Abstract
Various whole cell-based biosensors have been reported in the literature for the last 20 years and these reports have shown great potential for their use in the areas of pollution detection in environmental and in biomedical diagnostics. Unlike other reviews of this growing field, this mini-review argues that: (1) the selection of reporter genes and their regulatory proteins are directly linked to the performance of celllular biosensors; (2) broad enhancements in microelectronics and information technologies have also led to improvements in the performance of these sensors; (3) their future potential is most apparent in their use in the areas of medical diagnostics and in environmental monitoring; and (4) currently the most promising work is focused on the better integration of cellular sensors with nano and micro scaled integrated chips. With better integration it may become practical to see these cells used as (5) real-time portable devices for diagnostics at the bedside and for remote environmental toxin detection and this in situ application will make the technology commonplace and thus as unremarkable as other ubiquitous technologies.
1. Introduction
A biosensor is a type of sensor that can detect and identify a component within a cell or tissue. It is composed of synthetically made biomolecule recognition elements and various kinds of physical or chemical transducers [1]. Because of the cross-disciplinary nature of their construction, research on their development has been published in the fields of biology, chemistry, physics, and information science [2,3,4,5]. These biosensors can be classified into three types based on differences in their molecular, cellular and tissue sensing components [6]. The molecular-based biosensors use biological active substances such as enzymes, DNA, antigens, antibodies, and biofilms as the reporter elements [7]. The significant advantage of these molecular-based biosensors is their high selectivity [8]. However, the applicability of this type of biosensor has been limited by shortcomings such as expensive macromolecule isolation costs, limited detection capability and the short useable lifetime of the identifying molecules [1].
In contrast to molecular-based biosensors, sensors that are formed from cells or intact tisse have seen rapid development in novel methods of their microfabrication and immobilization, and these quite recent changes have provided these types of biosensors with unique and unexploited advantages [9]. Though whole cell-based biosensors are not as sensitive to environmental changes as molecular-based ones, these biosensors can be modified by simple genetic engineering methods so that they can then be used to detect a series of complex responses within a living cell. These biosensors can further provide information that molecule-based biosensors are not capable of, such as information related to the pharmacology, cell physiology, toxicology of a sample.
The main issues regarding the performance of a whole cell-based biosensor are: (1) the selection of reporter gene, as well as (2) the selectivity and sensitivity of the molecular recognition that occurs when regulator proteins bind to their target analytes [10]. As shown in Figure 1, the principal mechanism of a typical whole cell-based biosensor is the detection of a particular species of analyte and the amplification of this identification into an electrical and optical signal via a processor. This readout process is detectable through the immobilization and use of living cells or bacteria as the unit that provides the molecular recognition elements. Unlike a conventional biosensor, whole cell-based biosensors can detect a wider range of substances and thus are more sensitive to change in the electrochemical state of a tissue sample, other cells or in the environment, since they are capable of genetic modification and can operate over a broader range of conditions such as various temperature and pH values [11,12,13]. Because of the obvious advantages of whole cell-based biosensors such as their good sensitivity, high selectivity and their capability for high-throughput in situ detection, they have been applied successfully to fields such as environmental monitoring, food analysis, pharmacology and drug screening [10,14].
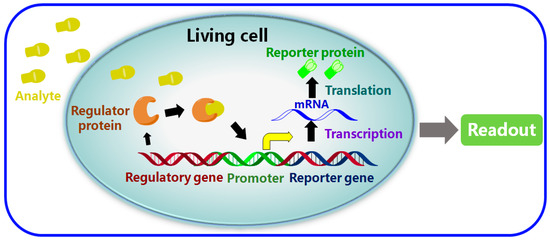
Figure 1.
A schematic diagram representing a typical whole cell-based biosensor.
Pollutants, such as heavy metals and biocides, are now commonly found in places where there is rapid industrial transformation and these pollutants are considered a global threat [15]. Toxins in the environment typically are associated with detrimental health outcomes and loss of ecological diversity [16,17]. Fast and accurate detection of pollutants is essential to reduce these threats. Although conventional detection techniques for environmental pollution using physical chemistry methods show a certain degree of sensitivity and specificity, there are still many challenges that limit their practical application such as the need to use expensive equipment, complicated and involved procedures, and long delays for detection to be complete [18,19,20]. Conventional approaches also typically over-evaluate the total amount of environmental pollutants and usually cannot measure the bioavailability of the contaminants themselves [21]. Measurements associated with bioavailability are better able to indicate the toxicity and effects of pollutants on living organisms and people. Since Sanseverino et al. reported on the design of a whole cell-based biosensor for naphthalene detection in 1990, it has become commonplace for whole cell-based biosensors to be used for bioavailability detection and toxicity assessment of contaminants [7,22], as these biosensors have been shown to be economic, simple, and efficient for the in situ detection of the bioavailability of a broad range of environmental pollutants.
On the other hand, development of biomedical sensor technologies at a cellular and molecular level has become important to clinical diagnostics especially where it is used for the detection of chronic pathologies. Cells consist of naturally evolved receptor, ion-channels, and enzymes that can be used as targets for biological or biologically active analytes. Thus, whole cell-based biosensors are able to measure functional information and the effects of the analyte on the physiological function of living cells. Parameters can include determining the impact of a compound drug composition on the physiological system, the inhibition or promotion effect relative to a given receptor, other influences that the analyte might have on the metabolism of cells (such as secondary messengers and their enzymes), and the toxicity and the side effects of the substances tested have on cells. Whole cell-based biosensors have therefore emerged as a dynamic technique for qualitative and quantitative analysis of different analytes for clinical diagnosis [23].
In this short-review article, we summarize what has occurred recently in the design and development of whole cell-based biosensors with an emphasis on their application for environmental pollution monitoring and biomedical diagnostics, two areas where these sensors are most commonly applied. We provide a literature summary on the selection of reporter genes, regulatory proteins, host cells, and multifunctionalization, for the detection of environmental pollutants such as heavy metals and organic waste. We also provide an outline of recent developments in the application of whole cell-based biosensors to the field of precision medicine, detection of micronutrients and the diagnosis of diseases. This work further provides a summary of the potential challenges and future prospects of the practical application of whole cell-based biosensors.
2. Using Cells That Can Act as Sensors Against Environmental Analytes
2.1. Detection of Bioavailability
The rapid detection of pollutants in an ecosystem and the evaluation of their impact on health is an important focus for research. Conventional physical and chemical-based analytical methods can be highly accurate and sensitive in determining the exact composition and content of toxins in samples, but only a small number of toxins can be tested for their bioavailability, toxicity and genotoxicity [24]. In most cases it becomes only possible for some important parameters to be measurable by using living cells [25]. Two unique advantages of whole cell-based biosensors are: (1) the ease with which they can be field-tested and (2) the ease with which they can identify those fractions that contain a bioavailable contaminant.
