3.1. Model Architecture
The different artifacts corrupting the ECG signal can be represented by mainly two components: additive and multiplicative components, respectively. For example, respiration motion causes the heart to change orientation and move from its original position (w.r.t. the measuring electrodes), causing amplitude modulation to the measured ECG [
61]. Furthermore, the change in conductivity of the skin tissues due to chest movement and blood volume shift in the area of measurement causes an effect known as baseline drift. Changes in body position, muscle tremors, and electrode motions can also cause similar effects [
62]. To model these effects, we consider these non-ECG components as either one of two components, i.e., multiplicative component
and additive component
, where those two components combine the rotation, scaling, baseline drift, and the additive Gaussian noise as shown in Equation (
3),
where
. The general aim would be to obtain the clean ECG signal
x from the noisy ECG signal
by suppressing the artifact components
and
. However, in this study, we are concerned with the additive component only, which represents the majority of different artifacts.
A denoising autoencoder (DAE) [
32] utilizing self-organized Operational Neural Network (self-ONN) [
48] layers is proposed to remove the additive artifacts in ECG signals. The DAE attempts to reconstruct the original clean data
x from a corrupted version of these data
, usually by applying a stochastic corruption process
such as applying Additive White Gaussian Noise (AWGN) or other means of partial destruction of the data. The encoder part of the DAE,
, tries to map the corrupted input data to a lower-dimensional manifold while ignoring the variations of the input data. The decoder part of the DAE,
, then tries to reconstruct the clean data from the learned manifold. The overall DAE model can be summarized by Equation (
4), where
is the estimated clean data:
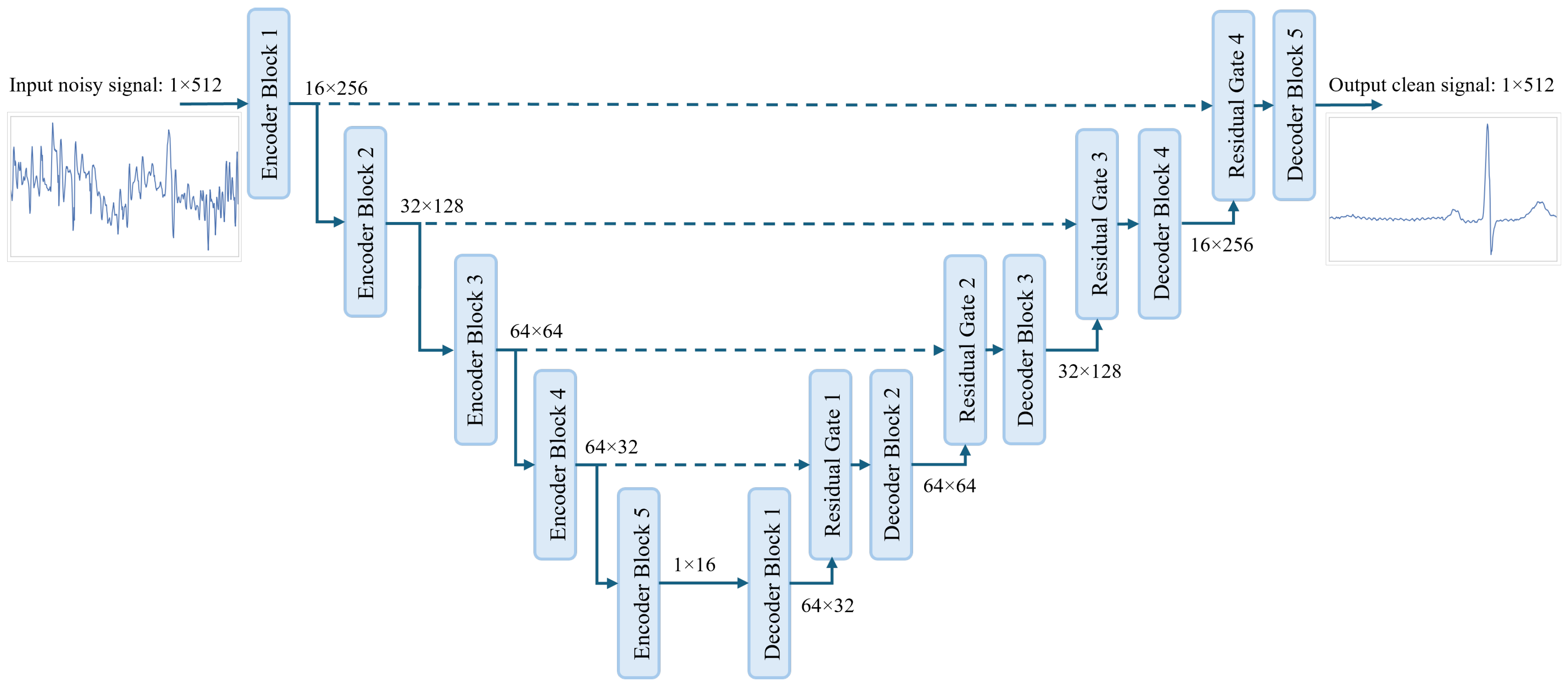
An overview of the proposed model is shown in
Figure 1. In our proposed model we utilize the powerful U-Net architecture [
