Abstract
Cercariae of Plagiorchis spp. are frequently reported in European freshwater snails, but their true diversity is difficult to estimate due to subtle differences in morphology. We molecularly characterized 67 isolates of Plagiorchis cercariae collected from four lymnaeid snail hosts, Ampullaceana balthica, Ampullaceana lagotis, Radix auricularia and Lymnaea stagnalis in freshwater ecosystems in the Czech Republic and Poland. Based on mitochondrial cox1 and nuclear 28S sequences, ten species or species-level lineages were identified, including the first molecular evidence of P. vespertilionis from snail hosts and two species-level lineages reported for the first time. Previously undescribed species and species-level lineages are characterized morphometrically. We confirm the overlapping spatial distribution of Plagiorchis spp. in their snail hosts from Central Europe with those from Western and sub-Arctic Europe. Our results increase the known diversity of Plagiorchis spp. in Europe to 25 species/lineages in snails, but further research is needed to establish links between life cycle stages and to assess the host specificity of these parasites.
1. Introduction
Biodiversity represents the wide range of life on Earth, encompassing different ecosystems, species and their genetic variations. It plays a crucial role in maintaining ecological processes, promoting interactions among species and contributing to the overall health and adaptability of ecosystems [1]. While research on understanding the biodiversity of free-living organisms has been ongoing for decades, even centuries [2], our understanding of the actual diversity of parasites has only recently begun to unfold thanks to rapid advances in genetic data (e.g., [3,4,5,6,7,8]). Despite the generally negative perception of parasites, largely justified by their potential harm to human and animal health, elucidating the true diversity of parasites is extremely important for several reasons. First, accurate species identification often translates into improved diagnosis, treatment and therapy of parasitic diseases [9]. Second, the diversity of parasites is inherently linked to the diversity, or at least the temporary presence, of their free-living hosts in an ecosystem [10,11,12,13,14,15,16]. For example, the occurrence of the same parasite species in different geographical regions may point to the migration routes of their highly mobile bird hosts [6,17,18]. Finally, parasite species richness is a strong indicator of overall ecosystem health [19,20,21]. As our understanding of parasite diversity expands through advances in genetic data, these important roles of parasites are becoming increasingly evident.
There are between 75,000 and 350,000 species of helminth parasites infecting vertebrates [22,23,24], among which trematodes (Platyhelminthes, Digenea) represent a substantial and noteworthy group with about 18,000 nominal species [25]. However, the actual number may be considerably higher, as far more cryptic species (morphologically uniform, but exhibiting significant molecular differences) have been discovered in trematodes compared to other helminth taxa [26,27,28,29,30]. Most trematode species are described based on the adult stage, as opposed to the larval stages, which lack sufficient morphological characteristics for unequivocal morphological identification [31]. Nevertheless, obtaining adult trematodes that parasitize vertebrates, primarily birds and mammals, proves challenging and unpredictable due to stringent ethical requirements [32], the need to obtain necessary permits, the high mobility and seasonal presence of some definitive hosts, e.g., migratory birds, and, most importantly, the serious ecological consequences resulting from the removal of vertebrate hosts from an ecosystem. As snails (obligate first intermediate hosts) are less mobile and easier to collect, their sampling and screening for most trematode infections represents a less demanding and invasive method to monitor the presence and diversity of trematodes and all their life cycle hosts in an ecosystem.
The genus Plagiorchis Lühe, 1899 is the type- and most species-rich genus of the family Plagiorchiidae [33,34] and comprises over 140 described species to date [35]. These parasites typically infect pulmonate freshwater snails of the family Lymnaeidae, especially Ampullaceana balthica (Linnaeus, 1758) (formerly reported as Radix balthica and R. ovata [36,37]) and Lymnaea stagnalis (Linnaeus, 1758), as their first intermediate hosts [6,17,18,38,39,40] (Supplementary Table S1). Various invertebrates, including aquatic insects, crustaceans and snails, can serve as second intermediate hosts. The parasites mature mainly in birds and mammals, less frequently in reptiles and amphibians [41]. This wide host spectrum suggests a potentially high diversity of Plagiorchis species in habitats with rich animal fauna. Plagiorchis species have also been identified as potential pathogens in humans, resulting in a disease known as plagiorchiasis, with 12 recorded cases mainly in Asia to date [42,43,44,45].
Cercariae of Plagiorchis, the free-living larvae emerging from a snail host, belong to the xiphidiocercariae morphotype, possessing a stylet to facilitate penetration into the next hosts, two suckers and a long, simple tail [46,47]. The distinguishing features of Plagiorchis species are inconspicuous and difficult to differentiate both in cercariae [6,40] and adults [33,48]. For example, the number and size of lipid droplets in the body parenchyma, the shape of the stylet, and body size can exhibit intraspecific variation [6,40], and some features, such as the presence of a slight base thickening on the stylet, can be very subtle and require a high level of morphological expertise. This is further exacerbated by vague morphological descriptions, limited access to type material and discrepancies in specimen measurements due to different fixation methods [6,40,49].
Before molecular analysis became common practice, cercariae of 11 named Plagiorchis species were reported in Europe, predominantly as P. elegans (Rudolphi, 1802), P. maculosus (Rudolphi, 1802) or P. neomidis Brendow, 1970 (see Supplementary Table S1 based on records in the Host–parasite Database of the Natural History Museum, London [50], and the literature data reviewed by Cichy et al. [38] and Faltýnková et al. [39], supplemented by more references). In Europe, Plagiorchis spp. have so far been detected in 11 lymnaeid snail species from 20 countries (six species in eight countries if only molecular studies are considered) (Figure 1, Supplementary Table S1). Recently, an unexpectedly high diversity of 17 species or species-level lineages of Plagiorchis in snails was discovered in various freshwater ecosystems of the sub-Arctic, Western and Central Europe using molecular or integrative taxonomic approaches [6,17,18,40,51] (Figure 1). In addition to molecular species delimitation, recent studies on this genus have focused on the characterization of the entire mitochondrial genome [45,52,53], which is highly useful for resolving phylogenetic relationships at the suprageneric level and provides a basis for future primer development. These findings emphasize the need to apply molecular methods to further explore the species diversity of the genus Plagiorchis, as the subtle and minor differences in cercarial morphology may lead to misidentifications and inaccurate assessments.
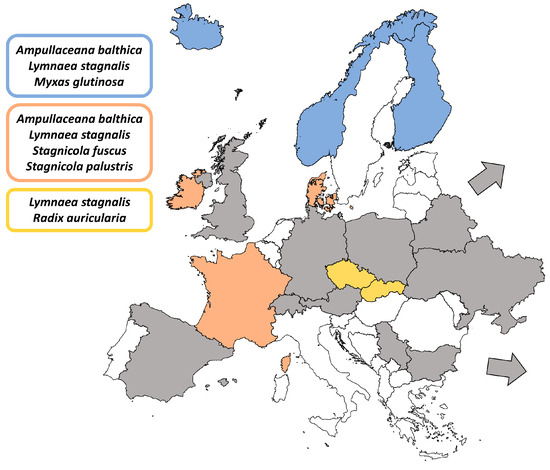
Figure 1.
Map showing records of Plagiorchis spp. in Europe based on Host–parasite Database of the Natural History Museum, London [50] and the literature data. Countries marked in color indicate records from molecular studies (DNA sequences available); sub-Arctic regions in blue, Central Europe in yellow and Western Europe in pink, with the snail hosts indicated for each region. Countries marked in grey indicate records based solely on morphological identification of cercariae; the grey arrows represent European Russia and Georgia. See Supplementary Table S1 for details.
Our study aimed to: (i) investigate the trematode diversity of Plagiorchis spp. in their first intermediate snail hosts in selected freshwater ecosystems in Central Europe, (ii) provide morphometric data for novel species/lineages, (iii) compare the distribution patterns of Plagiorchis species in Central Europe with those in sub-Arctic and Western European regions, and (iv) reassess the importance of Ampullaceana balthica as a primary first intermediate host of Plagiorchis spp. We expect to find a high diversity of Plagiorchis species, at least partly similar in composition to those from sub-Arctic and Western Europe, as several of the lakes studied hold significance as important nesting, breeding and wintering sites for migratory birds, which may serve as definitive hosts for trematodes, including Plagiorchis species.
2. Materials and Methods
2.1. Sample Collection and Material Processing
Snails infected with Plagiorchis spp. were collected from seven distinct localities in the Czech Republic and at one location in Poland between 2016 and 2023 (Table 1). Sampling was carried out from May to October, with all localities sampled at least once during the main summer months due to sufficient snail abundance in that period. The largest lakes (Medard, Milada and Most) were primarily targeted and sampled several times in consecutive years, often in all seasons (from May to October), as the pilot study on trematode communities in snails in recultivated post-mining lakes was initiated there in 2016 (Table 1). In the following years, other localities were gradually added as comparative sites for investigating trematode diversity, including Plagiorchis. In addition, the snails were part of an extensive sampling collection focused on exploring trematode diversity in the small lymnaeid hosts Ampullaceana spp. and Radix auricularia (Linnaeus, 1758). Therefore, the majority of sampled snails belong to these genera. The main reason for the narrow sampling focus was the fact that the data on trematode diversity and community composition in these snails are scarce [18,39,54,55], especially in comparison to the large lymnaeid snail L. stagnalis (reviewed by Żbikowska and Nowak [56]).
Sampling sites were selected to assess Plagiorchis diversity in habitats with abundant populations of snails and potential definitive hosts (former coal mining pits and Skulska Wieś) or to compare diversity in lakes with less diverse fauna due to higher anthropogenic pressure (Písník Dubina and Spůle). Most of the lakes represent former coal mining pits that have been revitalized into lakes for recreational purposes (namely Barbora, Medard, Milada, Most and Otakar) [57,58,59]. Lakes Medard and Most stand out as the largest among our study sites (496 and 309 ha, respectively) and serve as important ornithological habitats [60,61,62,63]. They provide secure nesting, feeding, and overwintering grounds for a variety of both native and migratory bird species, which may serve as major definitive hosts for Plagiorchis spp. (Supplementary Table S2). The lakes Barbora (63 ha), Milada (252 ha) and Otakar (9 ha) are situated in a close vicinity of Lake Most, suggesting a similar bird fauna due to the short flying distance. Lake Skulska Wieś in Poland (124 ha) is located within a Natura 2000 protected area, designated for the restoration and enlargement of habitats for successful bird feeding and nesting, thus probably supporting diverse bird populations as well [64]. Písník Dubina (10 ha) and Spůle (0.08 ha) are smaller waterbodies in adjacent urban areas that are mainly used for summer recreation and water sports.

Table 1.
Summary data for sampling locations, and lymnaeid snails examined and infected with Plagiorchis species. Abbreviations: CZ, Czech Republic; PL, Poland.
Table 1.
Summary data for sampling locations, and lymnaeid snails examined and infected with Plagiorchis species. Abbreviations: CZ, Czech Republic; PL, Poland.
| Sampling Site/Country | Coordinates | Sampling Period | Snail Species | Sampled Snails | Infected Snails | Plagiorchis Infected Snails | Plagiorchis Prevalence |
|---|---|---|---|---|---|---|---|
| Barbora | 50°38′35.6″ N, | 2021–2023 | Radix auricularia | 325 | 46 | 8 | 2.5% |
| CZ | 13°45′00.1″ E | Galba truncatula | 1 | — | — | — | |
| Stagnicola sp. | 2 | — | — | — | |||
| Medard | 50°10′44.0″ N, | 2016–2023 | Ampullaceana balthica 1 | 7995 | 877 | 141 | 1.8% |
| CZ | 12°35′50.1″ E | Lymnaea stagnalis | 251 | 72 | 5 | 2.0% | |
| Milada | 50°39′15.2″ N, | 2016–2021 | Ampullaceana lagotis 2 | 279 | 58 | 11 | 3.9% |
| CZ | 13°57′01.2″ E | Lymnaea stagnalis | 102 | 77 | 3 | 2.9% | |
| Galba truncatula | 11 | — | — | — | |||
| Most | 50°32′13.6″ N, | 2016–2023 | Ampullaceana lagotis | 2200 | 927 | 33 | 1.5% |
| CZ | 13°38′40.7″ E | Radix auricularia | 87 | 73 | — | — | |
| Otakar | 50°39′04.8″ N, | 2021–2023 | Radix auricularia | 438 | 80 | 2 | 0.5% |
| CZ | 13°44′23.1″ E | Galba truncatula | 16 | — | — | — | |
| Peregriana peregra | 1 | 1 | — | — | |||
| Stagnicola sp. | 3 | 2 | — | — | |||
| Písník Dubina | 50°11′20.8″ N, | 2023 | Radix auricularia | 326 | 99 | 3 | 0.9% |
| CZ | 15°47′09.7″ E | ||||||
| Skulska Wieś | 52°29′34.7″ N, | 2023 | Ampullaceana balthica 3 | 101 | 13 | 1 | 1.0% |
| PL | 18°18′57.5″ E | ||||||
| Spůle | 49°21′24.3″ N, | 2023 | Radix auricularia | 47 | 5 | 4 | 8.5% |
| CZ | 13°11′44.2″ E | ||||||
| Total | 12,185 | 2330 | 211 | 1.7% |
1 Nomenclature according to Aksenova et al. [37]; former synonym Radix balthica, 2 Nomenclature according to Aksenova et al. [37]; former synonym Radix lagotis, 3 Snail identified based on shell morphology [65,66].