An example of this is given in Figure 2, where a high-performance liquid chromatography (HPLC) and a whole cell-based biosensor Pseudomonas putida (BMB-PL) were tested and their ability to detect phenanthrene (PHE) added to red soil samples was compared [26]. The initial concentration of PHE was tested at various values from 10 to 60 mg/kg. It was found that around 80% PHE was detected by HPLC. In contrast, the application of BMB-PL was able to detect the bioavailable fraction at levels much lower than those tested with the total amount of PHE. This was mainly due to the sample extraction procedure during HPLC measurement. Besides the detection of organics, Pasi et al. also tested the bioavailability of Pb and Cu concentrations in natural soil using a similar bioluminescent whole cell biosensor. This result was again consistent with the results demonstrated by the PHE study; a much higher selectivity was obtained using the whole cell-based biosensor [27]. The chemical analysis was also found to be more expensive, time-consuming, and required a specialized laboratory to perform its procedure. Finally, whole cell-based biosensors were shown to be able to continually monitor the bioavailability and concentration of toxic compounds in situ to the sample and and in real time [28].
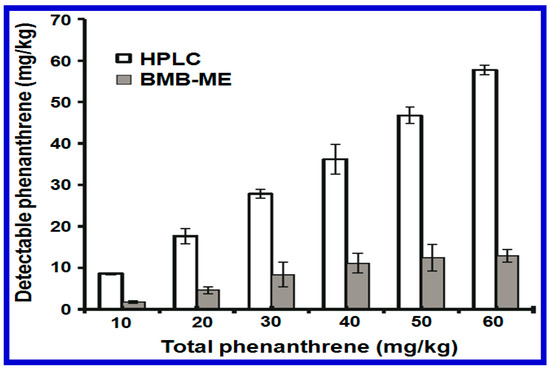
Figure 2.
A comparison of the PHE concentrations detected using a HPLC or using a BMB-PL whole cell biosensor approach is given. The values represent the mean ± standard error (n = 3) for the fluorescence and bioluminescence detected. Reprinted with permission from [26].
2.2. Reporter Genes
Typically, the performance of whole cell biosensors for the detection of environment pollutants is dependent on the reporter genes chosen for the control of transcriptional contaminants, and the type of regulatory protein associated with these promoters. A reporter gene in living cells used as sensors can convert its biological response into a signal detectable physicochemically. This process is critical to the sensitivity and selectivity of the whole cell biosensors. There are some widely used reporter genes that were shown to incorporate successfully into whole cell-based biosensors. These include lux (Bacterial lucberase), luc (firefly luciferase), gfp (green fluorescent protein) and lacZ (β-galactosidase). The selection of which reporter gene to use can be difficult since there is a large number from which to choose from. Table 1 lists some of the advantages and disadvantages of commonly reported reporter genes used to form a whole cell-based biosensor.

Table 1.
A summary of typical reporter genes used in whole cell-based sensors.
Gfp is a reporter gene coding for the green fluorescent protein (GFP) and it autofluorescences, so it does not require a substrate or an ATP to emit [34]. But with this reporter, the intrinsic fluorescence of certain host cells generally increases the background fluorescence and this can cause signal interference. So gfp-based biosensors are usually not able to detect with as much sensitivity as other lux- and lacZ based biosensors [35]. In addition, GFP requires a longer time to emit a stable fluorescence, reducing its maximum detection activity [36]. Thus, gfp-based whole cell biosensors are typically unsuitable for the rapid detection of containments. Likewise, the bacterial luciferases (lux) generally suffer from thermal lability and dimeric protein interference, and this also limits its use as a reporter gene in mammalian cells [37]. To avoid these limitations, the firefly luciferase (luc) reporter was frequently incorporated into mammalian cells considering its high sensitivity and broad linear range (up to 7–8 orders of magnitude) [38,39]. Another reporter, β-Galactosidase (lacZ), a well-characterized bacterial enzyme, was also frequently used in molecular biology as it is an excellent monitor of transfection efficiency. lacZ exhibits some unique advantages for detection using either colorimeric or fluorescent methods as its use with a sample is simple and rapid [40]. The wide availability of chemiluminescent and electrochemical substrates for lacZ also provides the advantages of ultra-high sensitivity, low detection limit (as low as 2 fg) and an extensive dynamic detection range [41,42].
In another reporter system, Fujimoto and coworkers developed a novel type reporter gene, crtA that is responsible for carotenoid synthesis [43]. When applied to a sample, the crtA-based whole cell-based biosensors change the color of the culture media from yellow to red without the addition of a supporting substrate, and is thus considered a good choice for rapid detection in emergency situations [44]. Table 2 compares the biosensing sensitivity of recently developed whole cell-based biosensors for the detection of environmental pollutants such as heavy metals and organic waste. A variety of cell lines and reporter genes were used in these systems. For instance, Sharma et al. utilized luxCDABE and E. coli to design whole cell-based biosensors and these were demonstrated to have a detection sensitivity of 0.74 μg/L when arsenic was added to water, a level well below the minimum safety standards for arsenic (10 μg/L) in the EU and in the United States [45]. In addition, the detection sensitivity of whole cell-based biosensors to organic waste was found to be excellent and showed great potential.

Table 2.
A comparison of the sensitivities for different types of whole cell-based biosensors.
2.3. Regulatory Proteins
The regulatory protein, which possesses complex interactions with the target analytes of the contaminants of interest, is essential for the specificity and sensitivity of the whole cell-based biosensors. In recent years, with the discovery of metallo-regulatory proteins, there are many reports elsewhere that utilized these biosensors for the detection of heavy metals in water and soil samples. These have shown higher selectivity, expanded detection ranges and enhanced sensitivity when compared to conventional biosensors. For example, GolS protein, a regulatory protein of the MerR family, was reported having a high selectivity for Au ions [56]. It was also found that with the introduction of a single amino acid at the position 77 in the GolS protein [57], the detection ranges of GolS-based whole cell biosensors were improved and became suitable for the detection of mercury (Hg), lead (Pd), cadmium (Cd) and/or gold (Au) ions (Figure 3). Metallothionein (MT), a cysteine-rich peptide, was reported having a high affinity to various heavy metals, and consisted of five isoforms encoded by T. thermophila genes [58,59]. In a report by Amro et al., a whole cell biosensor with MTT5 and MTT1 promoters (separated from MTs) was constructed, and it responded rapidly and strongly to the presence of heavy metal contaminants [10]. Whangsuk et al. discovered that the Sinorhizobium meliloti chpA promoter was highly induced by the pesticide chlorpyrifos (CPF) through the inclusion of the transcriptional activator ChpR [60].
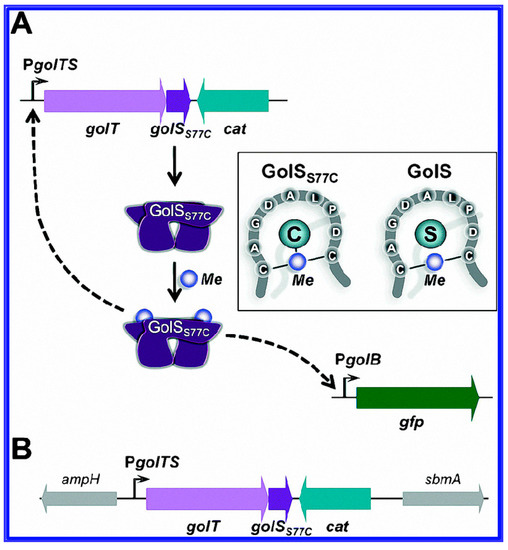
Figure 3.