57], which has proven to be efficient in ECG denoising. In U-Net architecture, the signal is passed through a few encoder layers that aim to find the latent representation of the input signal, which can be used by the decoder layers to reconstruct the clean ECG signal. The length of the signal after each encoder block is compressed, and the number of channels increases progressively. The opposite happens after each decoder layer, where the signal’s length increases and the number of channels decreases progressively. For the signal shape after each stage of the proposed model, we followed similar shapes as proposed by [
36], which we found to be well suited for our model.
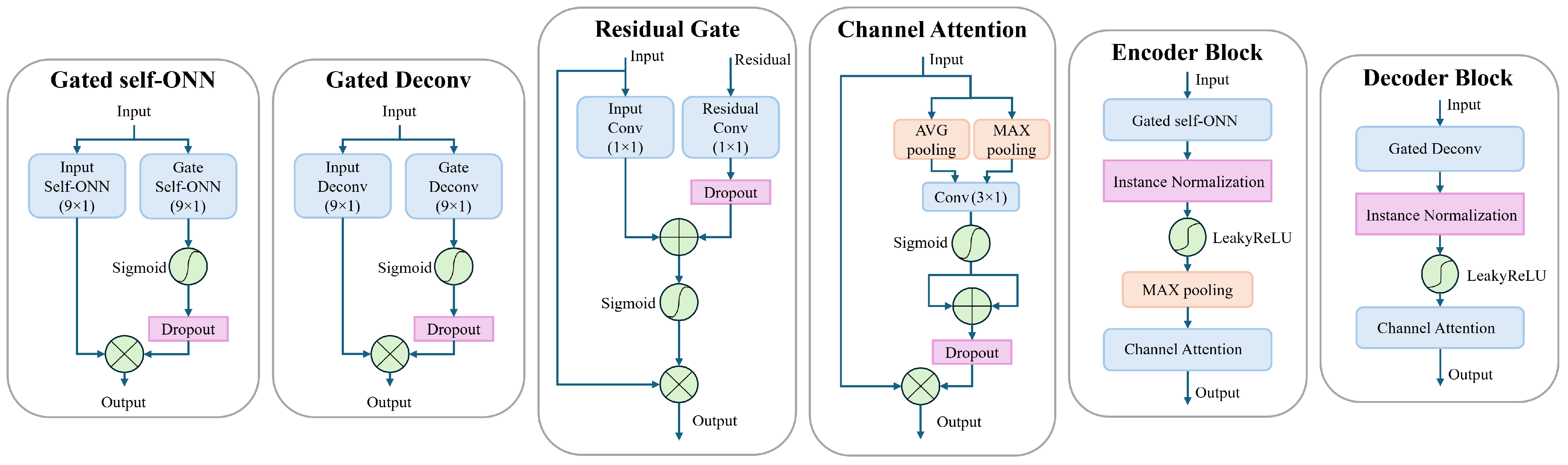
The structures of the encoder and decoder blocks are shown in
Figure 2. In the encoder block, we employ a gated self-ONN layer for feature extraction where two 1D self-ONN layers (kernel size = 9 × 1) parallelly process the input signal. One layer (input self-ONN) is extracting features from the input signal, while the other layer (gate self-ONN), followed by a sigmoid activation, is used to generate the gating mask to the extracted features. A dropout layer [
63] is introduced to the gating path after the sigmoid activation to force the gate self-ONN to produce a robust gating mask and prevent overfitting, which is inspired by [
64]. The dropout layer is by default active only during training, and we additionally limit it to be active only when the number of channels is more than 1 feature, which means that, for example, the dropout layer is not active in “Encoder block 5” shown in
Figure 1. That also applies to all the dropout layers used anywhere in the proposed model. The gated self-ONN layer can be expressed by Equation (
5),
where
x and
y are the input and output signals of the gated self-ONN layer, respectively. The ⊗ represents point-wise multiplication,
and
represent the input and gate self-ONN layers, respectively, generating input and gating feature maps, and
and
represent sigmoid activation and dropout layers, respectively.
The decoder block uses a similar feature extraction layer structure (Gated Deconv) where the only difference is that the 1D self-ONN layers are replaced by 1D transpose convolutional (a.k.a. Deconvolution) layers (kernel size = 9 × 1). Equation (
6) depicts the structure of the Gated Deconv block, where
and
represent the input and gate deconvolution layers, respectively, generating input and gating feature maps:
The feature extraction layers in encoder and decoder blocks are followed by a normalization layer where we employ the instance normalization [
