Abstract
Chalcopteryx rutilans Rambur, 1842 (Polythoridae, Odonata) is a species widely distributed in central Amazonia. Due to its sensitivity to environmental changes, it is a bioindicator species used to evaluate the environmental conditions of streams in federally protected areas. By sequencing C. rutilans mitogenome, we report the first whole mitogenome from the Polythoridae family and the first from an Odonata species endemic to South America. The entire mitogenome has 15,653 bp and contains 13 protein-coding, 22 tRNA, and two rRNA genes. The nucleotide composition of the mitogenome is 42.7%, T: 25.5%, C: 19.4%, and G: 12.4%. The AT and GC skews of the mitogenome sequence were 0.249 and −0.220, respectively. C. rutilans was recovered as a sister to Pseudolestes mirabilis Kirby, 1900 (Pseudolestidae), demonstrating the absence of mitogenomes of species from multiple families in the current literature. Mitogenome data from this study will provide useful information for further studies on the phylogeny and conservation of Polythoridae.
1. Introduction
Odonata (Hexapoda: Insecta) is an order of insects known as dragonflies and damselflies. It is divided into three suborders: Zygoptera (about 3000 species), Anisoptera (about 3000 species), and Anisozygoptera (only three species) [1]. Despite their attractive coloration and their importance in the food web and biomonitoring of aquatic environments, genomic knowledge of Odonata is still far behind that of the other insect orders [2]). The few whole genomes and mitogenomes available are concentrated in research groups and species found in countries with large economies, such as England, the USA, and China [3,4,5]. At the same time, they are rare or absent in poor or developing countries, such as those in South America.
The genus Chalcopteryx Selys, 1853 (Insecta: Odonata) is an endemic South-American damselfly of the family Polythoridae (61 species, [1]) that inhabits streams and rivers where it flies above the water or perches on aquatic macrophytes and on branches of marginal vegetation and is distinguished by the unique coloration of its hind wings [6]. Their species lays its eggs on plant debris in the streambed and is characterized by great environmental specificity. It is found only in streams and rivers with well-preserved riparian forests, good environmental conditions, and few anthropogenic disturbances. Because of its sensitivity to environmental changes, it is a bioindicator species used in the Amazon as part of the Chico Mendes Institute of Biodiversity Conservation (ICMBio) water monitoring program, in partnership with local communities, to assess the environmental conditions of rivers in federally protected areas [7].
Species representing the genus Chalcopteryx are distributed in the states of the central and northern regions of Brazil, especially in the Amazon area, as well as in Argentina, Bolivia, Colombia, Peru, and Venezuela [8,9]. Five species have been described: Chalcopteryx machadoi Costa, 2005; Chalcopteryx radians Ris, 1914; Chalcopteryx rutilans Rambur, 1842; Chalcopteryx scintillans McLachlan, 1870; and Chalcopteryx seabrai Santos and Machado, 1961. Chalcopteryx rutilans Rambur, 1842 is a species widely distributed in the central Amazon. It has a small body size, and both males and females have a conspicuous black-orange coloration on the pterothorax [10]. The hindwings are very conspicuous, with an iridescent multicolored reflection (mainly greenish, purple, reddish, and copper), and when they move, a brief and shiny metallic flash can be seen [11].
Whole mitogenome sequencing provides important information about evolution, genetic diversity, and species identification by offering higher taxonomic resolution than single gene regions used in conventional approaches (see [12]). Mitogenomes for the family Polythoridae are not available in the GenBank (data valid for 24th of April 2023). In this study, we performed detailed sequencing, annotation, and analysis of the mitogenome of C. rutilans. In addition, we acquired mitochondrial genome sequences from GenBank for other Odonata representatives to investigate the phylogenetic position of Polythoridae. This study is the first time a mitogenome has been reported from the family Polythoridae, and it is also the first from an Odonata species endemic to South America.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
Genomic libraries (one from a male and one from a female) were prepared from specimens of C. rutilans (Figure 1) collected in April 2021 in the Capim River watershed, Paragominas, Pará, Brazil. Total genomic DNA was extracted from the legs of an adult individual using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA), according to the supplier’s instructions, and subsequently quantified using the NanoDrop 1000 spectrophotometer and Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). The voucher specimens (CHR-01 and CHR-02) were deposited in the natural history collection of the Museology Course of the Universidade Federal do Pará (UFPA), Belém, Brazil. The voucher specimens were determined to species level by Leandro Juen, one of the authors of this article.