(A) The GolS77C-based biosensor platform is shown. The sensor protein GolSS77C is expressed using its chromosomally encoded gene with an operon with golT encoding the P1B-type Au (I) transporter (STM14_0413-STM14_0412); (B) Genetic organization of the site chosen for the insertion of the golTSS77C-cat locus in the E. coli chromosome is also given. Reprinted with permission from [57].
In another report, the biosensor with a chpA promoter was hosted in Escherichia coli and used for the detection of CPF over a linear response range of 25 to 500 nM CPF [61]. Elsewhere, Fujimoto et al. developed a highly sensitive whole cell biosensor consisting of the promoter region of the ars operon and a reporter gene, the crtA gene, and were able to efficiently detect arsenite [43]. The color change was clearly recognized by the naked eye, even when the concentration of arsenite present was at 5 μg/L. The ars operon was also reported having an association with arsenite resistance, while the crtA gene, which determines the carotenoid synthesis in Rhodovulum sulfidophilum, was responsible for the color of the culture changing from yellow to red as the concentration of arsenite changed [44,62].
DNA microarrays are a technology that can be used as a high-throughput method for biosensors to select for positive regulatory genes as these arrays contain many protein, one of which typically responds to presence of the pollutant of interest in a sample. For example, Byoung et al. utilized DNA microarray data to select appropriate biomarker genes that were highly induced after exposure to the toxin paraquat [63]. Through two rounds of directed evolution of the transcriptional regulator, AlkSp protein, an alkane-inducible biosensor for detection of short-chain alkanes was constructed and the authors were able to demonstrate a 5-fold emission increase in its reporter signal [64].
2.4. Host Cells
The selection of the type of host cell is also important. A biosensor’s specificity, sensitivity and time-response, can be greatly influenced by the type of host cells used as the vehicle for sensing. Since there is a high similarity in metabolism, genome, and cellular organization between eukaryotic-based biosensors and a host organism, about 85% of whole cell biosensors currently used for detection of metals are eukaryotes [65]. Hernández-Sánchez and their coworkers constructed different whole cell biosensors using various host cells, but the same recombinant regulatory system, for the detection of monocyclic aromatic compounds in different environmental samples [66]. The biosensor from Alcanivorax borkumensis SK2 was reported as having a lower tolerance towards pollutants, but a higher tolerance towards salinity, and it demonstrated the best performance for detecting pollutants at low concentrations in seawater samples. The biosensor hosted on Pseudomonas putida DOT-T1E was found to be the best option for heavily contaminated environments due to its highest solvent tolerance at high concentrations before it bacame saturated [64]. As shown in Figure 4a, it was hard to obtain small oil droplets in the E. coli-oil mixture due to the poor accessibility and emulsifying capability of E. coli (the circle in Figure 4a) towards oil droplets. However, Acinetobacter baylyi ADP1 and its derivative ADPWH-alk (the circles in Figure 4b–d) were naturally adhesive to an oil-water interface and can emulsify both mineral and crude oils into oil droplets that are about 10–80 μm in diameter (Figure 4b–d) [67]. These properties make ADP1 an excellent bacterial substrate for constructing whole cell biosensors for detecting a broad range of alkanes and alkenes with carbon chain lengths from C7 to C36 found in samples such as water, seawater and soils [67,68,69].
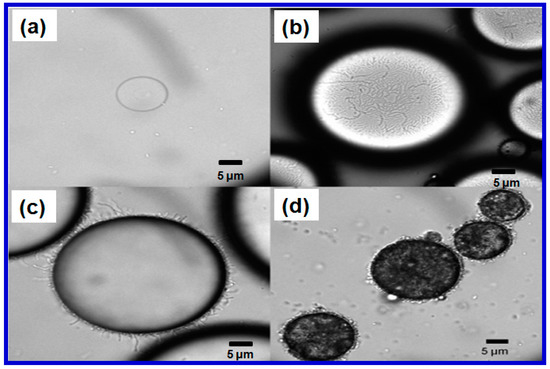
Figure 4.
(a) Poor accessibility and emulsifying capability of E. coli (the circle) towards oil droplets. Good affinity and emulsifying capability of acinetobacter baylyi ADPWH-alk (the circles) towards the surface of (b,c) the mineral oil droplets; and (d) the curde oil droplets. Reprinted with permission from [67].
A biosensor based on Alcanivorax borkumensis, specialized in assimilating linear alkanes, and exhibited a four-fold lower detection sensitivity towards the fuel octane (0.5 μM) when compared to biosensors that used Escherichia coli as a vehicle. This improvement in performance was most evident when measuring low concentrations of pure alkanes or petrol in samples [70]. Brutesco et al. prepared a functional biosensor based on Deinococcus deseri (an environmental, desiccation- and radiation-tolerant bacterium), and reported that these sensors were able to detect arsenite after 7 days of storage [71]. A series of whole cell biosensors were also prepared from several Escherichia coli strains for the detection of nickel in drinking water. In a typical example, the TD2158 wild-type E. coli showed a 10 times higher activity and sensitivity than its W3110 E. coli K12 equivalent, even though the same mechanism was used with RcnR Ni/Co metallo-regulator and a rcnA natural target promoter fused to the luc reporter genes was used [72]. In this work, the detection limit for Ni was reported to be as low as 80 nM, and so was found useful for meeting the required quality standards for most drinking water. These studies demonstrated that the choice of host bacteria has a significant impact on the performance of a fully constructed whole cell-based biosensor.
2.5. Multifunctionalization
Although whole cell biosensors typically exhibit better sensing performance than conventional chemical-based biosensors, the literature on the subject that has had a continual focus on improving the sensitivity, accuracy, and applicability of these sensors. This concern possibly reflects an understandable reaction to the widespread and increasing levels of pollution [73]. Recently, different types of functional whole cell biosensors were mixed together and were shown to be better at contaminant detection and measurement than a single biosensor type. For example, a combination of bioluminescent bacteria-based toxicity screening and a yeast-based estrogenic activity assay was applied to the monitoring of water samples [74]. Here, multiple biosensors were prepared using a combination of regulatory proteins such as CadC, ZntR, and ArsR, which then simultaneously responded when several different metals were added [75,76]. In another example, for the purpose of improving the specific and accurate detection of bioavailable Cd, Pb and As in a co-contaminated environment, a tailored whole cell biosensor set (Figure 5) was prepared by analyzing a E. coli sensor set using binary regression and binary linear equations to minimize the interference in signal generated when different metals (such as Cd, Pb and As) were presented [77].
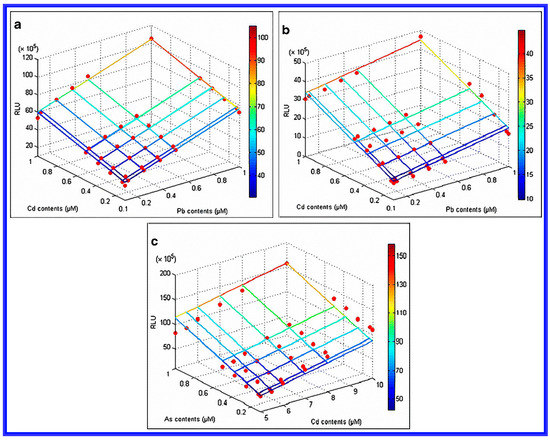
Figure 5.
