The First Mitochondrial Genome of an Odonata Endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and Its Implications for the Phylogeny of the Zygoptera
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Mitogenome Sequencing, Assembly, Annotation and Bioinformatic Analyses
2.3. Molecular Phylogenetic Analysis
3. Results and Discussion
3.1. Mitogenome Organization and Nucleotide Composition
3.2. Protein-Coding Genes (PCGs)
3.3. Transfer and Ribosomal RNA Genes
3.4. Control Region (CR)
3.5. Phylogenetic Relationships
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paulson, D.; Schorr, M.; Deliry, C. World Odonata List. Available online: https://www.pugetsound.edu/slater-museum-natural-history-0/biodiversity-resources/insects/dragonflies/world-odonata-list (accessed on 22 May 2023).
- Wellenreuther, M.; Dudaniec, R.Y.; Lancaster, L.T. Genomic Insights into Micro- and Macro-Evolutionary Processes in Odonata. In Dragonflies and Damselflies; Córdoba-Aguilar, A., Beatty, C.D., Bried, J.T., Eds.; Oxford University Press: Oxford, UK, 2022; pp. 7–20. [Google Scholar]
- Lorenzo-Carballa, M.O.; Thompson, D.J.; Cordero-Rivera, A.; Watts, P.C. Next Generation Sequencing Yields the Complete Mitochondrial Genome of the Scarce Blue-Tailed Damselfly, Ischnura Pumilio. Mitochondrial DNA 2014, 25, 247–248. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-N.; Yu, P.-P.; Zhang, L.-P.; Storey, K.B.; Gao, X.-Y.; Zhang, J.-Y. Increasing 28 Mitogenomes of Ephemeroptera, Odonata and Plecoptera Support the Chiastomyaria Hypothesis with Three Different Outgroup Combinations. PeerJ 2021, 9, e11402. [Google Scholar] [CrossRef] [PubMed]