Figure 1.
Example of a stream (A) and habitats (B) where the species Chalcopteryx rutilans (C) can be found in the wild (Capim River Basin, Pará, Brazil). Source: CMP.
2.2. Mitogenome Sequencing, Assembly, Annotation and Bioinformatic Analyses
The genomic libraries were prepared using the Illumina DNA Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions with a short insert size of 500 bp. Libraries were then sequenced on a NextSeq550 Illumina platform using a paired-end High Output Kit v2 (300 cycles) (Illumina).
Raw sequencing data were filtered to remove adapters and low-quality sequences, yielding approximately 14 Gb. Genome assembly was performed using MEGAHIT [13] and SOAPdenovo [14]. Protein-coding genes (PCGs), ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) locations were predicted by using MITOS Web Server 2 [15]. The AT and GC skews were calculated according to the following formulas: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(C + G) [16]. The tandem repeat units of the control region (CR) were identified with the Tandem Repeats Finder Server (http://tandem.bu.edu/trf/trf.advanced.submit.html; accessed on 13 June 2023; with default settings, except “Minimum Alignment Score To Report Repeat”= 40) [17]. The arrangement of genes encoding rRNA proteins, comparative relative synonymous codon usage (RSCU), and CR organization analysis were performed using an in-house Python script. For these analyses, we used the following mitogenomes deposited in Genbank: Euphaea formosa Hagen in Selys, 1869 (HM126547), Ischnura elegans (Vander Linden, 1820) (KU958378), Matrona basilaris Selys, 1853 (MK722304), Mesopodagrion tibetanum McLachlan, 1896 (MK951671), Platycnemis foliacea Selys, 1886 (KP233804), and Pseudolestes mirabilis Kirby, 1900 (FJ606784).
2.3. Molecular Phylogenetic Analysis
Phylogenetic analysis of concatenated PCGs from all available Zygoptera mitogenomes (GenBank, 10 April 2023) was performed using the MAFFT alignment program with default parameters (Geneious® version 9.0.5 software, [18]) and the IQ-TREE web server [19] using the ultra-fast bootstrap algorithm with 10,000 replicates and automatic model selection (GTR + F + I + G4). To show that there are no mitogenomes in South America, information from the deposited sequences was used to indicate the origin of the related specimens. The genome sequence data supporting the results of this study are freely available in NCBI’s GenBank (https://www.ncbi.nlm.nih.gov/; accessed on 13 June 2023) under the accession number OQ868370.
3. Results and Discussion
3.1. Mitogenome Organization and Nucleotide Composition
The complete mitochondrial genome of C. rutilans was sequenced and assembled (Figure 2). The mitogenome has 15,653 bp and contains 13 protein-coding, 22 tRNA, and two rDNA genes. Nucleotide compositions are 42.7%, T: 25.5%, C: 19.4%, and G: 12.4%. The AT and GC skewness of the mitogenome sequence was 0.249 and −0.220, showing the A-skew and C-skew.
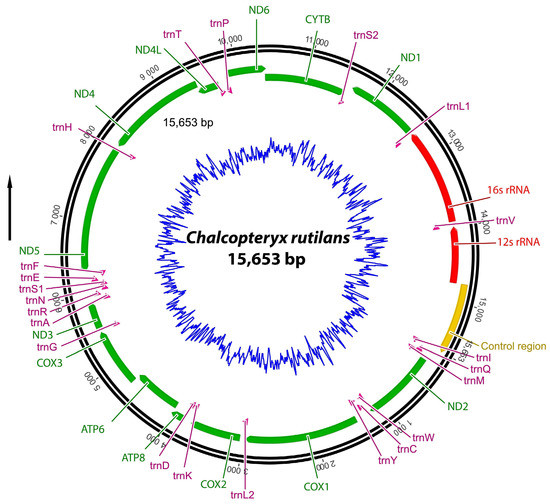
Figure 2.
Graphical representation of the circular mitogenome map of C. rutilans. Different colors indicate the groups of genes: 13 protein-coding genes (green), 2 rRNAs (red), 22 tRNAs (pink), and control region (yellow). The blue ring represents the GC content.
The start and termination codons were (respectively) assigned to ND2 (ATT/TAA), COX1 (ATG/T--), COX2 (ATG/T--), ATP8 (ATC/TAA), ATP6 (ATG/TAA), COX3 (ATG/T--), ND3 (ATA/TAG), ND5 (ATT/TAA), ND4 (ATG/TAG), ND4L (ATG/TAA), ND6 (ATA/TAA), Cytb (ATG/TAA), and ND1 (ATT/TAG) (Table 1).