Binary linear regressions of (a) a pcdCluc sensor in Group 1 to Cd-Pb mixtures; (b) a pzntRluc sensor in Group 1 to Cd-Pb mixtures; and (c) a parsRluc sensor in Group 2 to As-Cd mixtures are given. Reprinted with permission from [77].
In that work [77], sensor sets were classified into two groups according to their specific response to Cd, Pb and As. Group 1 was composed of pcadCluc and pzntRluc sensors to identify the bioavailability of Cd and Pb; Group 2 consisted of a parsRluc sensor to detect the specific bioavailable As. The increased concentration range of mixtures resulted in a linearly increase in the relative light unit (RLU). Three binary liner equations for two sensor groups were then used to determine the concentrations of bioavailable Cd, Pb, and As in soils sampled from a co-contaminated mine site. With Group 1, the overlapping concentration ranges (0.1–1.0 μM) of mixed Cd-Pt were determined using a linearly increased RLU. For Group 2, the overlapping concentration ranges were found to be 0.1–1.0 μM for As, and 5.0–10.0 μM for Cd. These results in this article showed that a conventional single target whole-cell sensor system tended to overestimate the bioavailability of the heavy metals detected. More accurate bioavailability data were obtained when a multiple whole cell biosensor were applied.
This review was, in part, born from increasing concerns with the so-called “antibiotics crisis” which relates to the excessive and unnecessary pesticides, antimicrobials use, and their uncontrolled environmental release [78]. There is an urgent demand for the development of whole cell-based biosensors for the stable and rapid detection of diverse antibiotics in samples sourced from extremely polluted environment. For example, microbial biosensors based on Pseudomonas putida DOT-T1E have shown a capability to detect a wide range of structurally diverse antibiotics due to its adaptability in the environmental and its resistance to a diverse number of toxic organic compounds [79].
Though whole cell-based biosensors have typically exhibited superior performance compared to their conventional chemical counterparts, their commercialize realization is still a major challenge. Reproducibility and stability during long-term storage and transport is still not efficient enough to meet the demands for their large-scale fabrication. Camanzi and colleagues, however found that freeze-dried bacteria emitted a stable luminous signal after their rehydration several months later [80]. Prévéral et al. developed an arsenite based whole cell biosensor that still retained its performance for the sensitive detection of arsenite seven months' after its lyophilization [81]. In that report, the entire bacterial luciferase lux operon (luxCDABE) with autonomous bioluminescence emission was used as the reporter gene, while lux a constitutive E. coli promoter PrpoD drove its expression. In another report, a flow-through biosensor based on disposable modular biochips incorporating agar-immobilized bioluminescent recombinant reporter bacteria, was proposed for the online and continuous monitoring of water toxicity [82]. This whole-cell biosensor was shown to work well in continuous flowing water over several days.
3. The Use of Sensors Based on Whole Cells in Medical Diagnostics
Cells possess and express a series of molecular recognition elements, such as receptors, ion channels, and enzymes. These molecules are usually sensitive to their corresponding analytes because of their native cellular mechanism [83]. Whole cell-based biosensors can therefore be used to continuously monitor and analyze a variety of physiological parameters under an external stimulation such as changes to the cell’s metabolism, impedance and action potential. These living cell-based sensors can thus be used to understand biological metabolic disorders and other diseases at a cellular level, and this has lead to these biosensors becoming widely applicable to many fields in biomedicine such as cellular physiological analysis, pharmaceutical evaluation and medical diagnosis.
3.1. Precision Medicine
A current research aim of precision medicine is to explain the genetic mutation of drug targets such as G protein-coupled receptors (GPCRs) and changes to their response to drugs . For instance, a novel label-free, whole cell-based biosensor was designed to characterize GPCR-mediated drug responses in lymphoblastoid cell lines (LCLs) [84]. This suggests their potential use as a cellular model system for the investigation of GPCR pharmacology in vitro in precision medicine. For example, Feng et al. fabricated a whole cell biosensor using a bullfrog fibroblast cell line (FT cells) expressing GPCRs as its basis, to evaluate the activity of GPCRs and determine the amount of adrenaline they secreted [85]. Here, the tumor suppressor gene p21 was used because it is a dominant downstream target gene of the activated p53 protein and is susceptible to carcinogens and can thus act as a sensor. In another report, Zager and co workers tested a human hepatoma cell-based biosensor that used a plasmid encoding enhanced green fluorescent protein (EGFP) under the control of a p21 promoter, for the simple and fast detection of genotoxic agents [86].
3.2. Detection of Micronutrients
Another area of application for whole cell sensors is in field of micronutrients. Riboflavin is an essential vitamin for human health and its lack of it can lead to serious diseases such as metabolism disorders, cataracts, and some cancers [87,88]. Excess intake of riboflavin can also be harmful and lead to oxidative damage in light-exposed tissue [89,90]. To address this, Si et al. reported a whole cell bioelectrochemical biosensor system for the amperometric detection of riboflavin [91]. As shown in Figure 6, a bioelectrochemical wire (BW) was made and consisted of riboflavin and cytochrome C strung between Shewanella oneidensis MR-1. An electrochemical response was obtained when riboflavin was added to the BW system. In comparison to conventional chemical biosensors, a 200 times enhancement in the electrochemical signal output was observed. The as-prepared whole cell biosensor had a high sensitivity (2.2 nM, S/N = 3), a wide linear range (5 nM–10 μM, 3 orders of magnitude) and a high resistance to signal interference.
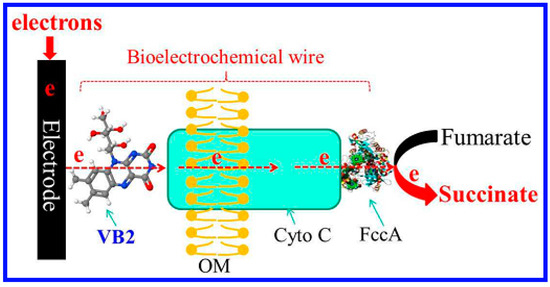
Figure 6.
A schematic diagram illustrating the electron transfer pathway used in bioelectrochemically-based whole cell biosensors. OM indicates the outer cell membrane; Cyto C represents cytochrome C proteins; and FccA indicates fumarate reductase. Reprinted with permission from [91].
3.3. Diagnosis of Diseases
Rapid and accurate detection of pathogens is an important goal in the diagnosis of diseases. Conventional microbiological methods can require several days to weeks since microbes in patient blood and urine samples have to first be precultured to sufficient numbers for their detection. A novel technique based on the whole cell biosensors was designed for the specific and direct detection of bacteria that did not require this lengthy culture step [92]. As seen in Figure 7, the whole cell biosensor, that included bacteriophages acting as recognition receptors, was immobilized covalently to a functionalized screen-printed carbon electrode (SPE) microarray for the direct impedimetric detection of E. coli. The results of the impedance measurement indicated that the biosensor acted as a rapid, direct, and label-free method for detecting specific bacteria, at the low cell numbers typically seen in patient samples.