- Grether, G.F.; Beninde, J.; Beraut, E.; Chumchim, N.; Escalona, M.; MacDonald, Z.G.; Miller, C.; Sahasrabudhe, R.; Shedlock, A.M.; Toffelmier, E.; et al. Reference Genome for the American Rubyspot Damselfly, Hetaerina americana. J. Hered. 2023, 114, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Resende, D.C.; De Marco, P., Jr. First Description of Reproductive Behavior of the Amazonian Damselfly Chalcopteryx Rutilans (Rambur) (Odonata, Polythoridae). Rev. Bras. Entomol. 2010, 54, 436–440. [Google Scholar] [CrossRef]
- Brasil, L.S.; Farias, D.D.; Marcolino, C.N.; Bassols, M.; Juen, L. Monitoramento Participativo Em Igarapés de Unidades de Conservação Da Amazônia Brasileira Utilizando Odonata. Hetaerina 2020, 2, 8–13. [Google Scholar]
- Koroiva, R.; Neiss, U.G.; Fleck, G.; Hamada, N. Checklist of Dragonflies and Damselflies (Insecta: Odonata) of the Amazonas State, Brazil. Biota Neotrop. 2020, 20, e20190877. [Google Scholar] [CrossRef]
- Calvão, L.B.; De Marco, P.J.; Batista, J.D. List Odonata (Insecta) from Nova Xavantina, Mato Grosso, Central Brazil: Information on Species Distribution and New Records. Check List 2014, 10, 299–307. [Google Scholar] [CrossRef]
- Santos, N.D.; Machado, A.B. Contribuição Ao Conhecimento Do Gênero Chalcopteryx Selys, 1853, Com a Descrição de Uma Nova Espécie. Bol. Mus. Para. Emilio Goeldi 1960, 24, 1–17. [Google Scholar]
- Guillermo-Ferreira, R.; Neiss, U.G.; Hamada, N.; Bispo, P.C. Behavior of the Amazonian Damselfly Chalcopteryx scintillans McLachlan (Zygoptera: Polythoridae) and Comments on Its Morphological Distinction from C. rutilans (Rambur). Int. J. Odonatol. 2014, 17, 251–258. [Google Scholar] [CrossRef]
