines involves mitochondrial apoptosis, G0/G1cell cycle arrest and regulation of STAT3 signalling pathway. Phytomedicine 2018, 39, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Kwon, Y.H. Regulation of apoptosis and autophagy by luteolin in human hepatocellular cancer Hep3B cells. Biochem. Biophys. Res. Commun. 2019, 517, 617–622. [Google Scholar] [CrossRef]
- Wei, G.; Huang, Y.; Li, F.; Zeng, F.; Li, Y.; Deng, R.; Lai, Y.; Zhou, J.; Huang, G.; Chen, D. XingNaoJing, prescription of traditional Chinese medicine, prevents autophagy in experimental stroke by repressing p53-DRAM pathway. BMC Complement. Altern. Med. 2015, 15, 377. [Google Scholar] [CrossRef] [Green Version]
- Delou, J.; Biasoli, D.; Borges, H.L. The complex link between apoptosis and autophagy: A promising new role for RB. An. Acad. Bras. Ciências 2016, 88, 2257–2275. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Jain, V.K.; Rizwanullah, M.; Ahmad, J.; Jain, K. PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: A review on drug discovery and future challenges. Drug Discov. Today 2019, 24, 2181–2191. [Google Scholar] [CrossRef]
- Chen, W.-L.; Jin, X.; Wang, M.; Liu, D.; Luo, Q.; Tian, H.; Cai, L.; Meng, L.; Bi, R.; Wang, L. GLUT5-mediated fructose utilization drives lung cancer growth by stimulating fatty acid synthesis and AMPK/mTORC1 signaling. JCI Insight 2020, 5, 5. [Google Scholar] [CrossRef]
- Huang, W.; Liang, Y.; Ma, X. Alpha-mangostin induces endoplasmic reticulum stress and autophagy which count against fatty acid synthase inhibition mediated apoptosis in human breast cancer cells. Cancer Cell Int. 2019, 19, 151. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Sureda, A.; Dehpour, A.R.; Shirooie, S.; Silva, A.S.; Devi, K.P.; Ahmed, T.; Ishaq, N.; Hashim, R.; Sobarzo-Sánchez, E. Regulation of autophagy by polyphenols: Paving the road for treatment of neurodegeneration. Biotechnol. Adv. 2018, 36, 1768–1778. [Google Scholar] [CrossRef]