65], which in our experiments, seemed to give a better performance than batch normalization [
66], for example. The instance normalization operates on each individual sample independently, rather than normalization across the entire batch as in the batch normalization. For non-linearity, we use LeakyReLU activation following the normalization layer. Then in the encoder only, a max pooling layer is utilized after the activation to divide the signal length in half as explained earlier. The decoder does not need a max-unpool layer since the stride is set to 2 in the “Gated Deconv” layers, which perform the needed unpooling effect. Finally, a channel attention module is utilized at the end of the encoder and decoder blocks. In our model, we use the efficient channel attention [
35]. Yet, upon testing, we found that incorporating the information obtained from max pooling into the original average pooling resulted in a better performance. This is similar to the channel attention proposed by [
37]. In the proposed model, we propose a small modification that enhances the performance of our model. Namely, the input signal is max-pooled and average-pooled simultaneously, the pooled features are passed through a shared 1D convolutional layer, and then each output is activated by a sigmoid activation function. The activation outputs are added together to form the attention mask. A dropout layer is applied to the attention mask to enhance generalization and prevent overfitting. The channel attention can be expressed as follows:
where
x and
y are the input and output signals of the channel attention block, respectively. The
operator represents the shared 1D convolutional layer, and
and
represent sigmoid activation and dropout layers, respectively. The attention mask is referred to as
, while
and
are the output features of applying global max pooling and global average pooling on the input signal, respectively. These are 1D features that have
C data points, where
C is the number of channels in the input signal
x. We follow the other configurations for the convolutional layer proposed by [
35], where, for the proposed model structure, the kernel size of the shared convolutional layer is 3 × 1, and “same padding” is used along all the used channel attention layers.