- Havird, J.C.; Santos, S.R. Performance of Single and Concatenated Sets of Mitochondrial Genes at Inferring Metazoan Relationships Relative to Full Mitogenome Data. PLoS ONE 2014, 9, e84080. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A Fast and Scalable Metagenome Assembler Driven by Advanced Methodologies and Community Practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de Novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of Nucleotide Composition at Fourfold Degenerate Sites of Animal Mitochondrial Genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Agunbiade, T.A.; Coates, B.S.; Sun, W.; Tsai, M.-R.; Valero, M.C.; Tamò, M.; Pittendrigh, B.R. Comparison of the Mitochondrial Genomes of the Old and New World Strains of the Legume Pod Borer, Maruca vitrata (Lepidoptera: Crambidae). Int. J. Trop. Insect Sci. 2017, 37, 125–136. [Google Scholar] [CrossRef]
- Ma, X.; Agudelo, P.; Richards, V.P.; Baeza, J.A. The Complete Mitochondrial Genome of the Columbia Lance Nematode, Hoplolaimus Columbus, a Major Agricultural Pathogen in North America. Parasit. Vectors 2020, 13, 321. [Google Scholar] [CrossRef]
- Dijkstra, K.D.B.; Kalkman, V.J.; Dow, R.A.; Stokvis, F.R.; Van Tol, J. Redefining the Damselfly Families: A Comprehensive Molecular Phylogeny of Zygoptera (Odonata). Syst. Entomol. 2014, 39, 68–96. [Google Scholar] [CrossRef]
- Bybee, S.M.; Kalkman, V.J.; Erickson, R.J.; Frandsen, P.B.; Breinholt, J.W.; Suvorov, A.; Dijkstra, K.-D.B.; Cordero-Rivera, A.; Skevington, J.H.; Abbott, J.C.; et al. Phylogeny and Classification of Odonata Using Targeted Genomics. Mol. Phylogenet. Evol. 2021, 160, 107115. [Google Scholar] [CrossRef]