- Park, M.H.; Kim, S.; Song, Y.-r.; Kim, S.; Kim, H.-J.; Na, H.S.; Chung, J. Rutin induces autophagy in cancer cells. Int. J. Oral. Sci. 2016, 41, 45–51. [Google Scholar] [CrossRef]
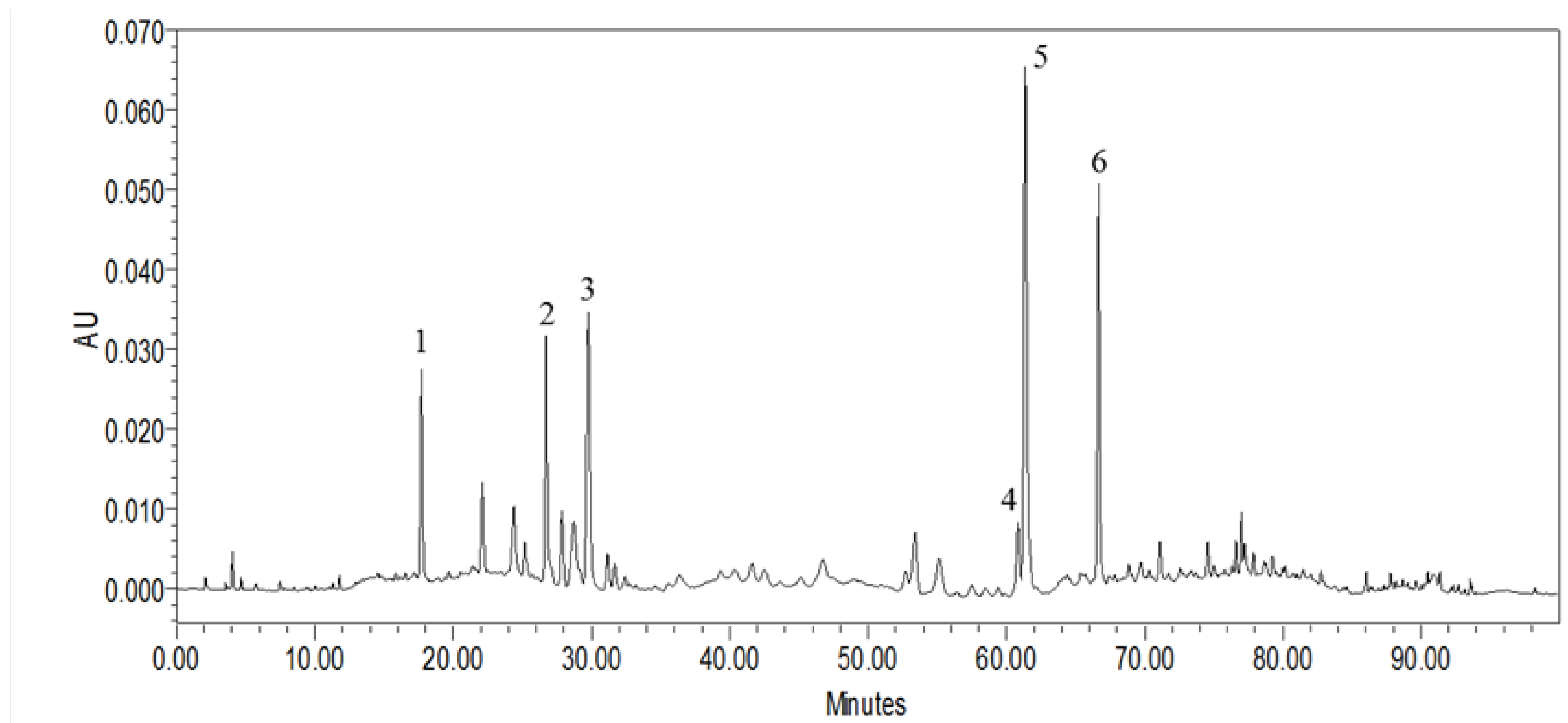
| Peak No. | RT (min) | [M + H]+ (m/z) | UV Band (nm) | Compounds Name | Concentration (μg/mg) |
|---|---|---|---|---|---|
| 1 | 17.72 | 153.1 [M − H]− | 259.7 (max), 291 | Protocatechuic acid | 6.25 ± 0.33 |
| 2 | 26.73 | 355.7 [M + H]+ | 241.9, 326.3 (max) | Chlorogenic acid | 12.06 ± 0.41 |
| 3 | 29.77 | 355.7 [M + H]+ | 238.4, 323.9 (max) | Cryptochlorogenic acid | 19.70 ± 1.68 |
| 4 | 60.84 | 595.9 [M + H]+ | 265.6 (max), 346.6 | Nicotiflorin | 8.34 ± 0.31 |
| 5 | 61.38 | 609.2 [M − H]− | 256.1 (max), 355.0 | Rutin | 43.35 ± 0.91 |
| 6 | 66.67 | 449.7 [M + H]+ | 265.6 (max), 346.6 | Astragalin | 24.67 ± 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, M.-H.; Tsai, M.-C.; Wang, C.-C.; Wu, S.-W.; Chang, Y.-J.; Wu, C.-H.; Wang, C.-J. Mulberry Leaf Polyphenol Extract and Rutin Induces Autophagy Regulated by p53 in Human Hepatoma HepG2 Cells. Pharmaceuticals 2021, 14, 1310. https://doi.org/10.3390/ph14121310
Yu M-H, Tsai M-C, Wang C-C, Wu S-W, Chang Y-J, Wu C-H, Wang C-J. Mulberry Leaf Polyphenol Extract and Rutin Induces Autophagy Regulated by p53 in Human Hepatoma HepG2 Cells. Pharmaceuticals. 2021; 14(12):1310. https://doi.org/10.3390/ph14121310
Chicago/Turabian StyleYu, Meng-Hsun, Ming-Chang Tsai, Chi-Chih Wang, Sheng-Wen Wu, Ya-Ju Chang, Cheng-Hsun Wu, and Chau-Jong Wang. 2021. "Mulberry Leaf Polyphenol Extract and Rutin Induces Autophagy Regulated by p53 in Human Hepatoma HepG2 Cells" Pharmaceuticals 14, no. 12: 1310. https://doi.org/10.3390/ph14121310
APA StyleYu, M.-H., Tsai, M.-C., Wang, C.-C., Wu, S.-W., Chang, Y.-J., Wu, C.-H., & Wang, C.-J. (2021). Mulberry Leaf Polyphenol Extract and Rutin Induces Autophagy Regulated by p53 in Human Hepatoma HepG2 Cells. Pharmaceuticals, 14(12), 1310. https://doi.org/10.3390/ph14121310