Table 1.
Mitochondrial genome organization and gene content of C. rutilans with detailed description of gene boundaries, gene length (in bp), as well as start and stop codons for protein-coding genes and anticodons for tRNA genes. Strand: heavy (H) and light (L).
Gene order and orientation, as well as the distribution of genes on the heavy and light strands, were identical to the mitogenome of the representants of other Zygoptera families, such as Megapodagrionidae, Euphaedidae, Pseudolestidae, Calopterygidae, Platycnemididae, and Coenagrionidae. Although not necessary in principle, we show a simple way to illustrate this similarity between families in Figure 3.
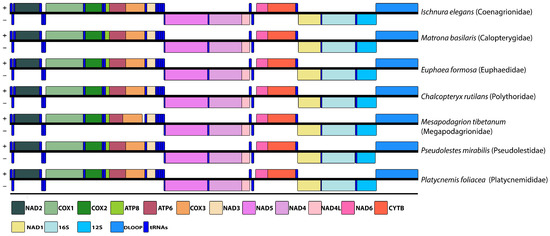
Figure 3.
Arrangement of genes encoding rRNAs and proteins of C. rutilans and other six species from different Zygoptera families. Heavy (+) and light (−) strands.
3.2. Protein-Coding Genes (PCGs)
The overall length of the PCGs in the C. rutilans mitogenome was 11,128 bp, ranging from 162 bp (ATP8) to 1729 bp (ND5). Most PCGs used the conventional start codon ATG and ended with the codon TAN (Table 1). As shown in Figure 4, the RSCU was biased for most amino acids. Comparative summaries of the RSCU of the mitogenomes for families of Zygoptera show that they are very similar. In addition, synonymous codon preferences were conserved for all seven species.
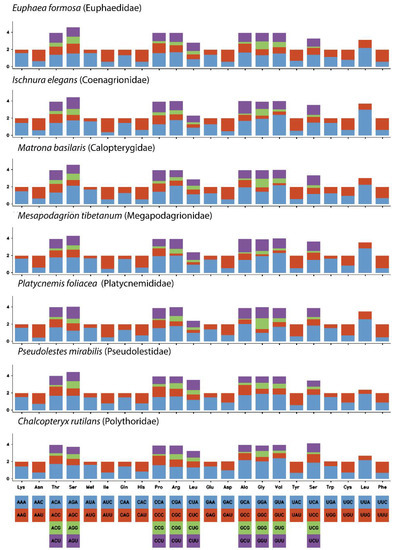
Figure 4.
Relative synonymous codon usage (RSCU) of protein-coding genes of C. rutilans and other six species from different Zygoptera families.
3.3. Transfer and Ribosomal RNA Genes
The mitogenome of C. rutilans has two rRNAs and 22 typical tRNAs. The 16S rRNA and 12S rRNA have sizes of 751 and 1302 bp, respectively. Compared with other Zygoptera mitogenomes, the tRNA genes are totally conserved. Among them, 14 tRNAs were encoded on the heavy strand and the remaining eight on the light strand. As seen in Figure 5, the 22 tRNAs have a typical cloverleaf secondary structure, and their sizes ranged from 63 bp (tRNA-Ser2) to 76 bp (tRNA-Ala). The total lengths of the 22 tRNAs were 1511 bp.
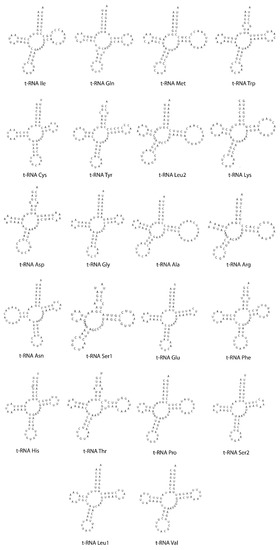
Figure 5.
Predicted secondary structures of 22 inferred tRNAs from the C. rutilans mitochondrial genome.