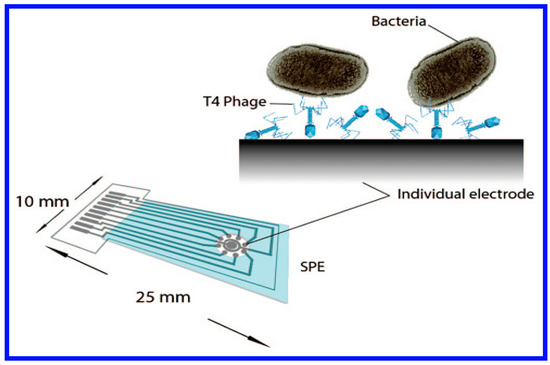
Figure 7.
A schematic diagram of the assay for immobilizing phages onto electrochemical electrodes is shown. Reprinted with permission from [92].
Whole cell-based biosensors can provide high-content screening and analysis in the diagnosis of diseases. For example, bacteria quorum-sensing molecules (QSMs) influenced 4% to 10% of bacterial genome and more than 20% of its proteome, indicating that quorum sensing was associated with both modulate virulence factor production and the basic metabolic processes [93]. Whole cell-based biosensors have also been used as the non-invasive methods to evaluate QSMs in physiological samples sourced from patients affected by bacterial gastrointestinal disorders [94,95]. The detection limits of QSMs were improved to the nanomolar level in biological matrices [96]. This was important, as invasive non-typhoid Salmonella (iNTS) is a main cause of human mortality and morbidity seen in young children and at-risk adults in sub-Saharan Africa [97]. Venkatesh et al. also reported a whole cell based electrochemical immunosensor, hosted in yeast, to detect iNTS antigens [98]. In that report, yeast cells were genetically modified to display both single chain variable fragment (scFv) antibodies and gold-binding peptide (GBP) on their surfaces. The resulting whole cell sensor showed a wide dynamic range with high nanomolar sensitivity and was capable of detecting iNTS OmpD antigens [98].
The unique advantages of whole cell-based biosensors have arisen in part because of their simplicity and fast application to result time for diagnosis of various diseases. Whole cell biosensor based biomedical diagnosis techniques thus show promise and great potential in the area of biomedical engineering.
4. A Summary on the Use of Whole Cell-Based Biosensors and Their Prospect
Biosensors formed from living cells have become an attractive field for researchers because of the rapid pace of innovations seen in microfabrication and in cell immobilization. Whole cell-based biosensors can quantitatively detect information relating to the status of live animal or bacteria cells, by converting signals that form part of their homeostasis, into electrical and optical outputs that can then be detected. Compared to conventional chemically based biosensors, these whole-cells can provide real-time, rapid and unique data streams of the homeostatic status of the cell and by inference, and its microenvironment, to a very high sensitivity and selectivity. This work reviewed recent progress in development and use of whole cell-based biosensors and their application across a range of fields with a particular focus on the area of environmental pollution monitoring and medical diagnostics.
Whole-cell based biosensors have many advantages, but there are also limitations to their use. Most environmental samples tested contain a large number of pollutants as well as other naturally occurring molecules and together these mask the signal coming from the analyte of interest [99]. Another issue is the toxic nature of the samples, as these can contain heavy metals or organic pollutants and their presence can limit the choice of cells used to those microbes resistant to their action. Finally, emission from whole cell-based sensors employed for extended periods of time can become unstable over time as these cells undergo leakage or diffusion. One solution reported elsewhere was to first immobilize cells onto a substrate before their application to a sample for testing [100].
The future prospects of whole cell-based biosensors may prove exciting. An early example of what this future might hold was demonstrated in a report on a portable whole cell-based optical biosensor named Luisens 2 developed to provide real-time online detection of pollutants [101]. The biosensor consisted of three main parts: (1) a central unit; (2) a disposable card where the bacteria were first immobilized and inserted; and (3) an acquisition unit to control the device. All kinds of recombinant bioluminescent bacteria were immobilized onto the disposable card. In another example, a portable whole cell biosensor system for environmental monitoring was demonstrated [102]. This system consisted of a lightweight mechanical housing, a temperature regulation system and a microfluidic housing bacterial inoculation channel. The system was used as a portable incubator for cell inoculation and as a remote monitor for testing at unreachable locations by its attachment to an unmanned aerial vehicle.
Another future focus for whole cell-based biosensors is the fabrication of specific and multifunctional biosensors for rapid and real-time detection in extreme unfriendly environments where there is high acidity, alkalinity, high salinity, extreme temperature, and toxic substrates [103]. It could be envisaged that many cell types, such as thermophiles, alkalophiles, halophiles and etc, could be applied as host-cells for their use as whole cell-based biosensors for extreme environmental analysis.
Acknowledgments
The Authors acknowledge financial supports for this work provided by Natural Science Foundation of Zhejiang Province (LR17H180001), National Natural Science Foundation of China (21374081, 51433005), and Xinmiao Talent Program of Zhejiang Province (2015R413062).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Daunert, S.; Barrett, G.; Feliciano, J.S.; Shetty, R.S.; Shrestha, S.; Smithspencer, W. Genetically engineered whole-cell sensing systems: Coupling biological recognition with reporter genes. Cheminform 2000, 100, 2705–2738. [Google Scholar]
- Zhang, W.; Li, X.; Zou, R.; Wu, H.; Shi, H.; Yu, S.; Liu, Y. Multifunctional glucose biosensors from Fe3O4 nanoparticles modified chitosan/graphene nanocomposites. Sci. Rep. 2015, 5, 11129. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Liu, Y.; Ou, S.; Pan, Y.; Zhang, S.; Chen, H.; Dai, L.; Qu, J. Optical turn-on sensors based on graphene oxide for selective detection of D-glucosamine. Anal. Chem. 2012, 84, 5641–5644. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Chen, W.; Ren, Q.; Hu, X.; Xiong, H.; Zhang, X.; Wang, S.; Zhao, Y. A highly sensitive nitric oxide biosensor based on hemoglobin–chitosan/graphene–hexadecyltrimethylammonium bromide nanomatrix. Sens. Actuators B Chem. 2012, 166–167, 444–450. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, C.; Yang, D.; Dong, G.; Jia, S.; Zhao, B.; Yan, L.; Yao, Q.; Sunna, A.; Liu, Y. Optical biosensors based on nitrogen-doped graphene functionalized with magnetic nanoparticles. Adv. Mater. Interfaces 2016, 3, 1600590. [Google Scholar] [CrossRef]