A total of 12,185 snails belonging to seven species were collected either by handpicking them from stones or by sieving the vegetation in the littoral zone of all lakes and identified morphologically according to the keys of Glöer [65,66] (Table 1). However, due to the overlapping shell characteristics of the genera Ampullaceana and Radix [67,68], two Plagiorchis-infected snails of each genus per locality were subjected to molecular identification. A single infected snail from Skulska Wieś was not molecularly identified due to loss of tissue material. In the laboratory, snails were placed individually in plastic beakers filled with lake water and left under a light source for at least 48 h to stimulate cercarial emergence. Snails shedding cercariae of Plagiorchis (identified to genus level using the key of Faltýnková et al. [47]) were then separated, live cercariae were photographed using a digital camera Promicam 3-5CP (Promicra, Prague, Czech Republic) attached to an Olympus BX51 light microscope (Olympus Optical Co., Ltd., Tokyo, Japan), and representative samples of cercariae were fixed in molecular-grade ethanol (Penta, Prague, Czech Republic). Several samples of cercariae were additionally fixed in a ‘cold’ (i.e., unheated) 4% formaldehyde solution (formalin) to obtain morphometric data. A total of 67 isolates were subjected to molecular analyses. All non-shedding snails were examined by pressing the tissue between two glass slides under a stereomicroscope to detect prepatent trematode intramolluscan stages (sporocysts or rediae). Sporocysts of xiphidiocercariae are uniform in morphology [46], and therefore were also fixed in molecular-grade ethanol for subsequent molecular identification. Molecular voucher material is deposited at the Laboratory of Helminthology, Biology Centre of the Czech Academy of Sciences, České Budějovice, Czech Republic.
2.2. Sequence Generation
Genomic DNA of Plagiorchis spp. was isolated from ethanol-fixed samples, using 10–20 cercariae released from an individual snail or sporocysts in case of prepatent infections. The Monarch® Genomic DNA Purification Kit (New England Biolabs®, Ipswich, MA, USA) was used for DNA isolation and purification. For molecular identification of Plagiorchis species, the mitochondrial cytochrome c oxidase subunit 1 gene (cox1) was used. Two sets of sequences were generated: (i) sequences compatible with data from Europe were amplified with the forward primer JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) and the reverse primer JB4.5 (5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′) [69] or CO1-R trema (5′-CAA CAA ATC ATG ATG CAA AAG G-3′) [70] according to the cycling conditions of Zikmundová et al. [40] and (ii) sequences compatible with data from North America were generated with the forward primer Dice1F (5′-ATT AAC CCT CAC TAA ATT WCN TTR GAT CAT AAG-3′) and the reverse primer Dice14R (5′-TAA TAC GAC TCA CTA TAC CHA CMR TAA ACA TAT GAT G-3′) under the cycling conditions described by Van Steenkiste et al. [71]. In several representatives of novel or previously unreported species/lineages, D1–D3 region of the large ribosomal subunit (28S rDNA) was additionally sequenced with the forward primer ZX-1 (5′-ACC CGC TGA ATT TAA GCA TAT-3) and the reverse 1500R (5′-GCT ATC CTG AGG GAA ACT TCG-3′) under the cycling conditions described by Tkach et al. [72].
For the snail samples, ITS2 rDNA region was amplified with the forward primer NEWS (5′-TGT GTC GAT GAA -GA ACG CAG-3′) and the reverse primer RIXO (5′-TTC TAT GCT TAA ATT CAG GGG-3′) [73] following the protocol of Bargues et al. [74]. The PCR amplicons for all samples were purified using ExoSAP-IT™ Express PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The purified samples were sequenced from both strands with the corresponding PCR primers. In addition, the following internal primers were used for 28S: ECD2 (5′-CCT TGG TCC GTG TTT CAA GAC GGG-3′) [75] and 300F (5ʹ-CAA GTA CCG TGA GGG AAA GTT G-3ʹ) [76]. Sequencing of the purified material was conducted utilizing ABI Big Dye™ v.3.1 chemistry in accordance with the manufacturer’s guidelines at the commercial enterprise SEQme (Dobříš, Czech Republic, https://www.seqme.eu, accessed on 20 December 2023) on an AB3730 × 1 capillary sequencer. Newly generated sequences of parasites and snail sequences were assembled and edited using Geneious Prime® 2023.2.1 and deposited in GenBank.
2.3. Species Identification and Phylogenetic Analyses
Identification of novel sequences was checked using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nih.gov/BLAST/, accessed on 28 December 2023) and thereafter they were aligned with the selected sequences from GenBank using MUSCLE implemented in Geneious Prime® 2023.2.1. Three alignments for Plagiorchis spp. including 116 novel sequences and published sequences were prepared and used for phylogenetic analyses. Alignment 1 (324 nt, cox1) included 49 sequences generated in the present study and 45 published sequences. The haematoloechid Haematoloechus sibiricus Issaitschikoff, 1927 (AB818363), a parasite of Pelophylax nigromaculatus (Hallowell, 1861), was used as an outgroup (Supplementary Table S3). Alignment 2 (371 nt, cox1) included 62 sequences generated in the present study and 74 published sequences. The haematoloechid Haematoloechus sp. (KM538096) was used as an outgroup (Supplementary Table S4). Both cox1 alignments were aligned with reference to the amino acid translation, using the trematode mitochondrial code (translation Table 21; https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG21, accessed on 28 December 2023) [77,78]. Alignment 3 (1119 nt, 28S) included five sequences generated in the present study and 51 published sequences. The haematoloechid Haematoloechus longiplexus Stafford, 1902 (AF387801), a parasite of Lithobates catesbeianus (Shaw, 1802) was used as an outgroup (Supplementary Table S5). The selection of the outgroups was based on the topology in the phylogenetic trees of Plagiorchis spp. provided by Kudlai et al. [6].
To produce the phylogenetic hypotheses for the alignments, maximum likelihood and Bayesian inference analyses were applied. The best-fitting evolutionary model was estimated using the AIC criterion in jModelTest 2.1.2. [79]. These were the TN93 + G + I (Alignment 1) and GTR + I + G (Alignment 2; Alignment 3). The maximum likelihood analysis was conducted using PhyML ver. 3.0 [80] run on the Geneious Prime® 2023.2.1. Nodal support for the ML analysis was generated by performing 100 bootstrap pseudoreplicates. The Bayesian inference analysis was conducted using MrBayes software ver. 3.2.3 [81]. Markov chain Monte Carlo chains were run for 5,000,000 generations, log-likelihood scores were recorded to estimate burn-in, and only the last 75% of trees were used to build the consensus tree. FigTree v. 1.4 software [82] was used for the tree visualization. Nucleotide differences between sequences were estimated using MEGA ver. 11 [83] using the following conditions: “Variance Estimation Method = None”, “Model/Method = p-distance or no. of differences”, “Substitutions to Include = d: Transitions + Transversions” and “Gaps/Missing Data Treatment = Pairwise deletion”.
2.4. Morphometric Characterization
Photomicrographs of live, ethanol- and formalin-fixed cercariae belonging to novel or previously undescribed species/lineages, Plagiorchis vespertilionis (Müller, 1780), Plagiorchis sp. 10 and Plagiorchis sp. 11, were subjected to morphometric analysis. A total of 232 cercariae (17 live, 167 ethanol-fixed and 48 formalin-fixed) were used to obtain morphometric data (provided in micrometers). Thirteen morphometric body parameters were measured (abbreviations) according to Zikmundová et al. [40]: total length as the sum of body and tail length (ToL), body length (BL), body width (BW), tail length (TL), tail width (TW), oral sucker length (OSL), oral sucker width (OSW), ventral sucker length (VSL), ventral sucker width (VSW), stylet length (SL), stylet width at the anterior thickening (SWantt), stylet width above the base thickening (SWabt) and stylet width at the base thickening (SWbt). The following ratio parameters were then calculated based on the measurements: oral sucker width to ventral sucker width (OSW/VSW), tail length to body length (TL/BL; %) and stylet width at the anterior thickening to stylet length (SWantt/SL; %). In contrast to formalin-fixed cercariae, stylet dimensions and the corresponding ratio could only be determined on live and ethanol-fixed cercariae (Supplementary Figure S1), the latter proving impossible in all specimens. For each new species-level lineage, measurements are summarized for all snail hosts, but are also provided for each snail host separately to assess possible distortions due to the potentially different dimensions of cercariae. The majority of specimens contributed data to all metrical variables.
Photomicrographs of live and fixed cercariae were made by placing cercariae on a microscopic glass, covering them with a coverslip and removing excess water with a paper tissue. The amount of water removed was assessed according to the speed of cercarial movement to obtain representative photomicrographs while avoiding excessive flattening of the cercariae. The magnification was set based on the specific morphological features observed and the scale for the measurements was calibrated accordingly (e.g., body size and suckers 50 μm, stylet 20 μm). All measurements were obtained with the ImageJ v.1.53e program [84]. The morphological description of cercariae (if observable) is based on photomicrographs of live specimens.
2.5. Analyses of Ecological Data
The prevalence of Plagiorchis spp. found in this study was calculated as the proportion of infected snails divided by the total number of snails examined in a population sample [85]. We then assessed the diversity and distribution of Plagiorchis species in relation to the lakes and species of snail hosts investigated in our study.
In order to identify the distribution and diversity patterns of Plagiorchis species in freshwater lymnaeid snails in Central Europe with those in sub-Arctic and Western European regions, the literature data from available faunistic and molecular studies were reviewed in combination with records in the Host–parasite Database of the Natural History Museum, London [50] (reviews by Cichy et al. [38] and Faltýnková et al. [39]; see Supplementary Table S1 for references). These records were then compared with the results of the present study. As a result, we provide an updated overview of the diversity of Plagiorchis species in their first snail intermediate hosts in freshwater ecosystems in Europe. The classification and nomenclature of the snail species of the family Lymnaeidae correspond to Horsák et al. [36] and Aksenova et al. [37].
3. Results
3.1. Sequence-Based Identification
In total, 116 sequences (n = 111, cox1; n = 5, 28S) were newly generated for 67 isolates (Table 2). Molecular evaluation of the trematodes obtained in the present study confirmed the presence of ten species and species-level lineages in the examined snails. Species delineation for each isolate and identification of samples were based on the results of phylogenetic analyses and pairwise sequence comparisons of the partial cox1 sequences in Alignment 1 and Alignment 2, as well as the 28S sequences in Alignment 3.

Table 2.
Summary data for the newly generated sequences of Plagiorchis spp. Abbreviations: C, cercariae; S, sporocysts.
The phylogenetic tree obtained from the maximum likelihood analysis based on Alignment 1 (cox1 mtDNA; 324 nt) is presented in Figure 2 and pairwise genetic distances of this dataset are presented in Supplementary Table S3. Novel cox1 sequences (n = 49) on Alignment 1 were positioned in different clades with the species of the genus Plagiorchis, demonstrating the presence of five species (P. elegans, P. koreanus, P. maculosus, P. muelleri and P. vespertilionis) and five species-level lineages (Plagiorchis sp. 3 sensu Soldánová et al. [17], Plagiorchis sp. 5 sensu Soldánová et al. [17], Plagiorchis sp. 8 sensu Kudlai et al. [6], Plagiorchis sp. 10 and Plagiorchis sp. 11) in our material, out of which the last two were not previously reported (Figure 2).