3.4. Control Region (CR)
Like other Zygoptera mitogenomes, the CR was located between 12S rRNA and tRNA-Leu (898 bp) (Figure 6). In C. rutilans, this region was strongly A + T rich, with a base composition of A = 50.4%, T = 32.1%, C = 11.0%, and G = 06.5%. The control region varied in size among species (Figure 6), ranging from 407 bp in P. mirabilis to 1512 bp in M. basilaris. All species had at least one tandem repeat region (Figure 6, Table S1). In C. rutilians, a 29 bp tandem repeat unit extends across one more copy in the first repeat region. In the second repeat region, a 33 bp tandem repeat unit extends across a copy with a third partial copy. In addition, there are other conserved elements such as poly-A stretches (positions: 14,980–15,009 bp; 15,460–15,476 bp; 15,478–15,488 bp) and microsatellite-like elements (TTA)3 (position: 15,326–15,334 bp) that are potentially useful markers for analyzing the geographic structure of populations. Although high A + T rich nucleotide content and tandemly repeated sequences are not discussed in the published mitogenomes of Zygoptera, the features may be involved in the initiation of replication in the mitochondrial genome, as has been suggested in other invertebrates (e.g., [20,21]).

Figure 6.
Organization of the control region in the complete mitogenomes of C. rutilans and six other species from different Zygoptera families.
3.5. Phylogenetic Relationships
Based on the collected mitogenomes, we reconstructed the phylogenetic relationships within Odonata, including 26 Zygoptera species and one species from Anisoptera (Anax imperator Leach in Brewster, 1815) and Anisozygoptera (Epiophlebia superstes Selys, 1889) as the outgroup (Figure 7). This mitogenome is not only the first mitogenome of the family Polythoridae but also the first of the so-called ‘Calopterygoidea Group 2’ defined by Dijkstra et al. [22] and continued by Bybee et al. [23], comprising nine families exclusively from the Neotropics and Afrotropics. The phylogeny resulting from our study (PCGs) is also consistent with that found by Kohli et al. [24] (transcriptome), with the superfamily Calopterygoidea recovered as monophyletic, which differs from the phylogeny proposed by Dijkstra et al. [22] (nuclear and mitochondrial genes) and Bybee et al. [23] (anchored hybrid enrichment-AHE).
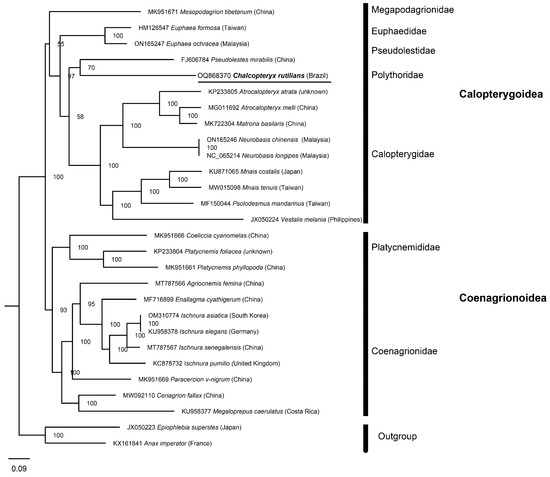
Figure 7.
Phylogenetic tree of Zygoptera based on 13 PCGs derived from IQ-tree, with information on the origin of the sequenced specimens. Values near the nodes are maximum likelihood ultrafast bootstrap values.
In this study, C. rutilans was recovered as a sister to Pseudolestes mirabilis (Insecta: Odonata), a species of the monospecific family Pseudolestidae endemic to China [25]. Following other phylogenies [17,18], several other families such as Philogeniidae, Protolestidae, and Dicteriadidae should be represented between these two named species in our phylogeny, but no genomic data are available. This highlights that mitogenomes (or even single gene sequences) for several tropical families are not yet available in global databases, as is the case for several insect orders, e.g., Trichoptera, Ephemeroptera, Plecoptera, and Diptera (e.g., [26,27]). As other authors have noted (e.g., [28]), more mitogenomes from other Odonata species are needed to provide a better estimate of phylogeny using this type of data.
4. Conclusions
Chalcopteryx rutilans is the first species of its family whose mitogenome has been sequenced. This mitogenome contains 13 protein-coding, 22 tRNA, and two rRNA genes. It has a similar structure and gene order to other deposited Zygoptera species. Phylogenetic analysis showed that it was recovered as a sister to Pseudolestes mirabilis (Odonata: Pseudolestidae), demonstrating the absence of mitogenomes from species in several families in the current literature. Mitogenome data from this study will provide useful information for further studies on the phylogeny of Zygoptera, databases for phylogeny, DNA barcoding and metabarcoding, etc.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15080908/s1, Table S1. Comparison of the control region in C. rutilans and six other species from different Zygoptera families.