- Pancrazio, J.J.; Whelan, J.P.; Borkholder, D.A.; Ma, W.; Stenger, D.A. Development and application of cell-based biosensors. Ann. Biomed. Eng. 1999, 27, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Bousse, L. Whole cell biosensors. Sens. Actuators B Chem. 1996, 34, 270–275. [Google Scholar] [CrossRef]
- Du, H.; Strohsahl, C.M.; Camera, J.; Miller, B.L.; Krauss, T.D. Sensitivity and specificity of metal surface-immobilized “molecular beacon” biosensors. J. Am. Chem. Soc. 2005, 127, 7932–7940. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, C.; Cai, H.; Hu, N.; Zhou, J.; Wang, P. Cell-based biosensors and their application in biomedicine. Chem. Rev. 2014, 114, 6423–6461. [Google Scholar] [CrossRef] [PubMed]
- Raut, N.; O’Connor, G.; Pasini, P.; Daunert, S. Engineered cells as biosensing systems in biomedical analysis. Anal. Bioanal. Chem. 2012, 402, 3147–3159. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yoav, H.; Biran, A.; Pedahzur, R.; Belkin, S.; Buchinger, S.; Reifferscheid, G.; Shacham-Diamand, Y. A whole cell electrochemical biosensor for water genotoxicity bio-detection. Electrochim. Acta 2009, 54, 6113–6118. [Google Scholar] [CrossRef]
- Tian, Y.; Lu, Y.; Xu, X.; Wang, C.; Zhou, T.; Li, X. Construction and comparison of yeast whole-cell biosensors regulated by two RAD54 promoters capable of detecting genotoxic compounds. Toxicol. Mech. Methods 2017, 27, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Behzadian, F.; Barjeste, H.; Hosseinkhani, S.; Zarei, A.R. Construction and characterization of Escherichia coli whole-cell biosensors for toluene and related compounds. Curr. Microbiol. 2011, 62, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Stüben, D.; Berner, Z.; Albers, J.; Hintsche, R.; Jantzen, E. Development of an ultramicroelectrode arrays (UMEAs) sensor for trace heavy metal measurement in water. Sens. Actuators B Chem. 2004, 97, 168–173. [Google Scholar] [CrossRef]
- Gardner, G.T.; Stern, P.C. Environmental Problems and Human Behavior; Allyn & Bacon: Boston, MA, USA, 1996. [Google Scholar]
- Pulido, M.D.; Parrish, A.R. Metal-induced apoptosis: Mechanisms. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2003, 533, 227–241. [Google Scholar] [CrossRef]
- Derraik, J.G. The pollution of the marine environment by plastic debris: A review. Mar. Pollut. Bull. 2002, 44, 842–852. [Google Scholar] [CrossRef]
- Turdean, G.L. Design and development of biosensors for the detection of heavy metal toxicity. Int. J. Electrochem. 2011. [Google Scholar] [CrossRef]
- Tüzen, M. Determination of heavy metals in soil, mushroom and plant samples by atomic absorption spectrometry. Microchem. J. 2003, 74, 289–297. [Google Scholar] [CrossRef]
- Noll, M.R. Trace elements in terrestrial environments: Biogeochemistry, bioavailability, and risks of metals. J. Environ. Qual. 2003, 32, 374. [Google Scholar] [CrossRef]
- Bosma, T.N.; Middeldorp, P.J.; Schraa, G.; Zehnder, A.J. Mass transfer limitation of biotransformation: Quantifying bioavailability. Environ. Sci. Technol. 1996, 31, 248–252. [Google Scholar] [CrossRef]
- Sanseverino, J.; Dunbar, P.; Larimer, F.; Sayler, G. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Biophys. J. 1986, 49, 79. [Google Scholar]
- Andreescu, S.; Sadik, O.A. Trends and challenges in biochemical sensors for clinical and environmental monitoring. Pure Appl. Chem. 2004, 76, 861–878. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef]
- Amaro, F.; Turkewitz, A.P.; Gutiérrez, J.C. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb. Biotechnol. 2011, 4, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Ze-Ling, S.; Le-Le, C.; Wen-Hui, Z.; Chuan-Chao, D. Specific detection of bioavailable phenanthrene and mercury by bacterium reporters in the red soil. Int. J. Environ. Sci. Technol. 2014, 11, 685–694. [Google Scholar] [CrossRef]
- Peltola, P.; Ivask, A.; Åström, M.; Virta, M. Lead and Cu in contaminated urban soils: Extraction with chemical reagents and bioluminescent bacteria and yeast. Sci. Total Environ. 2005, 350, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ecken, H.; Ingebrandt, S.; Krause, M.; Richter, D.; Hara, M.; Offenhäusser, A. 64-Channel extended gate electrode arrays for extracellular signal recording. Electrochim. Acta 2003, 48, 3355–3362. [Google Scholar] [CrossRef]
- Hakkila, K.; Maksimow, M.; Karp, M.; Virta, M. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 2000, 301, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.C.; Amaro, F.; Martín-González, A. Heavy metal whole-cell biosensors using eukaryotic microorganisms: An updated critical review. Front. Microbiol. 2015, 6, 1–8. [Google Scholar]
- Sagi, E.; Hever, N.; Rosen, R.; Bartolome, A.J.; Premkumar, J.R.; Ulber, R.; Lev, O.; Scheper, T.; Belkin, S. Fluorescence and bioluminescence reporter functions in genetically modified bacterial sensor strains. Sens. Actuators B Chem. 2003, 90, 2–8. [Google Scholar] [CrossRef]
- Mascher, T.; Zimmer, S.L.; Smith, T.A.; Helmann, J.D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004, 48, 2888–2896. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Ching, C.B. Development of colorimetric-based whole-cell biosensor for organophosphorus compounds by engineering transcription regulator DmpR. ACS Synth. Biol. 2016, 5, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Grimaud, R.; Hoskins, J.R.; Wickner, S.; Maurizi, M.R. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA 2000, 97, 8898–8903. [Google Scholar] [CrossRef] [PubMed]
- Stocker, J.; Balluch, D.; Gsell, M.; Harms, H.; Feliciano, J.; Daunert, S.; van der Meer, J.R. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 2003, 37, 4743–4750. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 73, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Naylor, L.H. Reporter gene technology: The future looks bright. Biochem. Pharmacol. 1999, 58, 749–757. [Google Scholar] [CrossRef]
- Joyeux, A.; Balaguer, P.; Germain, P.; Boussioux, A.M.; Pons, M.; Nicolas, J.C. Engineered cell lines as a tool for monitoring biological activity of hormone analogs. Anal. Biochem. 1997, 249, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Welsh, S.; Kay, S.A. Reporter gene expression for monitoring gene transfer. Curr. Opin. Biotechnol. 1997, 8, 617–622. [Google Scholar] [CrossRef]
- Jain, V.K.; Magrath, I.T. A chemiluminescent assay for quantitation of β-galactosidase in the femtogram range: Application to quantitation of β-galactosidase in lacZ-transfected cells. Anal. Biochem. 1991, 199, 119–124. [Google Scholar] [CrossRef]
- Biran, I.; Klimentiy, L.; Hengge-Aronis, R.; Ron, E.Z.; Rishpon, J. On-line monitoring of gene expression. Microbiology 1999, 145, 2129–2133. [Google Scholar] [CrossRef] [PubMed]