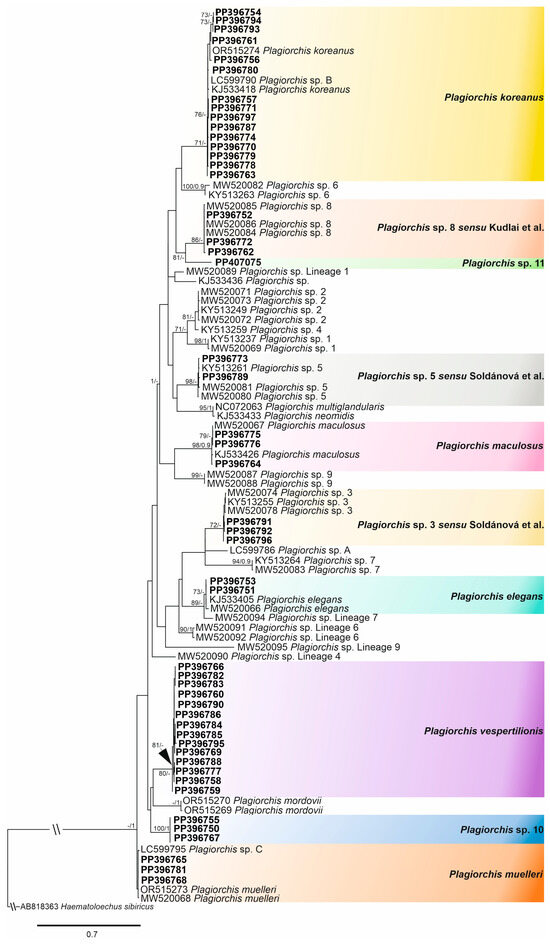
Figure 2.
The phylogenetic tree resulted from maximum likelihood (ML) analysis of the cox1 mtDNA sequences datasets of Plagiorchis spp. (compatible with data from Europe), with nodal support values shown at the node as ML/BI (Bayesian inference). Support values < 70 (ML) and 0.90 (BI) are not shown. Sequences generated in the present study are highlighted in bold (Kudlai et al. [6] and Soldánová et al. [17]).
The provisional names of novel lineages Plagiorchis sp. 10 and Plagiorchis sp. 11 were assigned following Soldánová et al. [17] and Kudlai et al. [6] to maintain continuity in the naming of novel European Plagiochis cercarial linages, and based on the fact that formal names are traditionally assigned only after describing the adult trematodes [25,31]. In Alignment 1, the intraspecific genetic variation rates were 0–3.1% (0–10 nt), with P. koreanus showing the highest divergence. The interspecific divergence rates were 0.9–18.9% (3–61 nt) with P. neomidis and P. multiglandularis Semenov, 1927 exhibiting the lowest interspecific divergence and Plagiorchis sp. 3 sensu Soldánová et al. [17] and P. maculosus exhibiting the highest interspecific divergence.
The phylogenetic tree obtained from the maximum likelihood analysis based on Alignment 2 (cox1 mtDNA; 371 nt) is presented in Figure 3 and pairwise genetic distances of this dataset are presented in Supplementary Table S4. Novel cox1 sequences (n = 62) are in accordance with Alignment 1 demonstrating the same species and species-level lineage in our material (Table 2). In Alignment 2, the intraspecific genetic variation rates were 0–3.2% (0–12 nt), with Plagiorchis sp. 2 sensu Soldánová et al. [17] showing the highest divergence. The interspecific divergence rates were 6.7–20.8% (21–77 nt) with Plagiorchis sp. 7 sensu Soldánová et al. [17] and P. elegans exhibiting the lowest interspecific divergence and Plagiorchis sp. Lineage 2 sensu Gordy and Hanington [86] and Plagiorchis sp. Lineage 8 sensu Gordy and Hanington [86] showed the highest sequence divergence. The phylogenetic tree obtained from the maximum likelihood analysis based on Alignment 3 (28S rDNA; 1119 nt) is presented in Figure 4 and pairwise genetic distances of this dataset are presented in Supplementary Table S5. Novel 28S sequences (n = 5) were generated for three species and species-level lineages found in the present study (P. vespertilionis, Plagiorchis sp. 10 and Plagiorchis sp. 11). In Alignment 3, the intraspecific genetic variation rates were 0–1.3% (0–10 nt), with P. muelleri exhibiting the highest divergence. The interspecific divergence rates were 0–3.1% (0–25 nt) with Plagiorchis sp. Lineage 7 sensu Gordy and Hanington [86] and P. elegans exhibiting the lowest divergence and P. elegans and Plagiorchis mordovii Schaldybin, 1958 exhibiting the highest divergence. The identification of P. vespertilionis was based on the analysis of 28S sequence data, as the cox1 sequences were not available for comparison in GenBank.
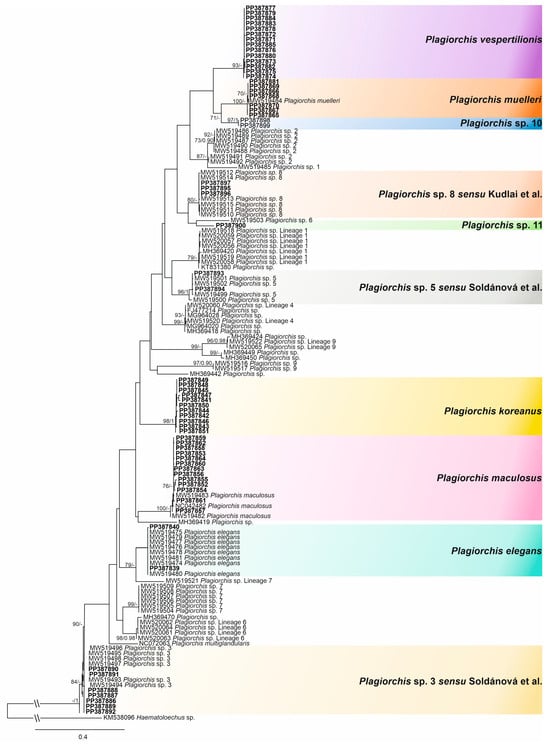
Figure 3.
The phylogenetic tree resulted from maximum likelihood (ML) analysis of the cox1 mtDNA sequences datasets of Plagiorchis spp. (compatible with data from North America), with nodal support values shown at the node as ML/BI (Bayesian inference). Support values < 70 (ML) and 0.90 (BI) are not shown. Sequences generated in the present study are highlighted in bold (Kudlai et al. [6] and Soldánová et al. [17]).
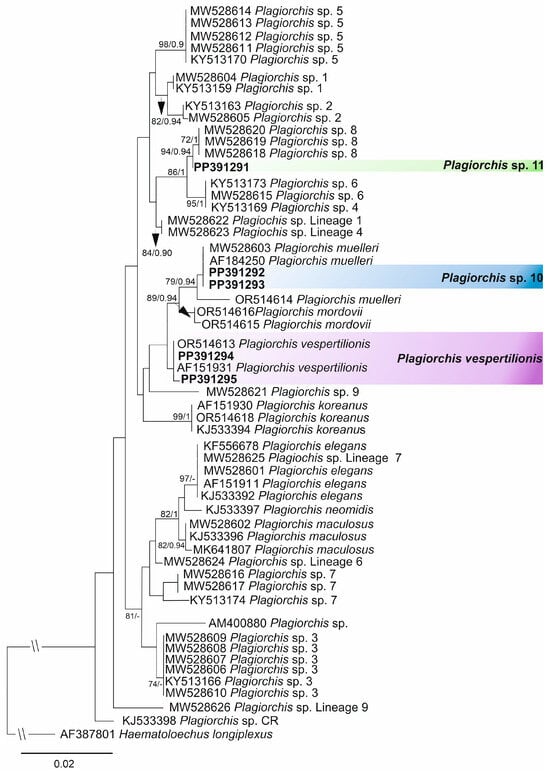
Figure 4.
The phylogenetic tree resulted from maximum likelihood (ML) analysis of the 28S sequences datasets of Plagiorchis spp. with nodal support values shown at the node as ML/BI (Bayesian inference). Support values < 70 (ML) and 0.90 (BI) are not shown. Sequences generated in the present study are highlighted in bold.
Fourteen novel sequences of the internal transcribed spacer 2 (ITS2) were generated in the present study for snails to confirm the morphological evaluation. The following species were molecularly identified: A. balthica (n = 2), Ampullaceana lagotis (Schrank, 1803) (n = 4), and R. auricularia (n = 8) (Supplementary Table S6). Molecular identification confirmed our preliminary identification based on morphology.
3.2. Morphometric Characterization
Photomicrographs of live cercariae of ten species or species-level lineages revealed in this study are presented in Figure 5 and Figure 6. Descriptions of cercariae in terms of metrical data and morphology are given for the three novel or undescribed species/lineages, P. vespertilionis, Plagiorchis sp. 10 and Plagiorchis sp. 11; the remaining seven species have been described in detail in previous studies [6,40].
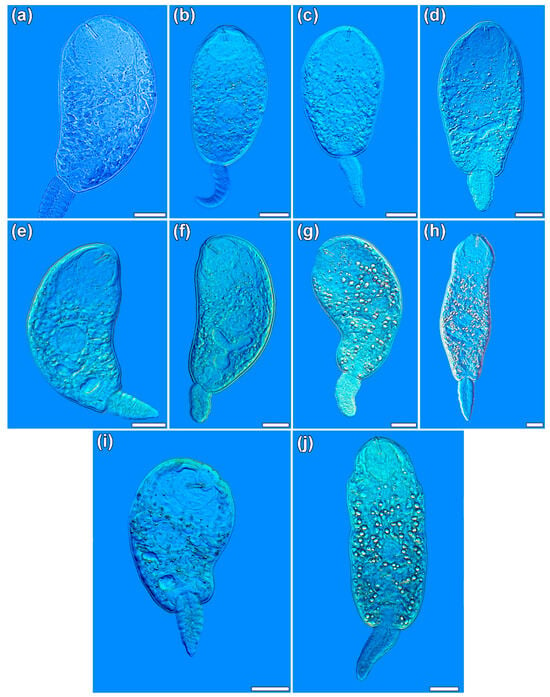
Figure 5.
Plagiorchis spp., photomicrographs of live cercariae: (a) P. elegans; (b) P. koreanus; (c) P. maculosus; (d) P. muelleri; (e) P. vespertilionis; (f) Plagiorchis sp. 3 sensu Soldánová et al. [17]; (g) Plagiorchis sp. 5 sensu Soldánová et al. [17]; (h) Plagiorchis sp. 8 sensu Kudlai et al. [6]; (i) Plagiorchis sp. 10 (present study); (j) Plagiorchis sp. 11 (present study). Scale-bars: 50 μm.
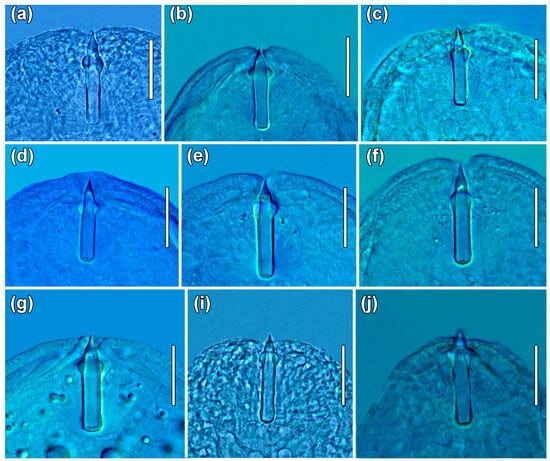
Figure 6.
Plagiorchis spp., photomicrographs of stylets of live cercariae: (a) P. elegans; (b) P. koreanus; (c) P. maculosus; (d) P. muelleri; (e) P. vespertilionis; (f) Plagiorchis sp. 3 sensu Soldánová et al. [17]; (g) Plagiorchis sp. 5 sensu Soldánová et al. [17]; (i) Plagiorchis sp. 10 (present study); (j) Plagiorchis sp. 11 (present study). Representative photomicrograph of the stylet of Plagiorchis sp. 8 sensu Kudlai et al. [6] (h) was not obtained. Scale-bars: 20 μm.
- Plagiorchis vespertilionis (Müller, 1780)
- First intermediate hosts: Ampullaceana balthica, Ampullaceana lagotis, Radix auricularia.
- Localities: Lakes Barbora, Medard, Most, Otakar.