Author Contributions
Conceptualization, L.J., L.F.d.A.M., T.S.M., R.L. and Â.R.-d.-S.; methodology and formal analysis, R.K., F.G.d.C., C.C.M.-P., J.d.S.B., L.B.C., V.R.S.F., C.S.S., S.G., G.C.C., L.M., J.E.S.d.S. and D.H.F.G.; data curation, R.K. and L.J.; writing—original draft preparation, L.F.d.A.M., T.S.M., R.L., Â.R.-d.-S., R.K., F.G.d.C., C.C.M.-P., J.d.S.B., L.B.C., V.R.S.F., C.S.S., S.G., G.C.C., L.M., J.E.S.d.S. and D.H.F.G.; writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
L.J., L.F.d.A.M. and R.L. thank the National Council for Scientific and Technological Development (CNPq) for a research productivity fellowship (Grants #304710/2019-9, 302881/2022–0, and 312531/2021-4, respectively). This study was partially funded by the Coordination of Improvement of Higher Education Personnel in Brazil (CAPES) under Financial Code 001. We are grateful for funding from authors’ grants. We also thank the Pró-Reitoria de Pesquisa e Pós-Graduação (PROPESP) from the Federal University of Pará (UFPA) (Editorial 02/2023).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The genome sequence data supporting the results of this study are freely available in NCBI’s GenBank (https://www.ncbi.nlm.nih.gov/, accessed on 13 June 2023) under the accession number OQ868370.
Acknowledgments
We thank the Biodiversity Research Consortium Brazil-Norway (BRC) and the Hydro Paragominas Company for their funding and logistical support for the project “Avaliando a integridade de ecossistemas aquáticos implementando um método de biomonitoramento baseado em sequenciamento de DNA de última geração”. This paper is number BRC0036 in the publication series of the BRC. We would like to thank the two anonymous reviewers for their suggestions and comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Paulson, D.; Schorr, M.; Deliry, C. World Odonata List. Available online: https://www.pugetsound.edu/slater-museum-natural-history-0/biodiversity-resources/insects/dragonflies/world-odonata-list (accessed on 22 May 2023).
- Wellenreuther, M.; Dudaniec, R.Y.; Lancaster, L.T. Genomic Insights into Micro- and Macro-Evolutionary Processes in Odonata. In Dragonflies and Damselflies; Córdoba-Aguilar, A., Beatty, C.D., Bried, J.T., Eds.; Oxford University Press: Oxford, UK, 2022; pp. 7–20. [Google Scholar]
- Lorenzo-Carballa, M.O.; Thompson, D.J.; Cordero-Rivera, A.; Watts, P.C. Next Generation Sequencing Yields the Complete Mitochondrial Genome of the Scarce Blue-Tailed Damselfly, Ischnura Pumilio. Mitochondrial DNA 2014, 25, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-N.; Yu, P.-P.; Zhang, L.-P.; Storey, K.B.; Gao, X.-Y.; Zhang, J.-Y. Increasing 28 Mitogenomes of Ephemeroptera, Odonata and Plecoptera Support the Chiastomyaria Hypothesis with Three Different Outgroup Combinations. PeerJ 2021, 9, e11402. [Google Scholar] [CrossRef] [PubMed]
- Grether, G.F.; Beninde, J.; Beraut, E.; Chumchim, N.; Escalona, M.; MacDonald, Z.G.; Miller, C.; Sahasrabudhe, R.; Shedlock, A.M.; Toffelmier, E.; et al. Reference Genome for the American Rubyspot Damselfly, Hetaerina americana. J. Hered. 2023, 114, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Resende, D.C.; De Marco, P., Jr. First Description of Reproductive Behavior of the Amazonian Damselfly Chalcopteryx Rutilans (Rambur) (Odonata, Polythoridae). Rev. Bras. Entomol. 2010, 54, 436–440. [Google Scholar] [CrossRef]
- Brasil, L.S.; Farias, D.D.; Marcolino, C.N.; Bassols, M.; Juen, L. Monitoramento Participativo Em Igarapés de Unidades de Conservação Da Amazônia Brasileira Utilizando Odonata. Hetaerina 2020, 2, 8–13. [Google Scholar]