- Kohli, M.; Letsch, H.; Greve, C.; Béthoux, O.; Deregnaucourt, I.; Liu, S.; Zhou, X.; Donath, A.; Mayer, C.; Podsiadlowski, L.; et al. Evolutionary History and Divergence Times of Odonata (Dragonflies and Damselflies) Revealed through Transcriptomics. iScience 2021, 24, 103324. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bu, W. A Description of the Remarkable Larva of Pseudolestes mirabilis Kirby (Odonata: Pseudolestidae). Int. J. Odonatol. 2011, 14, 105–110. [Google Scholar] [CrossRef]
- do Nascimento, B.L.S.; da Silva, F.S.; Nunes-Neto, J.P.; de Almeida Medeiros, D.B.; Cruz, A.C.R.; da Silva, S.P.; da Silva e Silva, L.H.; de Oliveira Monteiro, H.A.; Dias, D.D.; Vieira, D.B.R.; et al. First Description of the Mitogenome and Phylogeny of Culicinae Species from the Amazon Region. Genes 2021, 12, 1983. [Google Scholar] [CrossRef] [PubMed]
- Koroiva, R.; de Souza, M.S.; de Roque, F.O.; Pepinelli, M. DNA Barcodes for Forensically Important Fly Species in Brazil. J. Med. Entomol. 2018, 55, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Herzog, R.; Osigus, H.-J.; Feindt, W.; Schierwater, B.; Hadrys, H. The Complete Mitochondrial Genome of the Emperor Dragonfly Anax Imperator LEACH, 1815 (Odonata: Aeshnidae) via NGS Sequencing. Mitochondrial DNA Part B 2016, 1, 783–786. [Google Scholar] [CrossRef]

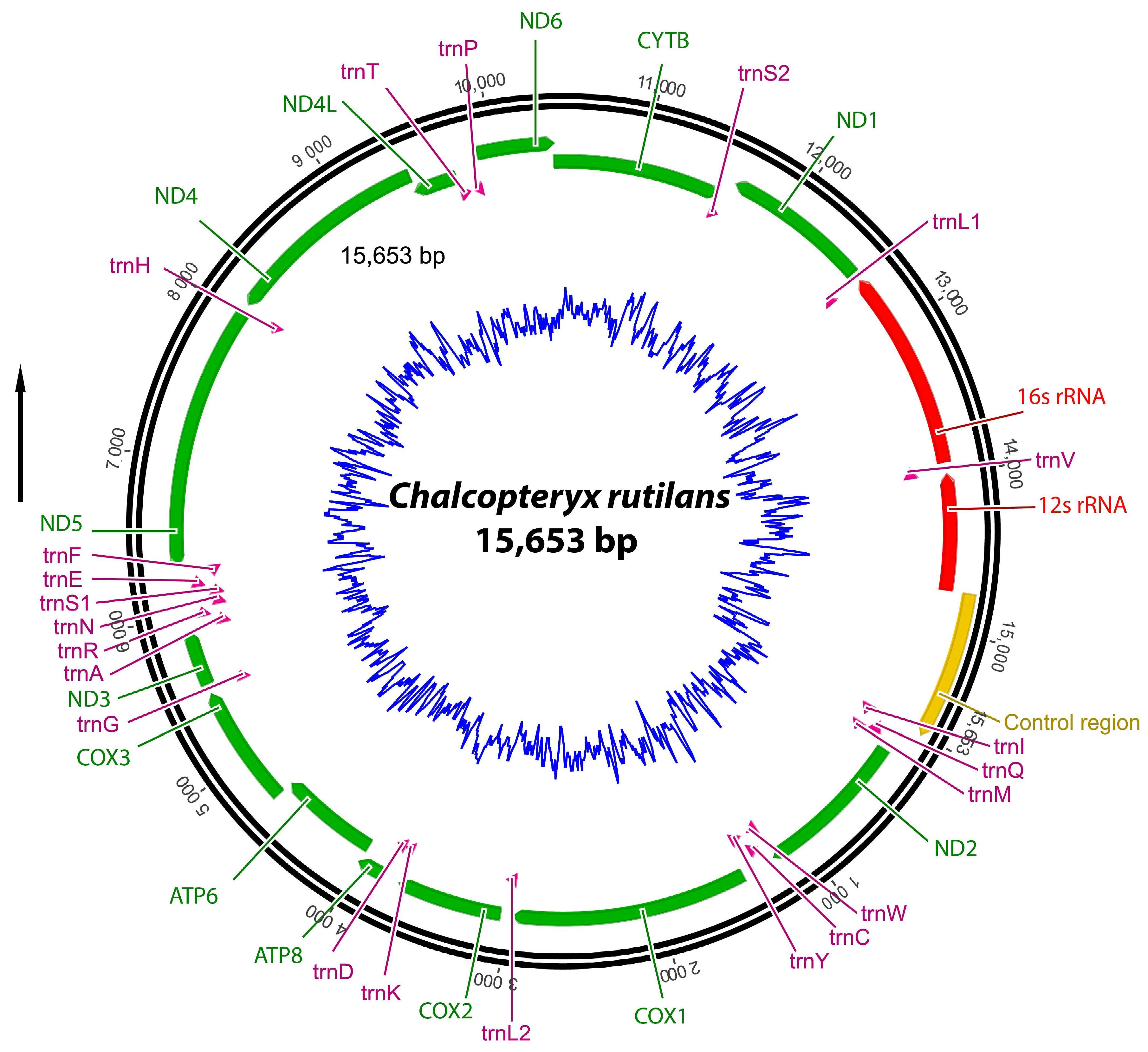

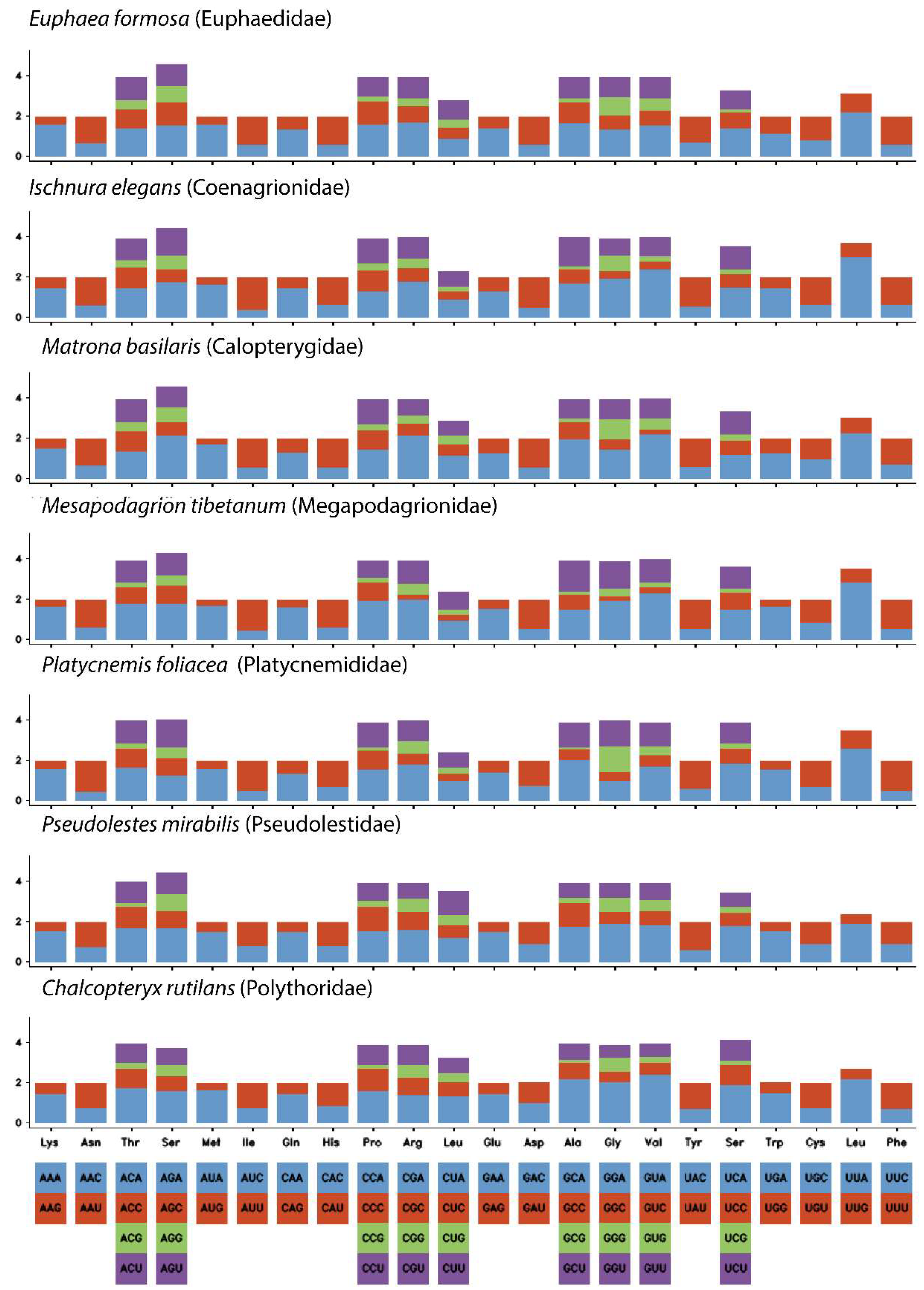
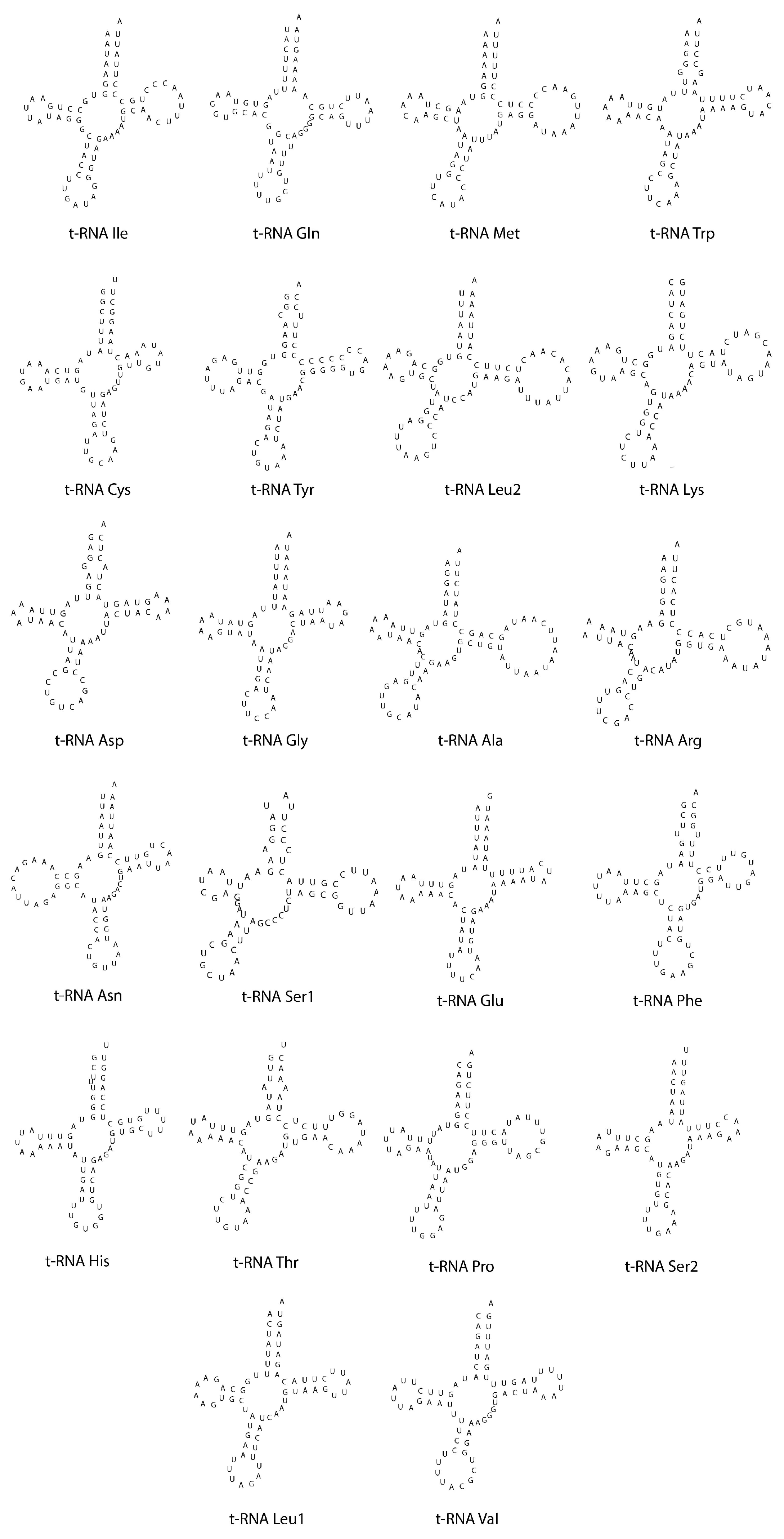


| Region | Start | Stop | Strand | Length | Intergenic Nucleotides | Anticodon and Start Codon/Stop Codon |
|---|---|---|---|---|---|---|
| tRNA-lle | 1 | 69 | H | 69 | −3 | GAT |
| tRNA-Gln | 67 | 134 | L | 68 | 0 | TTG |
| tRNA-Met | 135 | 206 | H | 72 | 0 | CAT |
| ND2 | 207 | 1202 | H | 996 | −2 | ATT/TAA |
| tRNA-Trp | 1201 | 1268 | H | 68 | −8 | TCA |
| tRNA-Cys | 1261 | 1324 | L | 64 | 0 | GCA |