- Karttunen, J.; Shastri, N. Measurement of ligand-induced activation in single viable T cells using the lacZ reporter gene. Proc. Natl. Acad. Sci. USA 1991, 88, 3972–3976. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, H.; Wakabayashi, M.; Yamashiro, H.; Maeda, I.; Isoda, K.; Kondoh, M.; Kawase, M.; Miyasaka, H.; Yagi, K. Whole-cell arsenite biosensor using photosynthetic bacterium Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 2006, 73, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Yeliseev, A.A.; Eraso, J.M.; Kaplan, S. Differential carotenoid composition of the B875 and B800-850 photosynthetic antenna complexes in Rhodobacter sphaeroides 2.4.1: Involvement of spheroidene and spheroidenone in adaptation to changes in light intensity and oxygen availability. J. Bacteriol. 1996, 178, 5877–5883. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Asad, S.; Ali, A. Bioluminescent bioreporter for assessment of arsenic contamination in water samples of India. J. Biosci. 2013, 38, 251–258. [Google Scholar] [CrossRef] [PubMed]
- De Mora, K.; Joshi, N.; Balint, B.L.; Ward, F.B.; Elfick, A.; French, C.E. A pH-based biosensor for detection of arsenic in drinking water. Anal. Bioanal. Chem. 2011, 400, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Joe, M.H.; Lee, K.H.; Lim, S.Y.; Im, S.H.; Song, H.P.; Lee, I.S.; Kim, D.H. Pigment-based whole-cell biosensor system for cadmium detection using genetically engineered Deinococcus radiodurans. Bioprocess Biosyst. Eng. 2012, 35, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Branco, R.; Cristóvão, A.; Morais, P.V. Highly sensitive, highly specific whole-cell bioreporters for the detection of chromate in environmental samples. PLoS ONE 2013, 8, e54005. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, S.; Ganesh, I.; Yoo, I.K.; Hong, S.H. Construction of a bacterial biosensor for zinc and copper and its application to the development of multifunctional heavy metal adsorption bacteria. Process Biochem. 2012, 47, 758–765. [Google Scholar] [CrossRef]
- Willardson, B.M.; Wilkins, J.F.; Rand, T.A.; Schupp, J.M.; Hill, K.K.; Keim, P.; Jackson, P.J. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microbiol. 1998, 64, 1006–1012. [Google Scholar] [PubMed]
- Tecon, R.; Beggah, S.; Czechowska, K.; Sentchilo, V.; Chronopoulou, P.M.; McGenity, T.J.; Van der Meer, J.R. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ. Sci. Technol. 2009, 44, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Shingler, V.; Moore, T. Sensing of aromatic compounds by the DmpR transcriptional activator of phenol-catabolizing Pseudomonas sp. strain CF600. J. Bacteriol. 1994, 176, 1555–1560. [Google Scholar] [CrossRef] [PubMed]
- Sticher, P.; Jaspers, M.C.; Stemmler, K.; Harms, H.; Zehnder, A.J.; Van Der Meer, J.R. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol. 1997, 63, 4053–4060. [Google Scholar] [PubMed]
- Korpela, M.T.; Kurittu, J.S.; Karvinen, J.T.; Karp, M.T. A recombinant Escherichia coli sensor strain for the detection of tetracyclines. Anal. Chem. 1998, 70, 4457–4462. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.I.; Oda, Y.; Shimada, T.; Oki, I.; Sugimoto, K. SOS-inducing activity of chemical carcinogens and mutagens in Salmonella typhimurium TA1535/pSK1002: Examination with 151 chemicals. Mutat. Res. Lett. 1987, 192, 239–246. [Google Scholar] [CrossRef]
- Pontel, L.; Audero, M.M.; Checa, S.; Soncini, F. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol. Microbiol. 2007, 66, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Cerminati, S.; Soncini, F.C.; Checa, S.K. A sensitive whole-cell biosensor for the simultaneous detection of a broad-spectrum of toxic heavy metal ions. Chem. Commun. 2015, 51, 5917–5920. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Song, X.; Bowen, J.; Corstanje, R.; Gao, Y.; Gaertig, J.; Gorovsky, M.A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 2002, 99, 3734. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Amaro, F.; Rico, D.; Campos, V.; Benítez, L.; Martíngonzález, A.; Hamilton, E.P.; Orias, E.; Gutiérrez, J.C. Tetrahymena metallothioneins fall into two discrete subfamilies. PLoS ONE 2007, 2, e291. [Google Scholar] [CrossRef] [PubMed]
- Whangsuk, W.; Dubbs, J.M.; Sallabhan, R.; Somsongkul, K.; Mongkolsuk, S.; Loprasert, S. ChpR is a chlorpyrifos-responsive transcription regulator in Sinorhizobium meliloti. J. Mol. Microbiol. Biotechnol. 2010, 18, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Whangsuk, W.; Thiengmag, S.; Dubbs, J.; Mongkolsuk, S.; Loprasert, S. Specific detection of the pesticide, chlorpyrifos, by a sensitive genetic-based whole cell biosensor. Anal. Biochem. 2015, 493, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Rosen, B.P. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 1991, 5, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Gross, M. Antibiotics in crisis. Curr. Biol. CB 2013, 23, R1063. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Urgel, M.; Serrano, L.; Ramos, J.L.; Fernández-Escamilla, A.M. Engineering biological approaches for detection of toxic compounds: A new microbial biosensor based on the pseudomonas putida TtgR repressor. Mol. Biotechnol. 2015, 57, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Magrisso, S.; Erel, Y.; Belkin, S. Microbial reporters of metal bioavailability. Microb. Biotechnol. 2008, 1, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Sánchez, V.; Molina, L.; Ramos, J.L.; Segura, A. New family of biosensors for monitoring BTX in aquatic and edaphic environments. Microb. Biotechnol. 2016, 9, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; He, Y.; Wang, Y.; Wang, H.; Wu, L.; Aries, E.; Huang, W.E. Whole-cell bacterial bioreporter for actively searching and sensing of alkanes and oil spills. Microb. Biotechnol. 2012, 5, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, A.; Geissdörfer, W.; Hillen, W. Expression of alkane hydroxylase from Acinetobacter sp. Strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J. Bacteriol. 1998, 180, 5822–5827. [Google Scholar] [PubMed]
- Sevilla, E.; Yuste, L.; Rojo, F. Marine hydrocarbonoclastic bacteria as whole-cell biosensors for n-alkanes. Microb. Biotechnol. 2015, 8, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Brutesco, C.; Prévéral, S.; Escoffier, C.; Descamps, E.C.T.; Prudent, E.; Cayron, J.; Dumas, L.; Ricquebourg, M.; Adryanczyk-Perrier, G.; Groot, A.D. Bacterial host and reporter gene optimization for genetically encoded whole cell biosensors. Environ. Sci. Pollut. Res. Int. 2016, 24, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cayron, J.; Prudent, E.; Escoffier, C.; Gueguen, E.; Mandrandberthelot, M.A.; Pignol, D.; Garcia, D.; Rodrigue, A. Pushing the limits of nickel detection to nanomolar range using a set of engineered bioluminescent Escherichia coli. Environ. Sci. Pollut. Res. 2017, 24, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Aisyah, W.N.; Jusoh, W.; Wong, L.S. Exploring the Potential of Whole Cell Biosensor: A Review in Environmental Applications. Int. J. Chem. Environ. Biol. Sci. 2014, 2, 52–56. [Google Scholar]