- Koroiva, R.; Neiss, U.G.; Fleck, G.; Hamada, N. Checklist of Dragonflies and Damselflies (Insecta: Odonata) of the Amazonas State, Brazil. Biota Neotrop. 2020, 20, e20190877. [Google Scholar] [CrossRef]
- Calvão, L.B.; De Marco, P.J.; Batista, J.D. List Odonata (Insecta) from Nova Xavantina, Mato Grosso, Central Brazil: Information on Species Distribution and New Records. Check List 2014, 10, 299–307. [Google Scholar] [CrossRef]
- Santos, N.D.; Machado, A.B. Contribuição Ao Conhecimento Do Gênero Chalcopteryx Selys, 1853, Com a Descrição de Uma Nova Espécie. Bol. Mus. Para. Emilio Goeldi 1960, 24, 1–17. [Google Scholar]
- Guillermo-Ferreira, R.; Neiss, U.G.; Hamada, N.; Bispo, P.C. Behavior of the Amazonian Damselfly Chalcopteryx scintillans McLachlan (Zygoptera: Polythoridae) and Comments on Its Morphological Distinction from C. rutilans (Rambur). Int. J. Odonatol. 2014, 17, 251–258. [Google Scholar] [CrossRef]
- Havird, J.C.; Santos, S.R. Performance of Single and Concatenated Sets of Mitochondrial Genes at Inferring Metazoan Relationships Relative to Full Mitogenome Data. PLoS ONE 2014, 9, e84080. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Agunbiade, T.A.; Coates, B.S.; Sun, W.; Tsai, M.-R.; Valero, M.C.; Tamò, M.; Pittendrigh, B.R. Comparison of the Mitochondrial Genomes of the Old and New World Strains of the Legume Pod Borer, Maruca vitrata (Lepidoptera: Crambidae). Int. J. Trop. Insect Sci. 2017, 37, 125–136. [Google Scholar] [CrossRef]
- Ma, X.; Agudelo, P.; Richards, V.P.; Baeza, J.A. The Complete Mitochondrial Genome of the Columbia Lance Nematode, Hoplolaimus Columbus, a Major Agricultural Pathogen in North America. Parasit. Vectors 2020, 13, 321. [Google Scholar] [CrossRef]
- Dijkstra, K.D.B.; Kalkman, V.J.; Dow, R.A.; Stokvis, F.R.; Van Tol, J. Redefining the Damselfly Families: A Comprehensive Molecular Phylogeny of Zygoptera (Odonata). Syst. Entomol. 2014, 39, 68–96. [Google Scholar] [CrossRef]
- Bybee, S.M.; Kalkman, V.J.; Erickson, R.J.; Frandsen, P.B.; Breinholt, J.W.; Suvorov, A.; Dijkstra, K.-D.B.; Cordero-Rivera, A.; Skevington, J.H.; Abbott, J.C.; et al. Phylogeny and Classification of Odonata Using Targeted Genomics. Mol. Phylogenet. Evol. 2021, 160, 107115. [Google Scholar] [CrossRef]
- Kohli, M.; Letsch, H.; Greve, C.; Béthoux, O.; Deregnaucourt, I.; Liu, S.; Zhou, X.; Donath, A.; Mayer, C.; Podsiadlowski, L.; et al. Evolutionary History and Divergence Times of Odonata (Dragonflies and Damselflies) Revealed through Transcriptomics. iScience 2021, 24, 103324. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bu, W. A Description of the Remarkable Larva of Pseudolestes mirabilis Kirby (Odonata: Pseudolestidae). Int. J. Odonatol. 2011, 14, 105–110. [Google Scholar] [CrossRef]
- do Nascimento, B.L.S.; da Silva, F.S.; Nunes-Neto, J.P.; de Almeida Medeiros, D.B.; Cruz, A.C.R.; da Silva, S.P.; da Silva e Silva, L.H.; de Oliveira Monteiro, H.A.; Dias, D.D.; Vieira, D.B.R.; et al. First Description of the Mitogenome and Phylogeny of Culicinae Species from the Amazon Region. Genes 2021, 12, 1983. [Google Scholar] [CrossRef] [PubMed]
- Koroiva, R.; de Souza, M.S.; de Roque, F.O.; Pepinelli, M. DNA Barcodes for Forensically Important Fly Species in Brazil. J. Med. Entomol. 2018, 55, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Herzog, R.; Osigus, H.-J.; Feindt, W.; Schierwater, B.; Hadrys, H. The Complete Mitochondrial Genome of the Emperor Dragonfly Anax Imperator LEACH, 1815 (Odonata: Aeshnidae) via NGS Sequencing. Mitochondrial DNA Part B 2016, 1, 783–786. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).