| tRNA-Tyr | 1325 | 1392 | L | 68 | 33 | GTA |
| COX1 | 1426 | 2959 | H | 1534 | 0 | ATG/T-- |
| tRNA-Leu2 | 2960 | 3033 | H | 74 | 0 | TAA |
| COX2 | 3034 | 3721 | H | 688 | 0 | ATG/T-- |
| tRNA-Lys | 3722 | 3793 | H | 72 | −1 | CTT |
| tRNA-Asp | 3793 | 3856 | H | 64 | 0 | GTC |
| ATP8 | 3857 | 4018 | H | 162 | −7 | ATC/TAA |
| ATP6 | 4012 | 4689 | H | 678 | −1 | ATG/TAA |
| COX3 | 4689 | 5475 | H | 787 | 0 | ATG/T-- |
| tRNA-Gly | 5476 | 5539 | H | 64 | 0 | TCC |
| ND3 | 5540 | 5893 | H | 354 | −2 | ATA/TAG |
| tRNA-Ala | 5892 | 5967 | H | 76 | −1 | TGC |
| tRNA-Arg | 5967 | 6036 | H | 70 | −3 | TCG |
| tRNA-Asn | 6034 | 6104 | H | 71 | 4 | GTT |
| tRNA-Ser | 6109 | 6176 | H | 68 | −1 | GCT |
| tRNA-Glu | 6176 | 6240 | H | 65 | −2 | TTC |
| tRNA-Phe | 6239 | 6306 | L | 68 | 0 | GAA |
| ND5 | 6307 | 8035 | L | 1729 | 0 | ATT/TAA |
| tRNA-His | 8036 | 8101 | L | 66 | 0 | GTG |
| ND4 | 8102 | 9439 | L | 1338 | −7 | ATG/TAG |
| ND4L | 9433 | 9726 | L | 294 | 2 | ATG/TAA |
| tRNA-Thr | 9729 | 9798 | H | 70 | 23 | TGT |
| tRNA-Pro | 9822 | 9891 | L | 70 | 11 | TGG |
| ND6 | 9903 | 10,400 | H | 498 | −1 | ATA/TAA |
| CYTB | 10,400 | 11,533 | H | 1134 | −2 | ATG/TAA |
| tRNA-Ser2 | 11,532 | 11,594 | H | 63 | 16 | TGA |
| ND1 | 11,611 | 12,546 | L | 936 | 16 | ATT/TAG |
| tRNA-Leu1 | 12,563 | 12,628 | L | 66 | 0 | TAG |
| 16S rRNA | 12,629 | 13,930 | L | 1302 | 4 | |
| tRNA-Val | 13,935 | 14,004 | L | 70 | 0 | TAC |
| 12S rRNA | 14,005 | 14,755 | L | 751 | 0 | |