It is known that using residual connections, as in ResNet architectures [
67], can help overcome issues like vanishing gradients and allows for the effective training of very deep networks. Recent works showed that residual connections in U-Net architectures have proven to be beneficial for ECG denoising. Furthermore, it has been shown by [
62] that using spatial attention with point convolution on the residual connections on similar architecture helps in retaining the key features of the encoded feature maps and helps in better reconstruction in the decoder layers. Hence, in our work, we use the idea of residual gating proposed by Oktay et al. [
68] to utilize the information from both the skip connection and the decoder output to form a gating for the decoder output as shown in
Figure 2. The proposed residual gating is more global and robust than using attention on pooled features as in [
62]. Both the spatial information and channel information are retained and utilized effectively to generate the required gating mask. In the proposed residual gating, two convolutional layers with a kernel size of 1 × 1 are used to generate features from the residual connection signal and the output of the preceding decoder block, respectively. A dropout layer is employed after the “Residual Conv” layer to put more emphasis on the output of the “Input Conv” layer while forcing more robust features from the residual connection. The rationale here is that the residual connection signal might still partially have noisy components, hence the need for more robustness. A sigmoid activation is used on the sum of the obtained features to generate the gating mask to the input signal of the residual gating block. The residual gate block can be described by Equation (
9),
where
,
, and
y are the input feature map to be gated (i.e., the output of the previous decoder block), the residual connection feature map, and the output of the residual gating block, respectively. The
and
operators represent the 1D convolutional layers (k = 1) that operate on input and residual feature maps, respectively. The
and
operators, as usual, represent sigmoid activation and dropout layers, respectively.
3.2. Training Loss Functions