Table 2 shows details of 15 isolates and their representative sequences and Figure 5e and Figure 6e show photomicrographs of live cercariae. Cercariae originating from each snail host separately did not differ in individual parameters; only the body and tail length showed higher variability in live specimens due to body contraction and elongation during movement (Supplementary Table S7). Measurements are therefore pooled across three snail hosts and based on 11 live, 113 ethanol-fixed and 29 formalin-fixed cercariae (presented as mean values ± standard deviation; see Table 3 for the range).

Table 3.
Metrical data for cercariae of Plagiorchis vespertilionis, Plagiorchis sp. 10, and Plagiorchis sp. 11 isolated from different snail hosts Ampullaceana balthica, A. lagotis and Radix auricularia (pooled together; see Supplementary Tables S7 and S8 for measurement similarities divided by snail species). Data are presented in micrometers as the minimum to maximum values (mean in parentheses). The width values correspond to maximum width measurements. See Materials and Methods for abbreviations of cercarial morphological parameters.
Live specimens: Body elongate-oval, maximum width anterior to ventral sucker, 254 ± 40.6 × 159 ± 23.4. Tail 125 ± 18.4 × 38 ± 9.8, short, comprising 51% of body length. Total length of cercaria 379 ± 36.7. Oral sucker more or less spherical, slightly subspherical 67 ± 9.6 × 64 ± 7.4, width exceeds width of ventral sucker (ratio 1.4). Ventral sucker distinctly smaller than the oral sucker, large, spherical 46 ± 6.5 × 47 ± 5.9. Stylet dorsal to mouth opening, robust, sharply pointed, 31.0 ± 9.8 long, with slightly thickened base (5.0 ± 1.7 wide at base and 4.8 ± 1.6 wide above base) and well-developed and pronounced anterior lateral thickening 7.8 ± 2.5 wide, comprising short but relatively wide blade; stylet width at anterior thickening 25.2% of stylet length. Penetration gland-cells large, asymmetric, in two groups of seven and eight on each side of body, antero- and postlateral to ventral sucker. Lipid droplets (also called spherical fat droplets, fat inclusions or refractile granules [40,47,87]) not numerous, small and medium-sized scattered throughout body parenchyma; small droplets form clusters in both oral (few present) and ventral suckers (more numerous).
Ethanol-fixed specimens: Body 211 ± 20.8 × 115 ± 13.9, tail 150 ± 21.7 × 29 ± 3.7, comprising 71% of body length; total length of cercaria 360 ± 38.2. Oral sucker more or less spherical, slightly wider than long 50 ± 4.5 × 54 ± 4.8, ventral sucker transversely oval 37 ± 4.2 × 44 ± 4.5; OSW/VSW ratio 1.2. Stylet 30 ± 1.4 long.
Formalin-fixed specimens: Body 190 ± 12.7 × 119 ± 6.9, tail 138 ± 43.2 × 32 ± 7.6, comprising 74% of body length; total length of cercaria 328 ± 39.0. Oral sucker transversely oval 48 ± 3.4 × 57 ± 3.5, ventral sucker transversely oval 38 ± 3.2 × 45 ± 3.0; OSW/VSW ratio 1.3.
- Plagiorchis sp. 10
- First intermediate hosts: Ampullaceana balthica, Ampullaceana lagotis.
- Localities: Lakes Medard, Most, Skulska Wieś.
Table 2 shows details of three isolates and their representative sequences and Figure 5i and Figure 6i show photomicrographs of live cercariae. Cercariae originating from each snail host separately did not differ in individual dimension parameters; only the body length and width and tail length showed higher variability in live specimens due to body contraction and elongation during movement (Supplementary Table S8). Measurements are therefore pooled across three snail hosts and based on five live, 49 ethanol-fixed and 19 formalin-fixed cercariae (presented as mean values ± standard deviation; see Table 3 for the range).
Live specimens: Body elongate-oval, maximum width anterior to ventral sucker, 298 ± 48.9 × 148 ± 11.5. Tail 119 ± 21.4 × 45 ± 6.1, short, comprising 41% of body length. Total length of cercaria 417 ± 61.1. Oral sucker slightly subspherical 63 ± 4.5 × 65 ± 4.8, width exceeds the width of ventral sucker (ratio 1.3). Ventral sucker distinctly smaller than oral sucker, large, transversely oval 46 ± 2.9 × 51 ± 4.8. Stylet dorsal to mouth opening, robust, sharply pointed, 29.5 ± 0.8 long, with distinctly thickened base (5.1 ± 0.1 wide at base and 4.4 ± 0.1 wide above base) and well-developed anterior lateral thickening 6.7 ± 0.1 wide, comprising short and relatively narrow blade; stylet width at anterior thickening 22.8% of stylet length. Penetration gland-cells large, asymmetric, in two groups of seven and eight on each side of body, antero- and postlateral to ventral sucker. Lipid droplets few, small and medium-sized scattered throughout body parenchyma mainly posterior to oral sucker; small droplets form clusters predominantly in ventral sucker or absent.
Ethanol-fixed specimens: Body 191 ± 17.0 × 88 ± 7.9, tail 125 ± 13.2 × 25 ± 2.4, comprising 66% of body length; total length of cercaria 317 ± 22.2. Oral sucker transversely oval 42 ± 3.8 × 48 ± 3.7, ventral sucker transversely oval 32 ± 3.6 × 38 ± 3.8; OSW/VSW ratio 1.3. Stylet 27 ± 0.8 long.
Formalin-fixed specimens: Body 147 ± 11.3 × 92 ± 7.0, tail 119 ± 37.5 × 23 ± 2.9, comprising 83% of body length; total length of cercaria 266 ± 31.8. Oral sucker transversely oval 38 ± 2.0 × 42 ± 1.7, ventral sucker transversely oval 30 ± 2.1 × 35 ± 2.0; OSW/VSW ratio 1.2.
- Plagiorchis sp. 11
- First intermediate host: Ampullaceana lagotis.
- Locality: Lake Most.
Table 2 shows details of the isolate and its sequence and Figure 5j and Figure 6j show photomicrographs of live cercaria. Measurements are based on one live and five ethanol-fixed cercariae (the latter presented as mean values ± standard deviation; see Table 3 for the range).
Live specimen: Body elongate-oval, maximum width anterior to ventral sucker, 302 × 158. Tail 138 × 44, short, comprising 46% of body length. Total length of cercaria 440. Oral sucker transversely oval (62 × 67), width distinctly exceeds the width of the ventral sucker (ratio 1.7). Ventral sucker distinctly smaller than oral sucker, small, spherical 39 × 38. Stylet dorsal to mouth opening, robust, sharply pointed, 31.1 long, lacking thickening at its base (5.3 wide at base and 5.3 wide above base), but with well-developed and robust anterior lateral thickening 8.9 wide, comprising short but wide blade; stylet width at anterior thickening 28.6% of stylet length. Penetration gland-cells small, antero- and postlateral to ventral sucker (number not observable). Lipid droplets numerous, conspicuous and very large, scattered throughout the body parenchyma posterior to pharynx, few medium-sized droplets also present in body; both large and medium-sized droplets absent in areas of oral and ventral suckers.
Ethanol-fixed specimens: Body 231 ± 9.8 × 129 ± 6.1, tail 137 ± 15.5 × 39 ± 1.7, comprising 60% of body length; total length of cercaria 368 ± 20.6. Oral sucker transversely oval 50 ± 5.8 × 56 ± 3.7, ventral sucker transversely oval 35 ± 4.9 × 39 ± 5.9; OSW/VSW ratio 1.4. Stylet 31 ± 1.5 long.
3.3. Prevalence, Distribution and Diversity of Plagiorchis Species
Of 12,185 snails collected, 2330 snails (19.1%) harbored trematode infections. In total, 211 (1.7%) snails were infected with intramolluscan stages of Plagiorchis spp. (Table 1). Four snail species (out of seven) were detected as the first intermediate hosts of Plagiorchis, namely A. balthica, A. lagotis, L. stagnalis and R. auricularia (Table 1). Prevalence levels of larval Plagiorchis spp. were generally low in all lakes, with an overall mean of 1.7%, varying between 0.5 and 8.5% (both for R. auricularia in Spůle and Otakar, respectively) (Table 1).
The highest species diversity of Plagiorchis spp. was found in Lake Medard (eight out of ten species/lineages), including three species-level lineages previously found in sub-Arctic and Western Europe [6,17] (Table 2). Lakes Barbora and Most supported four Plagiorchis species, followed by lakes Milada, Otakar and Písník Dubina (two species each) and Skulska Wieś and Spůle (one species each). The novel lineages Plagiorchis sp. 10 and Plagiorchis sp. 11 were both found in Lake Most, the former also in Medard and Skulska Wieś. The first molecular evidence of P. vespertilionis in European snails comes from three snail hosts sampled in four lakes closely situated to each other: R. auricularia from Barbora and Otakar, A. balthica from Medard and A. lagotis from Most. Ampulaceana balthica exhibited the highest Plagiorchis species richness (eight out of ten species/lineages), followed by A. lagotis and R. auricularia (four species/lineages each) (Table 2). In addition, A. lagotis is reported for the first time as a Plagiorchis spp. host in Europe. The largest lymnaeid L. stagnalis harbored only one species, namely P. elegans (Table 2).
Our review of the literature and other data sources on the distribution and diversity of Plagiorchis spp. in lymnaeid snail hosts revealed a relatively wide distribution and high species richness across various European freshwater ecosystems. A total of 23 species/lineages (species incertae sedis are not included) have been found to date in 11 species of lymnaeid snails belonging to seven genera (Supplementary Table S1). So far, 11 Plagiorchis species have been identified to the species level mainly based on cercarial morphology, but only five of them were confirmed molecularly (i.e., P. elegans, P. koreanus, P. maculosus, P. muelleri and P. neomidis [6,18,40]). The remaining 12 species represent previously undescribed species-level lineages discovered exclusively by molecular genetic methods [6,17,18,40,51] (Supplementary Table S1). The most frequent intermediate snail hosts are A. balthica and L. stagnalis, with 14 and 12 Plagiorchis species, respectively, while the remaining nine snail hosts harbor one to four species (Supplementary Table S1). Two species, P. elegans and Plagiorchis sp. sensu Duan et al. [18], from the planorbid snail Planorbarius corneus (Linnaeus, 1758) [18,88] likely represent erroneous identification and are not listed in the Plagiorchis species reported from European freshwater snails (Supplementary Table S1).
Plagiorchis species have been detected in 20 European countries, but studies from only eight countries provided molecular and/or morphological evidence for their accurate identification (Figure 1, Supplementary Table S1). In these, 17 species or species-level lineages of Plagiorchis have been detected, with the highest diversity found in sub-Arctic regions (ten species/lineages) and Western Europe (11 species/lineages), followed by Central Europe (five species/lineages) (Table 4). Our study revealed 10 Plagiorchis species/species-level lineages in four species of lymnaeid snails belonging to three genera (Table 2 and Table 4). Their distribution and molecular diversity to some extent overlap with that in sub-Arctic areas (five species/lineages), in Western Europe (six species/lineages) and in Central Europe (three species/lineages) (Table 4), which may be connected to the rich bird fauna of our region (Supplementary Table S2). Plagiorchis elegans and P. maculosus are the only species molecularly confirmed in all three European regions. In addition, three species/lineages identified by DNA sequences in our study, namely P. vespertilionis, Plagiorchis sp. 10 and Plagiorchis sp. 11, were reported for the first time.

Table 4.
Species of Plagiorchis spp. reported from European freshwater snails based on molecular data. The green color marks the presence of the species/lineages in a specific region (see Figure 1 for details on the countries of each region).
4. Discussion
The diversity of Plagiorchis spp. revealed by molecular genetic analysis in our study exceeded expectations, especially considering the generally low prevalence of these parasites in snails (1.7%). We detected ten Plagiorchis species and species-level lineages: five named species, three previously recorded lineages and two novel lineages in four intermediate snail hosts (A. balthica, A. lagotis, L. stagnalis and R. auricularia). This high diversity is twice as great as the diversity of Plagiorchis species so far known from Central Europe, mainly infecting the large lymnaeid L. stagnalis [40]. This emphasizes the importance of investigating the trematode fauna of the still poorly studied small lymnaeid snails. Our comprehensive survey of Plagiorchis spp. diversity in lymnaeids across large European geographical areas resulted in 23 species/lineages, with our new data accounting for nearly half of the total Plagiorchis diversity (44%), and more than half of the molecular diversity in Europe (59%). This increased the currently known molecular diversity of Plagiorchis species parasitizing snails in Europe from 17 to 20 (total diversity from 23 to 25 species). In addition, our study expanded the range of snail hosts for A. lagotis, the parasite fauna for P. vespertilionis and the newly discovered lineages Plagiorchis sp. 10 and Plagiorchis sp. 11, for which we provided the first molecular, morphological and morphometric data. This will facilitate future parasite identification and diversity assessment in faunistic and community studies of trematodes in snail populations.