| Control region | 14,756 | 15,653 | H | 898 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juen, L.; Koroiva, R.; Geraldo de Carvalho, F.; Mendoza-Penagos, C.C.; Brito, J.d.S.; Calvão, L.B.; Ferreira, V.R.S.; Ribeiro-dos-Santos, Â.; Silva, C.S.; Guerreiro, S.; et al. The First Mitochondrial Genome of an Odonata Endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and Its Implications for the Phylogeny of the Zygoptera. Diversity 2023, 15, 908. https://doi.org/10.3390/d15080908
Juen L, Koroiva R, Geraldo de Carvalho F, Mendoza-Penagos CC, Brito JdS, Calvão LB, Ferreira VRS, Ribeiro-dos-Santos Â, Silva CS, Guerreiro S, et al. The First Mitochondrial Genome of an Odonata Endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and Its Implications for the Phylogeny of the Zygoptera. Diversity. 2023; 15(8):908. https://doi.org/10.3390/d15080908
Chicago/Turabian StyleJuen, Leandro, Ricardo Koroiva, Fernando Geraldo de Carvalho, Cristian Camilo Mendoza-Penagos, Joás da Silva Brito, Lenize Batista Calvão, Victor Rennan Santos Ferreira, Ândrea Ribeiro-dos-Santos, Caio S. Silva, Sávio Guerreiro, and et al. 2023. "The First Mitochondrial Genome of an Odonata Endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and Its Implications for the Phylogeny of the Zygoptera" Diversity 15, no. 8: 908. https://doi.org/10.3390/d15080908
APA StyleJuen, L., Koroiva, R., Geraldo de Carvalho, F., Mendoza-Penagos, C. C., Brito, J. d. S., Calvão, L. B., Ferreira, V. R. S., Ribeiro-dos-Santos, Â., Silva, C. S., Guerreiro, S., Cavalcante, G. C., Magalhães, L., Souza, J. E. S. d., Gomes, D. H. F., Montag, L. F. d. A., Michelan, T. S., & Ligeiro, R. (2023). The First Mitochondrial Genome of an Odonata Endemic to South America, Chalcopteryx rutilans (Rambur, 1842) (Odonata: Polythoridae), and Its Implications for the Phylogeny of the Zygoptera. Diversity, 15(8), 908. https://doi.org/10.3390/d15080908