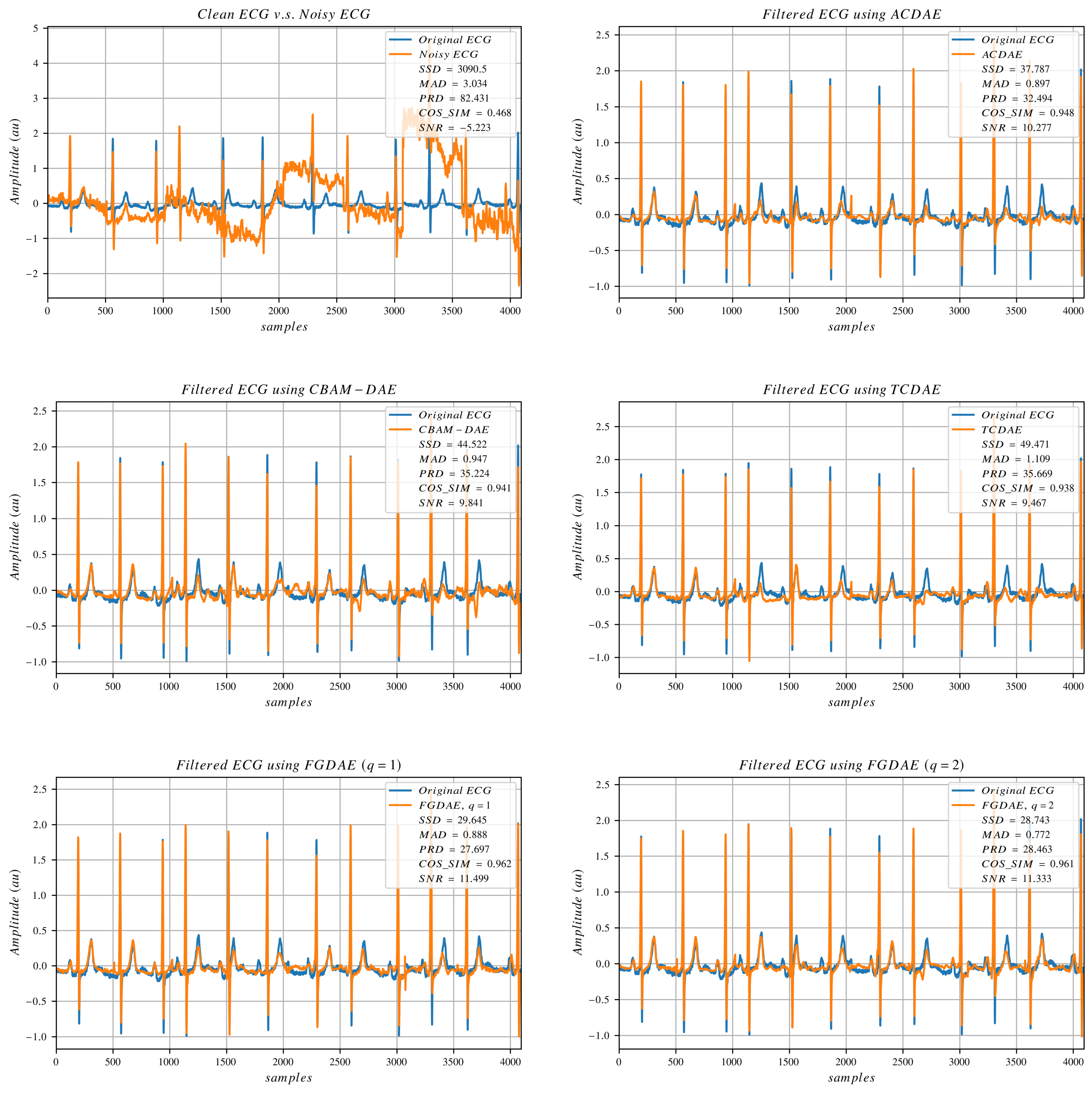
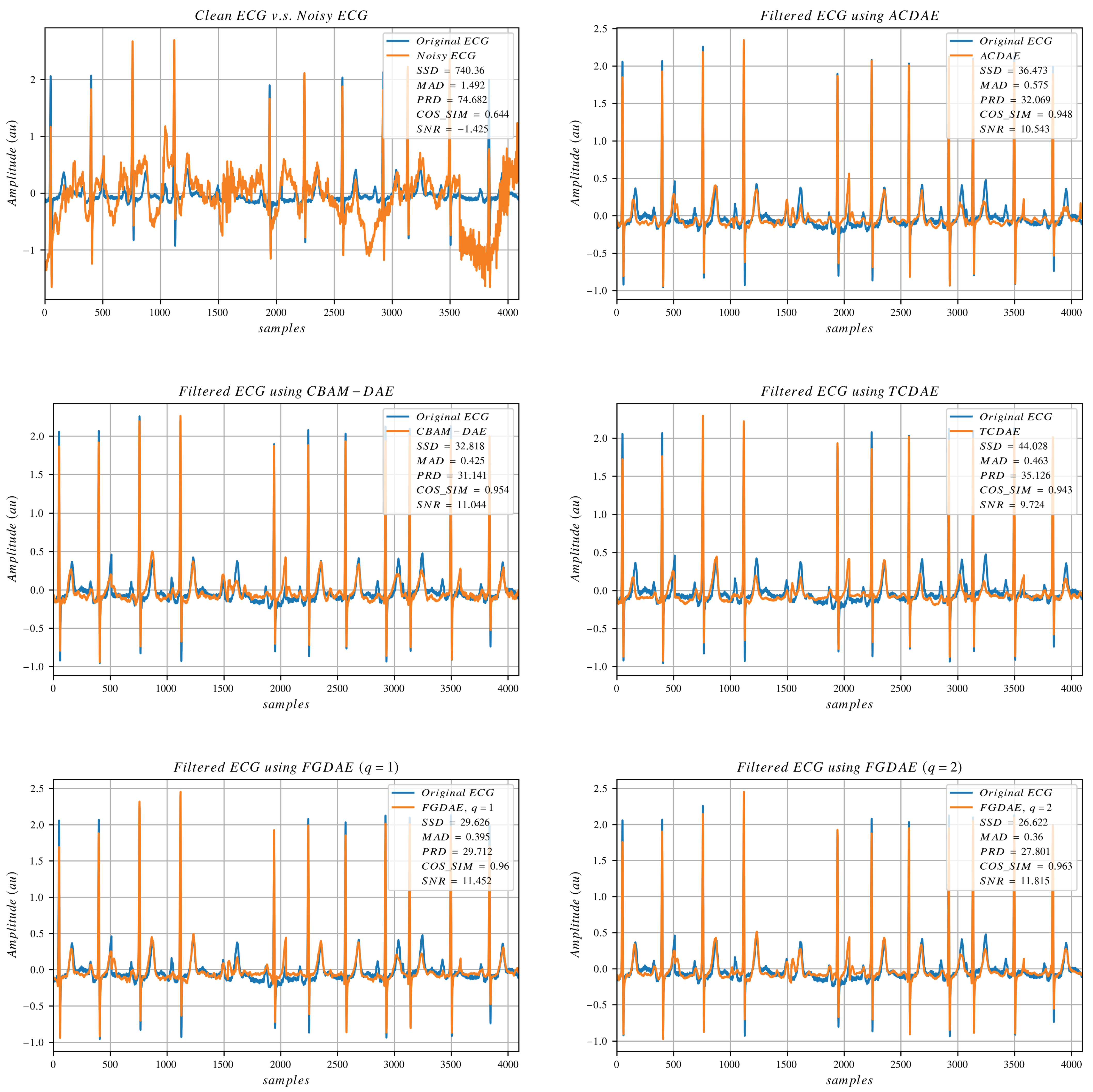
In this study, we aim to design a model that can benefit from the training process to gain the capability of performing effective denoising. Reducing the reconstruction error while keeping the morphology of the original signal as much as possible is of utmost priority. Hence, we designed a loss function that drives the model to learn the hidden features of the signal while retaining sparsity and generalization ability. As a reconstruction loss, we use the loss function proposed by [
28] as shown in Equation (
10),
where the sum of squared distance (SSD) (also defined in Equation (
17)) and maximum absolute distance (MAD) (also defined in Equation (
18)) are combined, and the tunable scaling coefficient
for MAD is set to 50, similar to [
28,
36]. This combined loss ensures that the overall signal looks similar to the target signal while also minimizing the point-wise errors and rejecting outliers.
To enhance the spatial resolution of the reconstructed signal and encourage the model to pay attention to the fine details, a gradient loss (G-Loss) [
69] is used as part of the overall model loss. The G-Loss was proposed by Ge et al. to improve the resolution of reconstructed images. In our case, it helps the model to follow the target signal closely and focus on subtle details that are not noise-related. The use of G-Loss also helps the model to learn a more detailed yet robust manifold of the ECG signals, which we believe would enhance the generalization ability of the model. We employed the idea of G-Loss, yet we calculated the MAD loss (instead of originally the sum of absolute error) between the gradients of the target signal
x and the estimated signal
. Following [
69], we did not add a scaling parameter to this loss component. This loss component aims to minimize the largest deviation between the gradients, preventing outliers and localized high-intensity noise. Equations (
11)–(
13) depict the proposed gradient loss:
To further enhance the morphological similitude between the target signal and the estimated signal, we employ a correlation loss using Pearson’s correlation coefficient to measure the correlation between the signals. Exponential transformation of correlation values was used in [