The first molecular study investigating the diversity of Plagiorchis spp. in snail hosts from Central Europe focused mainly on L. stagnalis and discovered five distinct lineages [40] using an integrative morphological and molecular approach, often regarded as the ‘best practice’ for trematode identification and systematics [89]. Later, an unexpectedly high diversity of seven novel lineages infecting A. balthica was found in sub-Arctic Norway, none of which overlapped with the Central European Plagiorchis species [17]. High-latitude ecosystems in various sub-Arctic and western European regions were also studied using the ‘best practice’ approach and confirmed the presence of 11 distinct species and lineages in four snail species (A. balthica, L. stagnalis, Stagnicola fuscus [Pfeiffer, 1821] and Myxas glutinosa [Müller, 1774]), two of which overlapped with the diversity captured in Central Europe [6].
Additional molecular data for cercariae of Plagiorchis spp. were provided by studies conducted in Western Europe investigating the structure of compound communities of trematodes and their molecular diversity in freshwater snails [18] or testing a new method for rapid identification of cercariae [51], including Plagiorchis spp. However, these two studies used the nuclear ribosomal internal transcribed spacer (ITS) for species delimitation, although nuclear loci such as 28S and ITS show a high degree of conservation and may not represent the true species divergence of Plagiorchis, e.g., [6]. Each trematode family requires a specific genetic marker for accurate species identification and subsequent phylogenetic comparisons between different taxa [89], and the preferred marker for Plagiorchis species delimitation is the mitochondrial cox1 region [6,40,86,90]. Therefore, the use of ITS regions does not comply with ‘best practice’, as it may lead to misidentification similar to that of Duan et al. [18], who presented ‘Plagiorchis sp.’ from the large planorbid Planorbarius corneus, despite the ample evidence from over 60 years of research that Plagiorchis spp. parasitize only lymnaeid snails in Europe (Supplementary Table S1). In addition, both the studies by Huguenin et al. [51] and Duan et al. [18] lack a morphological description and/or photodocumentation of cercariae, making a reliable conclusion about the identity of their ‘Plagiorchis’ species difficult. Future studies on the genus Plagiorchis should target two independent loci, mitochondrial and nuclear, to corroborate their own results [89].
The molecular delineation of Plagiorchis is further complicated by the confounding practice of sequencing incompatible regions of the cox1 marker, whereby sequence data from Europe [17,40] are not compatible with those from North America [86,90]. Therefore, in our study, we used two sets of primers and alignments based on cox1 for an individual sample to obtain data for both pertinent regions of the gene, thus providing a complete picture of species diversity and phylogenetic relationships. One of the primer sets used, targeting the cox1 Folmer region (compatible with data from North America), has proved efficient in mitochondrial barcoding for at least 23 digenean families [71], highlighting the importance of using the most appropriate sequencing region in broader phylogenetic comparisons of trematodes. In the future, a unified approach to sequencing both pertinent regions of the cox1 gene would greatly facilitate the accurate identification and comparison of Plagiorchis species, especially in the case of the discovery of putatively novel lineages. Migratory birds flying via the East Atlantic flyway [91,92,93] can transfer parasites between continents [7], but without data on both regions of cox1 and the deposition of reliable sequences in public DNA databases, it is impossible to accurately identify species, compare their phylogenetic relationships and assess the inter-continental fauna of parasites.
Our novel genetic dataset confirms the presence of cercariae of P. elegans, P. koreanus and P. maculosus in Central Europe [40], which is complemented by the species P. muelleri, previously reported only from Ireland [6], and by P. vespertilionis recorded in the present study for the first time. Our study is also the first to demonstrate the overlap in geographical distribution of six Plagiorchis species with sub-Arctic (Norway) and Western Europe (Ireland and Denmark), namely P. elegans, P. maculosus, P. muelleri, Plagiorchis sp. 3 and 5 sensu Soldánová et al. [17] and Plagiorchis sp. 8 sensu Kudlai et al. [6] (Table 4). The latter three lineages represent the first records in Central Europe and, at the same time, their southernmost distribution in Europe. In addition, phylogenetic analysis of our molecular data provided strong evidence for the existence of two novel species-level lineages, Plagiorchis sp. 10 and Plagiorchis sp. 11.
The life cycles of the five lineages reported here, namely Plagiorchis sp. 3 and 5 sensu Soldánová et al. [17], Plagiorchis sp. 8 sensu Kudlai et al. [6] and Plagiorchis sp. 10 and Plagiorchis sp. 11, are not yet elucidated, as all available sequences originate from snail hosts [6,17] (present study). The abundance and distribution of definitive hosts are generally considered to be the most important factors determining the spatial and temporal variability in the distribution, prevalence and diversity of trematodes in snails [94,95,96]. While a wide range of vertebrates can serve as definitive hosts of Plagiorchis (including mammals such as bats, rodents and carnivores, as well as amphibians and reptiles) [41,50], it is reasonable to assume that the long-distance distribution of Plagiorchis is accomplished by its most vagile definitive hosts, the birds. Their high mobility and long migration routes from breeding to wintering grounds make birds the main agents of parasite dispersal [13,95,97].
Although it is known that several Plagiorchis species from our study can complete their life cycle in bats (especially P. koreanus, P. muelleri and P. vespertilionis) or other small mammals [41], several facts support our hypothesis that mainly bird definitive hosts are responsible for the diversity of Plagiorchis species in snail populations inhabiting the studied lakes. First, the presence of three species-level lineages has so far only been reported from high-latitude European countries (Finland, Iceland, Ireland and Norway) [6,17], i.e., regions with absent, low or declining populations of bat colonies, insectivorous mammals and other rare hosts due to physiological limitations caused by unfavorable cold conditions [98,99,100]. Second, the most diverse fauna of Plagiorchis spp. was found in the largest lakes in post-mining areas, Medard and Most (Czech Republic), as well as in the nature reserve covering Lake Skulska Wieś (Poland). These ecosystems are important ornithological and stop-over sites with a high abundance and diversity of birds [60,61,62,63,64], including many species identified as hosts of Plagiorchis spp. (Supplementary Table S2). A large proportion are migratory birds capable of distributing parasites across and beyond Europe. Lakes Barbora, Milada and Otakar also belong to the reclaimed post-industrial habitats, located 25–100 km from each other and from Lake Medard, suggesting a similar avifauna and thus similar hosts likely influencing the overlapping occurrence of Plagiorchis species (Table 2). Third, the first intermediate snail hosts of Plagiorchis occur in high densities in these lakes, with A. balthica standing out as the most common and abundant. Ampullaceana balthica is recognized as a primary host for a variety of trematodes in Europe, including Plagiorchis spp. [18,38,39,88]. Hence, there is a causal link between the presence of A. balthica, the abundant populations and wide spectrum of birds and the high diversity of Plagiorchis spp. detected in this study (see below).
Plagiorchis elegans is the most frequently recorded Plagiorchis species in Europe, occurring in seven snail hosts from 12 countries (Supplementary Table S1). Despite our extensive collection of small lymnaeids, we were only able to detect P. elegans in L. stagnalis. This is consistent with other European molecular studies [6,18,40], suggesting that the occurrence of this parasite can be restricted to snails of the genera Lymnaea and Stagnicola and that previous faunistic records from other snail hosts could be misidentifications (see references in Supplementary Table S1). However, more molecular evidence is necessary for confirmation. Plagiorchis maculosus is the second most frequent species in Europe, representing, together with P. elegans, the only two molecularly valid species in all three European regions (sub-Arctic, Western and Central Europe) and thus confirming their wide geographical distribution. In contrast, P. neomidis and Plagiorchis sp. CR sensu Zikmundová et al. [40] were reliably confirmed only from one host, L. stagnalis, in Slovakia and Czech Republic, respectively [40]. In addition, the complete mitochondrial genome of P. multiglandularis infecting L. stagnalis from Asian Russia has been characterized [53], but this species has not yet been molecularly confirmed from snails in Europe. Therefore, further sampling efforts on a large geographical and snail-host scale are required to corroborate or refute the rare occurrences of valid Plagiorchis species.
Four species from European snails, namely Plagiorchis laricola Skjarbin, 1924, Plagiorchis megalorchis Rees, 1952, Plagiorchis mutationis Panova, 1927 and Plagiorchis nanus (Rudolphi, 1802), have been identified only based on the morphology of cercariae and adults (references in Supplementary Table S1). Moreover, P. laricola could be a senior synonym of P. mutationis and P. megalorchis [41]. Plagiorchis fastuosus Szidat, 1924 (not listed in our overview of Plagiorchis distribution) represents an erroneous record in European snails [101], as it has been revised and reassigned to the genus Plagioglyphe Krasnolobova, 1973, based on the morphology of the adult worms [25,41]. Further sequencing of larval and especially adult forms of Plagiorchis is urgently needed to confirm the validity of these species as well as to link the life cycle stages of the newly recorded lineages from European snails with those from second intermediate and definitive hosts.
The greatest diversity of Plagiorchis spp. was found in A. balthica sampled in Lake Medard, with five named species, two novel lineages and three lineages previously known only from sub-Arctic Europe [6,17]. As indicated by previous molecular studies, A. balthica is recognized as an important first intermediate host of Plagiorchis in high-latitude ecosystems, with seven lineages reported in Norway [17] and eight species/lineages detected in sub-Arctic Europe and Ireland [6]. The present study not only confirms these findings, but also shows that the high diversity of Plagiorchis spp. in A. balthica is not exclusively restricted to northern European regions. Furthermore, we report for the first time A. lagotis as an intermediate host of Plagiorchis, namely P. koreanus, P. vespertilionis and two novel lineages, Plagiorchis sp. 10 and Plagiorchis sp. 11. The absence of A. lagotis as a host for Plagiorchis in previous faunistic and community studies is surprising, considering its wide distribution in European freshwater ecosystems [66]. A possible reason lies in the similarity of snail morphology (shell shape and coloration) commonly used to distinguish the closely related genera Ampullaceana and Radix, likely being confused with A. balthica and Peregriana peregra (Rossmässler, 1835) (former synonym Radix labiata; Aksenova et al. [37]) [102]. The use of molecular methods combined with dissections of the reproductive organs is the best solution to avoid misidentification [37,68,102,103].
Three species, P. koreanus, P. muelleri and P. vespertilionis, were previously considered oioxenous, i.e., infecting only one snail host species [6,40,104]. Our study extends the snail host range to three species (A. balthica, A. lagotis and R. auricularia) for P. koreanus, two species (A. balthica and R. auricularia) for P. muelleri and four species (A. balthica, A. lagotis, L. stagnalis and R. auricularia) for P. vespertilionis. In general, Plagiorchis spp. have a wide range of snail hosts distributed over a large geographical area in Europe. Eleven snail species belonging to seven genera serve as first intermediate hosts, but DNA sequences of Plagiorchis spp. are available for only six of them (Figure 1). Several snail species, such as those of the genera Stagnicola or Galba, have not yet been sufficiently studied with regard to Plagiorchis diversity and distribution in Europe. In addition, molecular identification of snail host species is rarely provided [6,17] (present study), but it is highly important for a reliable assessment of snail host range.
Perhaps the most significant finding in our study, besides the record of two novel lineages, is the detection of P. vespertilionis from several snail hosts. Since no cox1 sequences were available in GenBank, our study benefited from the molecular data of Kirillova et al. [48], allowing us to characterize one of the putative novel lineages as P. vespertilionis based on 28S sequences of adult worms. Thus, we were able to partially elucidate the life cycle of P. vespertilionis, as it was documented in three snail species, A. balthica, A. lagotis and R. auricularia, in our lakes. The primary definitive hosts of P. vespertilionis are bats, but the parasite can also be found in lagomorphs, rodents and carnivores [41]. The Czech Republic harbors a diverse bat fauna with 26 reported species, of which at least 17 occur in northern Bohemia, where infected snails were sampled [105]. Bats are often found near water bodies due to abundant food sources [106]. This is consistent with our observation that P. vespertillionis was found exclusively in four post-mining lakes, which support remarkable animal biodiversity [107], and that most of the isolates were assigned to this species. The second intermediate hosts of P. vespertilionis remain be identified, but the host range is likely to be similar to that documented for other Plagiorchis species, including aquatic insects, crustaceans and snails [6,17,108,109].