- Bazin, I.; Seo, H.B.; Suehs, C.M.; Ramuz, M.; De, W.M.; Gu, M.B. Profiling the biological effects of wastewater samples via bioluminescent bacterial biosensors combined with estrogenic assays. Environ. Sci. Pollut. Res. 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tauriainen, S.; Karp, M.; Chang, W.; Virta, M. Recombinant luminescent bacteria for measuring bioavailable arsenite and antimonite. Appl. Environ. Microbiol. 1997, 63, 4456–4461. [Google Scholar] [PubMed]
- Anne, K.; Taisia, R.; Angela, I. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009, 9, 41. [Google Scholar]
- Hou, Q.; Ma, A.; Wang, T.; Lin, J.; Wang, H.; Du, B.; Zhuang, X.; Zhuang, G. Detection of bioavailable cadmium, lead, and arsenic in polluted soil by tailored multiple Escherichia coli whole-cell sensor set. Anal. Bioanal. Chem. 2015, 407, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.C.; Youn, C.H.; Ahn, J.M.; Gu, M.B. Screening of Target-Specific Stress-Responsive Genes for the Development of Cell-Based Biosensors Using a DNA Microarray. Anal. Chem. 2005, 77, 8020–8026. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.; Blazeck, J.; Alper, H. Evolution of an alkane-inducible biosensor for increased responsiveness to short-chain alkanes. J. Biotechnol. 2012, 158, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Camanzi, L.; Bolelli, L.; Maiolini, E.; Girotti, S.; Matteuzzi, D. Optimal conditions for stability of photoemission and freeze drying of two luminescent bacteria for use in a biosensor. Environ. Toxicol. Chem. 2011, 30, 801. [Google Scholar] [CrossRef] [PubMed]
- Prévéral, S.; Brutesco, C.; Descamps, E.C.T.; Escoffier, C.; Pignol, D.; Ginet, N.; Garcia, D. A bioluminescent arsenite biosensor designed for inline water analyzer. Environ. Sci. Pollut. Res. 2017, 24, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Elad, T.; Almog, R.; Yagur-Kroll, S.; Levkov, K.; Melamed, S.; Shacham-Diamand, Y.; Belkin, S. Online monitoring of water toxicity by use of bioluminescent reporter bacterial biochips. Environ. Sci. Technol. 2011, 45, 8536–8544. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Mascher, T. The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: Expression systems and whole-cell biosensors. Appl. Microbiol. Biotechnol. 2016, 100, 4817–4829. [Google Scholar] [CrossRef] [PubMed]
- Hillger, J.M.; Schoop, J.; Boomsma, D.I.; Slagboom, P.E.; Ijzerman, A.P.; Heitman, L.H. Whole-cell biosensor for label-free detection of GPCR-mediated drug responses in personal cell lines. Biosens. Bioelectron. 2015, 74, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Castracane, J.; Tokranova, N.; Gracias, A.; Lnenicka, G.; Szaro, B. A living cell-based biosensor utilizing G-protein coupled receptors: Principles and detection methods. Biosens. Bioelectron. 2007, 22, 3230–3237. [Google Scholar] [CrossRef] [PubMed]
- Zager, V.; Cemazar, M.; Hreljac, I.; Lah, T.T.; Sersa, G.; Filipic, M. Development of human cell biosensor system for genotoxicity detection based on DNA damage-induced gene expression. Radiol. Oncol. 2010, 44, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Powers, H.J. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 2003, 77, 1352. [Google Scholar] [PubMed]
- Prchal, J.T.; Conrad, M.E.; Skalka, H.W. Association of presenile cataracts with heterozygosity for galactosaemic states and with riboflavin deficiency. Lancet 1978, 1, 12. [Google Scholar] [CrossRef]
- Cardoso, D.R.; Libardi, S.H.; Skibsted, L.H. Riboflavin as a photosensitizer. Effects on human health and food quality. Food Funct. 2012, 3, 487. [Google Scholar] [CrossRef] [PubMed]
- Besaratinia, A.; Kim, S.I.; Bates, S.E.; Pfeifer, G.P. Riboflavin activated by ultraviolet A1 irradiation induces oxidative DNA damage-mediated mutations inhibited by Vitamin C. Proc. Natl. Acad. Sci. USA 2007, 104, 5953–5958. [Google Scholar] [CrossRef] [PubMed]
- Si, R.; Yang, Y.; Yu, Y.; Han, S.; Zhang, C.; Sun, D.; Zhai, D.; Liu, X.; Yong, Y. Wiring bacterial electron flow for sensitive whole-cell amperometric detection of riboflavin. Anal. Chem. 2016, 88, 11222–11228. [Google Scholar] [CrossRef] [PubMed]
- Shabani, A.; Zourob, M.; Allain, B.; Marquette, C.A.; Lawrence, M.F.; Mandeville, R. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal. Chem. 2008, 80, 9475–9482. [Google Scholar] [CrossRef] [PubMed]
- Sifri, C.D. Healthcare epidemiology: Quorum sensing: Bacteria talk sense. Clin. Infect. Dis. 2008, 47, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. Fems Microbiol. Lett. 1998, 163, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Pasini, P.; Deo, S.K.; Flomenhoft, D.; Shashidhar, H.; Daunert, S. Biosensing systems for the detection of bacterial quorum signaling molecules. Anal. Chem. 2006, 78, 7603–7609. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A second N-acylhomoserine lactone signal produced by pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Susan, C.; Morpeth, H.O.R.; Crump, J.A. Invasive non-Typhi Salmonella disease in Africa. Clin. Infect. Dis. 2009, 49, 606–611. [Google Scholar]
- Venkatesh, A.G.; Sun, A.; Brickner, H.; Looney, D.; Hall, D.A.; Aronoff-Spencer, E. Yeast dual-affinity biobricks: Progress towards renewable whole-cell biosensors. Biosens. Bioelectron. 2015, 70, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Bereza-Malcolm, L.T.; Mann, G.; Franks, A.E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synth. Biol. 2004, 4, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.B.M.; Yoshinobu, T.; Iwasaki, H.; Sugihara, H.; Yukimasa, T.; Hirata, I.; Iwata, H. Investigation on light-addressable potentiometric sensor as a possible cell–semiconductor hybrid. Biosens. Bioelectron. 2003, 18, 1509–1514. [Google Scholar] [CrossRef]
- Horry, H.; Charrier, T.; Durand, M.J.; Vrignaud, B.; Picart, P.; Daniel, P.; Thouand, G. Technological conception of an optical biosensor with a disposable card for use with bioluminescent bacteria. Sens. Actuators B Chem. 2007, 122, 527–534. [Google Scholar] [CrossRef]
- Lu, Y.; Macias, D.; Dean, Z.S.; Kreger, N.R. A UAV-mounted whole cell biosensor system for environmental monitoring applications. IEEE Trans. Nanobiosci. 2015, 14, 811–817. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, S.F. Microbial biosensors. Biosens. Bioelectron. 2001, 16, 337–353. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).