38] to enhance the correlation between power spectrums of the target and the estimated signals, while the absolute of the correlation was used as auxiliary loss in [
70] to enhance the morphology of ECG generated from given photoplethysmography (PPG) signals. In this study, we wanted to leverage the overall morphological resemblance between the target and the estimated signals, and hence we penalize the minimum correlation as shown in Equation (
14):
The rationale here is that the minimum correlations usually come from outliers or difficult pathological cases, and we wanted our model to be able to retain the morphology even for these challenging samples. The correlation loss is computed as
, where
and
are the
target and estimated samples. This means that the possible range of this loss component is from 0 to 2. The tunable scaling factor
is used to balance the significance of this component to other components of the overall loss. Empirically, we tested the values of
in the range of 5 to 30 and found 10 to be suitable for our model, where it is neither too large to dominate other loss components nor small enough to be negligible.
To promote the model’s sparsity, we employ
regularization on the weights of the encoder part of the model to ensure sparse and robust manifold learning. L1 regularization adds a penalty equal to the absolute value of the magnitude of the coefficients. L1 regularization can lead to sparse models where some weights become exactly zero, effectively selecting features and preventing overfitting [
71]. The
-regularization cost function (
) is described in Equation (
15),
where
is the set of weights of the
layer of the encoder, while the tunable weight decay factor
is used to control the strength of regularization. Empirically, we tested values for
in the range of 0.005 to 1 and found that 0.01 was suitable for our model. We noticed that higher values of
negatively affect the model performance and prevent it from converging due to over-regularization. A very low value of
makes the loss value very small, to a limit that it doesn’t affect the performance of the model.