In Europe, cercariae of P. vespertilionis have been documented in L. stagnalis from Belarus [104], but the certainty of the correct identification is questionable due to the lack of supporting data on the cercarial morphology. To the best of our knowledge, the only description comes from an earlier study in the USA in which the xiphidiocercariae with refractile granules were found in L. stagnalis and described as the subspecies Plagiorchis vespertilionis parorchis by Macy [110]. However, later studies demonstrated that adult worms of this ‘subspecies’ do not morphologically match adult P. vespertilionis from European bats [33,41]. In the absence of comparative data, we present here the first comprehensive morphometric analysis of the cercariae of P. vespertilionis, potentially aiding future identification efforts. By integrating the genetic and morphological approaches, we thus confirm for the first time the occurrence of P. vespertilionis cercariae in Europe. Plagiorchis vespertilionis belongs to the ‘Plagiorchis vespertilionis group’, comprising also P. koreanus and P. muelleri, all of which have been confirmed molecularly as distinct species parasitizing bats in Europe [33]. The adults of the group are morphologically very similar [33,48], and their cercariae appear to be uniform to a certain extent, especially with regard to the high number of small and medium-sized lipid droplets (see Figure 5 and descriptions of live cercariae in the present study and those of P. koreanus and P. muelleri in Zikmundová et al. [40] and Kudlai et al. [6], respectively). Nevertheless, cercariae of P. vespertilionis are larger in almost all body parameters than cercariae of P. koreanus described by Zikmundová et al. [40], but at the same time do not differ significantly from a single cercaria of P. muelleri described by Kudlai et al. [6]. When comparing the size of the stylet of P. vespertilionis with those of P. muelleri described by Kudlai et al. [6] and P. koreanus described by Zikmundová et al. [40], the former two have a similar size of stylet with a slightly thickened base, in contrast to the much smaller stylet and lack of base thickening in P. koreanus. However, we observed a high variability in the stylet length between P. koreanus and P. muelleri, making them more similar in this parameter (data not shown) (Figure 6b,d). The shape of the stylet can be another distinguishing feature, which in P. vespertilionis has a robust appearance, a pronounced anterior lateral thickening and a short but relatively wide blade, whereas the stylet in the remaining two species is more slender with less pronounced anterior thickening and a longer and narrower blade. Based on these data, we assume that the cercariae of P. vespertilionis can be distinguished from the other two species of the ‘Plagiorchis vespertilionis group’, mainly due to the size and shape of the stylet and body size.
Although the description of all identified species/lineages of Plagiorchis cercariae was beyond the scope of this study, we emphasize that future molecular research should include detailed morphological and morphometric analysis of all possible parameters, which would allow reliable comparison at both intra- and interspecific levels. It is also worth noting that analyzing metric data for cercariae is complex, as it largely depends on the number of specimens and the fixation method influencing the dimensions. While measurements made on photographs of live cercariae may vary due to their rapid movements, fixed specimens may be affected differently depending on the method used (hot or cold formalin, hot water, or ethanol), as has been demonstrated for other helminth parasites [111]. Therefore, it is necessary to standardize the use of different fixation methods to detect and properly evaluate the subtle morphometric differences between cercariae of Plagiorchis species, especially in situations where molecular analysis is not feasible.
5. Conclusions
Even though Plagiorchis species are common and ecologically important parasites of snails in Europe, their diversity has been underestimated both before and during the ‘molecular era’. Besides the molecular evidence of high Plagiorchis diversity in snails from Central European waterbodies, we confirmed a broad host range of lymnaeid snails, with Ampullaceana balthica being the most important species, and a wide geographical distribution of the parasite in Europe. Therefore, sampling and investigating the diversity of Plagiorchis species and trematodes in general should not be neglected, as understanding the richness and biogeographic patterns of parasite diversity is crucial to grasping the impact of global change [24]. The use of inconsistent fixation methods to obtain metric data from cercariae leads to challenges in morphological comparison, raising the need for standardized processing of cercariae samples. Furthermore, it is crucial to continue the study of parasite biodiversity using the ‘best practice’ approach in order to link the sequences of different developmental stages, elucidate parasite life cycles and host specificity, and assign formal names to novel species-level lineages. Finally, several Plagiorchis species are still awaiting molecular confirmation of their validity or occurrence in poorly studied snail hosts and European countries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16030158/s1, Figure S1: Comparison of Plagiorchis cercariae fixed in ethanol and cold formalin used for the measurements; Table S1: Plagiorchis species reported from European freshwater snails; Table S2: The composition of birds in Lakes Medard and Most based on literature data and online databases; Table S3: Nucleotide comparison of cox1 mtDNA sequences of Plagiorchis spp. (compatible with data from Europe) based on 324 nt long alignment; Table S4: Nucleotide comparison of cox1 mtDNA sequences of Plagiorchis spp. (compatible with data from North America) based on 371 nt long alignment; Table S5: Nucleotide comparison of the partial 28S rDNA sequences of Plagiorchis spp. based on 1119 nt long alignment; Table S6: Overview of newly generated sequences of snail hosts infected with Plagiorchis spp.; Table S7: Comparative metrical data for cercariae of Plagiorchis vespertilionis isolated from different snail hosts; Table S8: Comparative metrical data for cercariae of Plagiorchis sp. 10 isolated from different snail hosts. Note: the reference numbering in the Supplementary Materials (Tables S1 and S2) does not follow that in the main text.
Author Contributions
Conceptualization, P.K. and M.S.; methodology, P.K., C.P. and M.S.; formal analysis, P.K., C.P., K.J. and M.S.; investigation, P.K., K.J. and M.S.; data curation, M.S.; writing—original draft preparation, P.K. and M.S.; writing—review and editing, P.K., C.P., K.J. and M.S.; visualization, C.P. and M.S.; supervision, M.S.; project administration, M.S.; funding acquisition, P.K. and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Czech Science Foundation (project no. 19-28399X), the Czech Academy of Sciences (the program “Strategy AV 21: Land Conservation and Restoration”) and the Erasmus+ program of the European Union.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials. The genetic sequence data reported in this paper are deposited in the GenBank database (see accession numbers in Table 2 and Supplementary Table S6). Molecular voucher material is deposited at the Laboratory of Helminthology, Biology Centre of the Czech Academy of Sciences in České Budějovice, Czech Republic. Uncorrected p-distances from the DNA sequences are provided online as electronic Supplementary Materials.
Acknowledgments
We are grateful to Anna Stanicka (Nicolaus Copernicus University, Toruń), Jana Baboučková, Šárka Kreslová (Faculty of Science, University of South Bohemia, České Budějovice), Martina Borovková, Caroline Kibet, Blanka Škoríková (Institute of Parasitology, Biology Centre CAS) and Tereza Vyhlídalová for their help during field collection and sample processing; Blanka Škoríková for technical assistance with figures (Institute of Parasitology, Biology Centre CAS, Czech Republic); Markéta Pravdová for assistance with cercarial measurements; Tomáš Scholz for his valuable comments and suggestions; the two anonymous reviewers for their ideas and suggestions which helped to improve the manuscript; the companies Fuel Combine Company Ústí (PKU), Sokolovská uhelná, a.s. and Lesy České Republiky, s.p. for granting us access to the collection sites.
Conflicts of Interest
The authors declare no conflicts of interests.
References
- Rands, M.R.; Adams, W.M.; Bennun, L.; Butchart, S.H.; Clements, A.; Coomes, D.; Entwistle, A.; Hodge, I.; Kapos, V.; Scharlemann, J.P.; et al. Biodiversity conservation: Challenges beyond 2010. Science 2010, 329, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Ali, A. Biodiversity-ecosystem functioning research: Brief history, major trends and perspectives. Biol. Conserv. 2023, 285, 110210. [Google Scholar] [CrossRef]
- Beck, H.P.; Blake, D.; Dardé, M.L.; Felger, I.; Pedraza-Díaz, S.; Regidor-Cerrillo, J.; Gómez-Bautista, M.; Ortega-Mora, L.M.; Putignani, L.; Shiels, B.; et al. Molecular approaches to diversity of populations of apicomplexan parasites. Int. J. Parasitol. 2009, 39, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Lacorte, G.A.; Felix, G.M.; Pinheiro, R.R.; Chaves, A.V.; Almeida-Neto, G.; Neves, F.S.; Leite, L.O.; Santos, F.R.; Braga, E.M. Exploring the diversity and distribution of neotropical avian malaria parasites—A molecular survey from Southeast Brazil. PLoS ONE 2013, 8, e57770. [Google Scholar] [CrossRef] [PubMed]
- Morand, S.; Krasnov, B.R.; Littlewood, D.T.J. Parasite Diversity and Diversification: Evolutionary Ecology Meets Phylogenetics; Cambridge University Press: Cambridge, UK, 2015; ISBN 110-703-765-4. [Google Scholar] [CrossRef]
- Kudlai, O.; Pantoja, C.; O’Dwyer, K.; Jouet, D.; Skírnisson, K.; Faltýnková, A. Diversity of Plagiorchis (Trematoda: Digenea) in high latitudes: Species composition and snail host spectrum revealed by integrative taxonomy. J. Zoolog. Syst. Evol. Res. 2021, 59, 937–962. [Google Scholar] [CrossRef]
- Pantoja, C.; Faltýnková, A.; O’Dwyer, K.; Jouet, D.; Skírnisson, K.; Kudlai, O. Diversity of echinostomes (Digenea: Echinostomatidae) in their snail hosts at high latitudes. Parasite 2021, 28, 59. [Google Scholar] [CrossRef]
- Scholz, T.; Kuchta, R. Fish tapeworms (Cestoda) in the molecular era: Achievements, gaps and prospects. Parasitology 2022, 149, 1876–1893. [Google Scholar] [CrossRef]
- Cháves-González, L.E.; Morales-Calvo, F.; Mora, J.; Solano-Barquero, A.; Verocai, G.G.; Rojas, A. What lies behind the curtain: Cryptic diversity in helminth parasites of human and veterinary importance. Curr. Res. Parasitol. Vector. Borne Dis. 2022, 2, 100094. [Google Scholar] [CrossRef]
- MacKenzie, K. Parasites as indicators of host populations. Int. J. Parasitol. 1987, 17, 345–352. [Google Scholar] [CrossRef]
- Vaughn, C.C.; Taylor, C.M. Macroecology of a host-parasite relationship. Ecography 2000, 23, 11–20. [Google Scholar] [CrossRef]
- Krasnov, B.R.; Shenbrot, G.I.; Khokhlova, I.S.; Allan Degen, A. Relationship between host diversity and parasite diversity: Flea assemblages on small mammals. J. Biogeogr. 2004, 31, 1857–1866. [Google Scholar] [CrossRef]
- Hechinger, R.F.; Lafferty, K.D. Host diversity begets parasite diversity: Bird final hosts and trematodes in snail intermediate hosts. Proc. R. Soc. B 2005, 272, 1059–1066. [Google Scholar] [CrossRef]
- Hechinger, R.F.; Lafferty, K.D.; Huspeni, T.C.; Brooks, A.J.; Kuris, A.M. Can parasites be indicators of free-living diversity? Relationships between species richness and the abundance of larval trematodes and of local benthos and fishes. Oecologia 2007, 151, 82–92. [Google Scholar] [CrossRef]
- Hechinger, R.F.; Lafferty, K.D.; Kuris, A.M. Trematodes indicate animal biodiversity in the Chilean intertidal and Lake Tanganyika. J. Parasitol. 2008, 94, 966–968. [Google Scholar] [CrossRef]
- Kuris, A.M.; Hechinger, R.F.; Shaw, J.C.; Whitney, K.L.; Aguirre-Macedo, L.; Boch, C.A.; Dobson, A.P.; Dunham, E.J.; Fredensborg, B.L.; Huspeni, T.C.; et al. Ecosystem energetic implications of parasite and free-living biomass in three estuaries. Nature 2008, 454, 515–518. [Google Scholar] [CrossRef]
- Soldánová, M.; Georgieva, S.; Roháčová, J.; Knudsen, R.; Kuhn, J.A.; Henriksen, E.H.; Siwertsson, A.; Shaw, J.C.; Kuris, A.M.; Amundsen, P.-A.; et al. Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake. Int. J. Parasitol. 2017, 47, 327–345. [Google Scholar] [CrossRef]
- Duan, Y.; Al-Jubury, A.; Kania, P.W.; Buchmann, K. Trematode diversity reflecting the community structure of Danish freshwater systems: Molecular clues. Parasit. Vectors 2021, 14, 43. [Google Scholar] [CrossRef]
- Hudson, P.J.; Dobson, A.P.; Lafferty, K.D. Is a healthy ecosystem one that is rich in parasites? Trends Ecol. Evol. 2006, 21, 381–385. [Google Scholar] [CrossRef]
- Gómez, A.; Nichols, E.S.; Perkins, S.L. Parasite conservation, conservation medicine, and ecosystem health. In New Directions in Conservation Medicine: Applied Cases of Ecological Health; Aguirre, A.A., Ostfeld, R., Daszak, P., Eds.; Oxford University Press: Oxford, UK, 2012; pp. 67–81. ISBN 978-019-973-147-3. [Google Scholar]
- Gagne, R.B.; Crooks, K.; Craft, M.E.; Chiu, E.S.; Fountain-Jones, N.M.; Malmberg, J.L.; Carver, S.; Funk, C.; VandeWoude, S. Parasites as conservation tools. Conserv. Biol. 2021, 36, e13719. [Google Scholar] [CrossRef]
- Poulin, R.; Morand, S. Parasite Biodiversity; Smithsonian Institution Scholarly Press: Washington, DC, USA, 2005; ISBN 158-834-170-4. [Google Scholar]
- Dobson, A.; Lafferty, K.D.; Kuris, A.M.; Hechinger, R.F.; Jetz, W. Homage to Linnaeus: How many parasites? How many hosts? Proc. Natl. Acad. Sci. USA 2008, 105, 11482–11489. [Google Scholar] [CrossRef]
- Carlson, C.J.; Dallas, T.A.; Alexander, L.W.; Phelan, A.L.; Phillips, A.J. What would it take to describe the global diversity of parasites? Proc. R. Soc. B 2020, 287, 20201841. [Google Scholar] [CrossRef]
- Bray, R.A.; Gibson, D.I.; Jones, A. Keys to the Trematoda, Volume 3; CABI: Wallingford, UK, 2008; ISBN 978-0-85199-588-5. [Google Scholar] [CrossRef]
- Nadler, S.A.; Pérez-Ponce de León, G. Integrating molecular and morphological approaches for characterizing parasite cryptic species: Implications for parasitology. Parasitology 2011, 138, 1688–1709. [Google Scholar] [CrossRef]
- Poulin, R. Uneven distribution of cryptic diversity among higher taxa of parasitic worms. Biol. Lett. 2011, 7, 241–244. [Google Scholar] [CrossRef]
- Georgieva, S.; Faltýnková, A.; Brown, R.; Blasco-Costa, I.; Soldánová, M.; Sitko, J.; Scholz, T.; Kostadinova, A. Echinostoma ‘revolutum’ (Digenea: Echinostomatidae) species complex revisited: Species delimitation based on novel molecular and morphological data gathered in Europe. Parasit. Vectors 2014, 7, 520. [Google Scholar] [CrossRef][Green Version]
- Selbach, C.; Soldánová, M.; Georgieva, S.; Kostadinova, A.; Sures, B. Integrative taxonomic approach to the cryptic diversity of Diplostomum spp. in lymnaeid snails from Europe with a focus on the ‘Diplostomum mergi’ species complex. Parasit. Vectors 2015, 8, 300. [Google Scholar] [CrossRef]
- Pérez-Ponce de León, G.; Poulin, R. An updated look at the uneven distribution of cryptic diversity among parasitic helminths. J. Helminthol. 2018, 92, 197–202. [Google Scholar] [CrossRef]
- Blasco-Costa, I.; Poulin, R. Parasite life-cycle studies: A plea to resurrect an old parasitological tradition. J. Helminthol. 2017, 91, 647–656. [Google Scholar] [CrossRef]
- EUR-Lex. Available online: https://eur-lex.europa.eu/eli/dir/2010/63/oj (accessed on 21 December 2023).
- Tkach, V.V.; Pawlowski, J.; Sharpilo, V.P. Molecular and morphological differentiation between species of the Plagiorchis vespertilionis group (Digenea, Plagiorchiidae) occurring in European bats, with a re-description of P. vespertilionis (Müller, 1780). Syst. Parasitol. 2000, 47, 9–22. [Google Scholar] [CrossRef]
- Tkach, V.V. Family Plagiorchiidae Lühe, 1901. In Keys to the Trematoda, Volume 3; Bray, R.A., Gibson, D.I., Jones, A., Eds.; CABI: Wallingford, DC, UK, 2008; pp. 295–325. [Google Scholar] [CrossRef]
- Greani, S.; Quilichini, Y.; Foata, J.; Greiman, S.E.; Ndiaye, P.I.; Tkach, V.V.; Marchand, B. Vitellogenesis of the digenean Plagiorchis elegans (Rudolphi, 1802) (Plagiorchioidea, Plagiorchiidae). Parasitol. Int. 2014, 63, 537–543. [Google Scholar] [CrossRef]
- Horsák, M.; Juřičková, L.; Beran, L.; Čejka, T.; Dvořák, L. Annotated list of mollusc species recorded outdoors in the Czech and Slovak Republics. Malacol. Bohemoslov. 2010, 1, 1–37. (In Czech) [Google Scholar] [CrossRef]
- Aksenova, O.V.; Bolotov, I.N.; Gofarov, M.Y.; Kondakov, A.V.; Vinarski, M.V.; Bespalaya, Y.V.; Kolosova, Y.S.; Palatov, D.M.; Sokolova, S.E.; Spitsyn, V.M.; et al. Species richness, molecular taxonomy and biogeography of the Radicine pond snails (Gastropoda: Lymnaeidae) in the Old World. Sci. Rep. 2018, 8, 11199. [Google Scholar] [CrossRef]
- Cichy, A.; Faltýnková, A.; Żbikowska, E. Cercariae (Trematoda, Digenea) in European freshwater snails—A checklist of records from over one hundred years. Folia Malacol. 2011, 19, 165–189. [Google Scholar] [CrossRef]
- Faltýnková, A.; Sures, B.; Kostadinova, A. Biodiversity of trematodes in their intermediate mollusc and fish hosts in the freshwater ecosystems of Europe. Syst. Parasitol. 2016, 93, 283–293. [Google Scholar] [CrossRef]
- Zikmundová, J.; Georgieva, S.; Faltýnková, A.; Soldánová, M.; Kostadinova, A. Species diversity of Plagiorchis Lühe, 1899 (Digenea: Plagiorchiidae) in lymnaeid snails from freshwater ecosystems in central Europe revealed by molecules and morphology. Syst. Parasitol. 2014, 88, 37–54. [Google Scholar] [CrossRef]
- Krasnolobova, T.A. Trematodes of the Fauna of USSR. Genus Plagiorchis; Nauka: Moscow, Russia, 1987. (In Russian) [Google Scholar]
- Guk, S.M.; Kim, J.L.; Park, J.H.; Chai, J.Y. A human case of Plagiorchis vespertilionis (Digenea: Plagiorchiidae) infection in the Republic of Korea. J. Parasitol. 2007, 93, 1225–1227. [Google Scholar] [CrossRef]
- Boyce, K.; Hide, G.; Craig, P.S.; Reynolds, C.; Hussain, M.; Bodell, A.J.; Bradshaw, H.; Pickles, A.; Rogan, M.T. A molecular and ecological analysis of the trematode Plagiorchis elegans in the wood mouse Apodemus sylvaticus from a periaquatic ecosystem in the UK. J. Helminthol. 2014, 88, 310–320. [Google Scholar] [CrossRef][Green Version]
- Chai, J.Y. Plagiorchids. In Human Intestinal Flukes; Springer: Dordrecht, The Netherlands, 2019; pp. 463–489. ISBN 978-94-024-1702-9. [Google Scholar]
- Suleman, M.J.; Khan, M.S.; Tkach, V.V.; Muhammad, N.; Zhang, D.; Zhu, X.Q. Characterization of the complete mitochondrial genome of Plagiorchis maculosus (Digenea, Plagiorchiidae) representative of a taxonomically complex digenean family. Parasitol. Int. 2019, 71, 99–105. [Google Scholar] [CrossRef]
- Našincová, V. Trematode Developmental Stages in Czech Aquatic Snails and Life-Cycles of Selected Species of the Family Omphalometridae and Echinostomatidae. Ph.D. Thesis, Institute of Parasitology, Czechoslovak Academy of Sciences, České Budějovice, Czech Republic, 1992. (In Czech). [Google Scholar]
- Faltýnková, A.; Našincová, V.; Kablásková, L. Larval trematodes (Digenea) of the great pond snail, Lymnaea stagnalis (L.), (Gastropoda, Pulmonata) in Central Europe: A survey of species and key to their identification. Parasite 2007, 14, 39–51. [Google Scholar] [CrossRef]
- Kirillova, N.Y.; Kirillov, A.A.; Shchenkov, S.V.; Knyazev, A.E.; Vekhnik, V.A. Morphological and molecular characterization of plagiorchiid trematodes (Plagiorchis: Plagiorchiidae, Digenea) from bats with redescription of Plagiorchis mordovii Shaldybin, 1958. J. Helminthol. 2024, 98, e2. [Google Scholar] [CrossRef]
- Pinto, H.A. Describing formally larval trematodes: Some reflections in the taxonomic integrative era. Trends Parasitol. 2023, 39, 889–890. [Google Scholar] [CrossRef]
- Gibson, D.I.; Bray, R.A.; Harris, E.A. (Compiler) Host-Fresent Study Are Highlighted in bMuseum. 2005. Available online: https://www.nhm.ac.uk/research-curation/scientific-resources/taxonomy-systematics/host-parasites/index.html (accessed on 22 January 2024).
- Huguenin, A.; Depaquit, J.; Villena, I.; Ferté, H. MALDI-TOF mass spectrometry: A new tool for rapid identification of cercariae (Trematoda, Digenea). Parasite 2019, 26, 11. [Google Scholar] [CrossRef]
- Li, R.; Wang, H.M.; Liu, G.H.; Tu, Y.; Deng, Y.P. Characterization of the complete mitochondrial genome of the fluke of turdus, Plagiorchis elegans, and phylogenetic implications. Exp. Parasitol. 2022, 242, 108387. [Google Scholar] [CrossRef]
- Gacad, J.L.J.; Yurlova, N.I.; Ponomareva, N.M.; Urabe, M. Characterization of the complete mitochondrial genome of Plagiorchis multiglandularis (Digenea, Plagiorchiidae): Comparison with the members of Xiphidiatan species and phylogenetic implications. Parasitol. Res. 2023, 122, 1545–1556. [Google Scholar] [CrossRef]
- Soldánová, M.; Selbach, C.; Sures, B.; Kostadinova, A.; Pérez-del-Olmo, A. Larval trematode communities in Radix auricularia and Lymnaea stagnalis in a reservoir system of the Ruhr River. Parasit. Vectors 2010, 3, 56. [Google Scholar] [CrossRef]
- Selbach, C.; Soldánová, M.; Feld, C.K.; Kostadinova, A.; Sures, B. Hidden parasite diversity in a European freshwater system. Sci. Rep. 2020, 10, 2694. [Google Scholar] [CrossRef]
- Żbikowska, E.; Nowak, A. One hundred years of research on the natural infection of freshwater snails by trematode larvae in Europe. Parasitol. Res. 2009, 105, 301–311. [Google Scholar] [CrossRef]
- Přikryl, I.; Kabrna, M. Findings from flooding residual pits remaining after coal mining in the Czech Republic. In Mining Meets Water—Conflicts and Solutions, Proceedings of the International Mine Water Association Symposium, Leipzig, Germany, 11–15 July 2016; Drebenstedt, C., Paul, M., Eds.; Technische Universität Bergakademie: Freiberg, Germany, 2016; pp. 201–208. [Google Scholar]
- Žižka, L.; Burda, J. Lake Most. In Post Exploitation Lakes, Risk Assessment of Final pits during Flooding; Burda, J., Bajcar, A., Eds.; Zpravodaj Hnědé Uhlí: Most, Czech Republic, 2020; pp. 11–46. [Google Scholar]
- Žižka, L.; Valvoda, P.; Burda, J. Lake Medard. In Post Exploitation Lakes, Risk Assessment of Final pits during Flooding; Burda, J., Bajcar, A., Eds.; Zpravodaj Hnědé Uhlí: Most, Czech Republic, 2020; pp. 47–59. [Google Scholar]
- Bažant, J. Waterbirds on lake Most 2013–2015. Sbor. Obl. Muz. Most. ř. př. 2015, 37, 61–80. (In Czech) [Google Scholar]
- Bažant, J. Interesting ornithological observations in the Most district (Northwestern Bohemia). Sbor. Obl. Muz. Most. ř. př. 2018, 39, 143–153. (In Czech) [Google Scholar]
- Bažant, J. Birds of the sandpit in Polerady village (Most county, Northwestern Bohemia). Sbor. Obl. Muz. Most. ř. př. 2020, 40, 122–134. (In Czech) [Google Scholar]
- Pozorování Ptáků. Available online: https://pozorovaniptaku.cz/ (accessed on 22 January 2024). (In Czech).