The overall loss function (
) is the sum of all the discussed terms as shown in Equation (
16), promoting better reconstruction, morphological similitude, and sparsity:
3.3. Datasets and Experimental Setup
In this study, we follow the benchmark experiment guidelines proposed by [
28]. The benchmark tests the model performance on a dataset created by combining signals from two openly available databases: QT Database (QTDB) [
72] and the MIT-BIH Stress Test Database (NSTDB) [
73]. The QTDB provides the clean ECG samples collected from 7 other databases. A total of 105 records of dual-channel ECG with a duration of 15 min each with a sampling rate of 250 Hz are available from this database collected from different leads and containing different pathological cases. The NSTDB provides three artifact types (namely, baseline wanders (BW), muscle artifacts (MA), and electrode motions (EM)) that resemble ambulatory ECG noise. Each available artifact type has a duration of 30 min and a sampling rate of 360 Hz. The samples from QTDB are resampled to 360 Hz to match the sampling rate of noise records. In the original benchmark experiment, the ECG records from the QT database are corrupted by the noise in NSTDB that contains baseline wander (BW) only. The contamination noise samples are scaled randomly to have maximum amplitudes in the range [0.2:2] times the ECG sample maximum amplitude. Since NSTDB has noise records in two channels, each channel is split in half (where the first and second halves are referred to as “a” and “b”, respectively), and one half from each channel is used for either training or testing as shown in
Table 1.
The selected models are trained on ECG signals corrupted with “channel 1_a” and tested on ECG signals corrupted with “channel 2_b” (this noise version is referred to as “Noise v1 (nv1)” elsewhere in this study) and then trained on ECG signals corrupted with “channel 2_a” and tested on ECG signals corrupted with “channel 1_b” (this noise version is referred to as “Noise v2 (nv2)” elsewhere in this study). This ensures that each time the model is trained and tested on different uncorrelated noises. The combined results of the two experiments are then reported, as in the following section. The test datasets use selected subjects from the QT database as shown in
Table 2. The selected test set represents 13% of the total data amount. Using that inter-patient scheme for data division ensures better evaluation of the model’s generalization capability and adapts to real-world scenarios.
However, the original experiment assumes non-realistic assumptions for the data preparation, such as having one beat per signal (data sample) centered with zero-padding to ensure size 512 per data sample. In most real-life situations it is not possible to perform that segmentation process before denoising, especially when the signal is too corrupted. Furthermore, assuming spatial consistency in the input samples is impractical. Also, using only one noise type as in [
28] or all three noise types combined as in [
36] does not cover the possibilities of real noisy signals. Hence, we follow the proposed data preparation scheme proposed by [
38], where a sliding window of length 512 and overlap of 256 is used to segment the signals into data samples of length 512 each with no prior assumptions of the beat length or its spatial position in the obtained signal. Additionally, we noticed that the clean ECG signals are not entirely clean, as some of the segments include very slow baseline variations. We removed these variations by subtracting the mean of the segments. Following [
28], no amplitude normalizations of the clean ECG signals were done. Furthermore, we utilize the same Random Mixed Noise (RMN) scheme proposed by [
38], where 8 random combinations of noise are created in the training and testing datasets (shown in
Table 3), giving more variation to the datasets with more realistic scenarios.
The random noise scaling and random noise type combinations are independent of each other. An important point in this combination is that it provides clean signal samples, which is essential to have in the training datasets for the deep learning models to influence the denoising functionality only on noisy signals rather than blindly performing reduction in innate features of the signal. For each noise split, we end up with 91,062 pairs of noisy (input) and clean (reference) ECG samples for training data and 15,535 pairs for testing data.