- Ptasie Wyspy 22. Available online: https://alauda.org.pl/ptasie-wyspy-22/obszary-objete-projektem/ (accessed on 8 January 2024). (In Polish).
- Glöer, P. Die Süßwassergastropoden Nord- und Mitteleuropas. Bestimmungschlüssel; Lebensweise, Verbreitung; ConchBooks: Harxheim, Germany, 2002; ISBN 392-591-960-0. [Google Scholar]
- Glöer, P. The Freshwater Gastropods of the West-Palaearctis, Volume 1: Fresh- and Brackish Waters except Spring and Subterranean Snails; Biodiversity Research Lab: Hetlingen, Germany, 2019. [Google Scholar]
- Schniebs, K.; Glöer, P.; Vinarski, M.V.; Hundsdoerfer, A.K. Intraspecific morphological and genetic variability in Radix balthica (Linnaeus 1758) (Gastropoda: Basommatophora: Lymnaeidae) with morphological comparison to other European Radix species. J. Conchol. 2011, 40, 657–677. [Google Scholar]
- Huňová, K.; Kašný, M.; Hampl, V.; Leontovyč, R.; Kuběna, A.; Mikeš, L.; Horák, P. Radix spp.: Identification of trematode intermediate hosts in the Czech Republic. Acta Parasitol. 2012, 57, 273–284. [Google Scholar] [CrossRef]
- Bowles, J.; Blair, D.; McManus, D.P. A molecular phylogeny of the human schistosomes. Mol. Phylogenet. Evol. 1995, 4, 103–109. [Google Scholar] [CrossRef]
- Koehler, A.V.; Springer, Y.P.; Keeney, D.B.; Poulin, R. Intra-and interclonal phenotypic and genetic variability of the trematode Maritrema novaezealandensis. Biol. J. Linn. Soc. 2011, 103, 106–116. [Google Scholar] [CrossRef]
- Van Steenkiste, N.; Locke, S.A.; Castelin, M.; Marcogliese, D.J.; Abbott, C.L. New primers for DNA barcoding of digeneans and cestodes (Platyhelminthes). Mol. Ecol. Resour. 2015, 15, 945–952. [Google Scholar] [CrossRef]
- Tkach, V.V.; Littlewood, D.T.J.; Olson, P.D.; Kinsella, J.M.; Swiderski, Z.P. Molecular phylogenetic analysis of the Microphalloidea Ward, 1901 (Trematoda: Digenea). Syst. Parasitol. 2003, 56, 1–15. [Google Scholar] [CrossRef]
- Almeyda-Artigas, R.J.; Bargues, M.D.; Mas-Coma, S. ITS-2 rDNA sequencing of Gnathostoma species (Nematoda) and elucidation of the species causing human gnathostomiasis in the Americas. J. Parasitol. 2000, 86, 537–544. [Google Scholar] [CrossRef]
- Bargues, M.D.; Vigo, M.; Horák, P.; Dvořák, J.; Patzner, R.A.; Pointier, J.P.; Jackiewicz, M.; Meier-Brook, C.; Mas-Coma, S. European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences. Infect. Genet. Evol. 2001, 1, 85–107. [Google Scholar] [CrossRef]
- Littlewood, D.T.; Rohde, K.; Clough, K.A. Parasite speciation within or between host species? Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997, 27, 1289–1297. [Google Scholar] [CrossRef]
- Littlewood, D.T.; Curini-Galletti, M.; Herniou, E.A. The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Mol. Phylogenet. Evol. 2000, 16, 449–466. [Google Scholar] [CrossRef]
- Garey, J.R.; Wolstenholme, D.R. Platyhelminth mitochondrial DNA: Evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989, 28, 374–387. [Google Scholar] [CrossRef]
- Ohama, T.; Osawa, S.; Watanabe, K.; Jukes, T.H. Evolution of the mitochondrial genetic code IV. AAA as an asparagine codon in some animal mitochondria. J. Mol. Evol. 1990, 30, 329–332. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.; Xie, W.; Drummond, A. Tracer v. 1.6. University of Edinburgh, Institute of Evolutionary Biology. Available online: https://beast.bio.ed.ac.uk/Tracer (accessed on 25 January 2024).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef]
- Gordy, M.A.; Hanington, P.C. A fine-scale phylogenetic assessment of digenean trematodes in central Alberta reveals we have yet to uncover their total diversity. Ecol. Evol. 2019, 9, 3153–3238. [Google Scholar] [CrossRef]
- Bušta, J.; Našincová, V. Record of Plagiorchis neomidis Brendow, 1970 (Trematoda: Plagiorchidae) in Czechoslovakia and studies on its life cycle. Folia Parasitol. 1986, 33, 123–129. [Google Scholar]
- Schwelm, J.; Selbach, C.; Kremers, J.; Sures, B. Rare inventory of trematode diversity in a protected natural reserve. Sci. Rep. 2021, 11, 22066. [Google Scholar] [CrossRef]
- Blasco-Costa, I.; Cutmore, S.C.; Miller, T.L.; Nolan, M.J. Molecular approaches to trematode systematics: ‘best practice’ and implications for future study. Syst. Parasitol. 2016, 93, 295–306. [Google Scholar] [CrossRef]
- Gordy, M.A.; Kish, L.; Tarrabain, M.; Hanington, P.C. A comprehensive survey of larval digenean trematodes and their snail hosts in central Alberta, Canada. Parasitol. Res. 2016, 115, 3867–3880. [Google Scholar] [CrossRef]
- Cramp, S.; Simmons, K.E.L. Handbook of the Birds of Europe, the Middle East and North Africa. The Birds of the Western Palearctic (Vol. 3). Waders to Gulls; Oxford University Press: Oxford, UK, 1983; ISBN 978-0-19-857506-1. [Google Scholar]
- Smit, C.J.; Piersma, T. Numbers, midwinter distribution, and migration of wader populations using the East Atlantic flyway. In Flyways and Reserve Networks for Water Birds; Boyd, H., Pirot, J.-Y., Eds.; Canadian Wildlife Service: Ottawa, ON, Canada, 1989; pp. 24–63. ISBN 978-066-217-104-1. [Google Scholar]
- Bairlein, F.; Norris, D.R.; Nagel, R.; Bulte, M.; Voigt, C.C.; Fox, J.W.; Hussell, D.J.T.; Schmaljohann, H. Cross-hemisphere migration of a 25 g songbird. Biol. Lett. 2012, 8, 505–507. [Google Scholar] [CrossRef]
- Esch, G.W.; Fernandez, J.C. Snail-trematode interactions and parasite community dynamics in aquatic aystems: A review. Am. Midl. Nat. 1994, 131, 209–237. [Google Scholar] [CrossRef]
- Smith, N.F. Spatial heterogeneity in recruitment of larval trematodes to snail intermediate hosts. Oecologia 2001, 127, 115–122. [Google Scholar] [CrossRef]
- Byers, J.E.; Blakeslee, A.M.; Linder, E.; Cooper, A.B.; Maguire, T.J. Controls of spatial variation in the prevalence of trematode parasites infecting a marine snail. Ecology 2008, 89, 439–451. [Google Scholar] [CrossRef]
- Fredensborg, B.L.; Mouritsen, K.N.; Poulin, R. Relating bird host distribution and spatial heterogeneity in trematode infections in an intertidal snail—From small to large scale. Mar. Biol. 2006, 149, 275–283. [Google Scholar] [CrossRef]
- Denk, T.; Grímsson, F.; Zetter, R.; Símonarson, L.A. Introduction to the Nature and Geology of Iceland. In Late Cainozoic Floras of Iceland: 15 Million Years of Vegetation and Climate History in the Northern North Atlantic. Topics in Geobiology; Springer: Dordrecht, The Netherlands, 2011; Volume 35, pp. 1–29. ISBN 978-94-007-0371-1. [Google Scholar] [CrossRef]
- Frafjord, K. Influence of night length on home range size in the northern bat Eptesicus nilssonii. Mamm. Biol. 2013, 78, 205–211. [Google Scholar] [CrossRef]
- Petersen, A.; Jensen, J.K.; Jenkins, P.; Bloch, D.; Ingmarsson, F. A Review of the occurrence of bats (Chiroptera) on Islands in the North East Atlantic and on North Sea installations. Acta Chiropt. 2014, 19, 169–195. [Google Scholar] [CrossRef]
- Krasnolobova, T.A. Validity of the species Plagiorchis fastuosus Szidat, 1924 and its life-cycle. Trudy Gel’mintol. Labor. 1973, 23, 86–96. (In Russian) [Google Scholar]
- Schniebs, K.; Glöer, P.; Vinarski, M.V.; Hundsdoerfer, A.K. Intraspecific morphological and genetic variability in the European freshwater snail Radix labiata (Rossmaessler, 1835) (Gastropoda: Basommatophora: Lymnaeidae). Contrib. Zool. 2013, 82, 55–68. [Google Scholar] [CrossRef]
- Schniebs, K.; Georgiev, D.; Glöer, P.; Hundsdoerfer, A.K. A molecular genetic evidence of the occurrence of the freshwater snail Radix lagotis (Schrank, 1803) (Gastropoda: Lymnaeidae) in Bulgaria. Ecol. Monten. 2015, 3, 29–39. [Google Scholar] [CrossRef]
- Akimova, L.N.; Shimalov, V.V.; Bychkova, E.I. Species diversity of trematode larvae of gastropods in water bodies of Belarus. Parazitologiya 2011, 45, 287–305. (In Russian) [Google Scholar]
- Hanák, V.; Anděra, M.; Uhrin, M.; Danko, Š.; Horáček, I. Bats of the Czech Republic and Slovakia: Distributional status of individual species. In A Tribute to Bats; Horáček, I., Uhrin, M., Eds.; Lesnická práce: Kostelec nad Černými lesy, Czech Republic, 2010; pp. 143–254. ISBN 978-80-87154-44-1. [Google Scholar]
- Ancillotto, L.; Bosso, L.; Salinas-Ramos, V.B.; Russo, D. The importance of ponds for the conservation of bats in urban landscapes. Landsc. Urban Plan. 2019, 190, 103607. [Google Scholar] [CrossRef]
- Pešout, P.; Porteš, M.; Pixová, K.Č.; Hendrychová, M.; Kříž, P.; Lacina, D. Ecosystem restoration of brown coal open-pit mines. Nat. Conserv. J. 2022, 77, 34–39. [Google Scholar]
- Gorman, A.M. Studies on the Biology of Plagiorchis elegans (Rudolphi, 1802), (Trematoda: Digenea) in Its Mammalian and Molluscan Hosts. Doctoral Thesis, University of Leeds, Leeds, UK, 1980. [Google Scholar]
- Ponomareva, N.M.; Popova, O.N.; Yurlova, N.I. Odonata (Insecta) larvae as the second intermediate hosts of the trematodes of genus Plagiorchis in the Basin of Chany Lake, Western Siberia. Contemp. Probl. Ecol. 2022, 15, 631–641. [Google Scholar] [CrossRef]
- Macy, R.W. The Life Cycle of Plagiorchis vespertilionis parorchis, n. ssp., (Trematoda: Plagiorchiidae), and observations on the effects of light on the emergence of the cercaria. J. Parasitol. 1960, 46, 337–345. [Google Scholar] [CrossRef]
- Chervy, L. Manual for the study of tapeworms (Cestoda) parasitic in ray-finned fish, amphibians and reptiles. Folia Parasitol. 2024, 71, 001. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).