Abstract
In this study, we used genetic approaches to assess the conservation status of a protected amphibian species, the Macedonian crested newt, Triturus macedonicus, in Northern Pindos National Park (Epirus, Greece). Mitochondrial DNA sequences and multilocus genotypes of individuals from 38 breeding sites were used to infer their phylogenetic position and to detect and measure genetic variation patterns, population genetic structure, and levels of gene flow. The examined individuals fell within two major clades of the Macedonian crested newt phylogeny, being geographically separated by the Aoos River valley and Vikos Gorge. Both groups constitute separate gene pools, bearing private haplotypes and alleles, and the groups were found to be highly differentiated in both their mitochondrial and microsatellite markers. Thus, they meet all of the criteria needed to be characterized as evolutionary significant units (ESUs) that deserve a separate conservation status. Within each ESU, the particularly high inter-population differentiation and low migration rates imply a lack of connectivity between breeding sites and local isolation in favorable habitats. Our results can inform future programs and actions towards the conservation and management of the Macedonian crested newt in the area.
1. Introduction
Conservation genetics encompasses the management of natural populations and aims to retain high levels of genetic diversity and minimize inbreeding, which reduces the risk of population and species extinction [1]. The successful conservation of a species takes into consideration its evolutionary potential by resolving taxonomic uncertainties and delineating conservation units [2]. Phylogeographic and population genetic analyses, apart from them significantly contributing to the resolution of taxonomic uncertainties, can also reveal areas of intraspecific diversity that are important for the conservation of a species [3]. By recognizing and delineating conservation units, such as evolutionary significant units (ESUs) and management units (MUs) [4,5,6], the evolutionary context of a species can be effectively utilized in the implementation of targeted conservation actions [2].
Amphibians are often targeted for conservation and management programs because of worldwide population declines [7,8]. Amphibians are particularly vulnerable to climate change and are likely to face serious survival problems; for example, 41% of species have been included in the IUCN Red List and are estimated to face some form of population decline in the future [9]. Especially, pond-breeding amphibians face severe declines and extirpations due to extensive droughts and extreme temperatures that are caused by climate change, as water availability is linked to their life cycle and survival [10,11,12]. In the Mediterranean region, due to the extended periods of high temperatures and droughts [13], amphibians suffer local population extinctions and geographical shifts [14]. Functional connectivity, conducted through migration, is a process of major importance with respect to the maintenance of the structure, development, and longevity of amphibians’ spatially structured population and metapopulation systems [15,16,17,18]. Locally isolated populations are also at risk of further loss of genetic variation, genetic degradation due to random genetic drift, inbreeding, and reduced population fitness [19,20,21,22]. Local isolation and loss of functional connectivity are crucial factors that affect the viability of pond-breeding amphibian populations and should be seriously considered in the design and implementation of effective conservation actions [23].
Among Mediterranean biodiversity hotspots, Northern Pindos National Park (Epirus prefecture, NW Greece) covers an area of 197,010 hectares of high landscape and geological complexity and hosts a large number of species of fauna and flora [24]. Within the national park, numerous natural and artificial ponds constitute ideal breeding sites for several amphibian species [25]. Among them, the Macedonian crested newt, Triturus macedonicus (Karaman, 1922), is an endemic salamandrid of the Balkan Peninsula [26,27,28,29] that shows a fragmented distribution; this is because it is usually associated with permanent ponds with rich aquatic vegetation [26] and prefers wetter conditions compared with its related species [28,30]. Previous phylogenetic analyses recognized three distinct mitochondrial clades with relatively clear geographic distributions [31], all of which are represented in Greece: Clade 1, located in the northern distribution of the species, includes parts of northern Greece and Corfu Island; Clade 2 is located in a geographically restricted area in northern Greece and southern Albania; and Clade 3 is located in the eastern part of the species distribution in Greece [31]. The distribution of species local breeding populations within the national park is highly fragmented and divided by an uninhabited area that is bounded by the Aoos River valley and Vikos Gorge [25]. However, the phylogenetic position of these populations has not yet been assessed.
As a member of the Triturus cristatus complex, the Macedonian crested newt is included in Annex II of the Directive 92/43/EEC as a species of Community interest, whose conservation requires the establishment of special conservation areas (SCAs); meanwhile, a European action plan has been drawn up (Action Plan for the Conservation of the Crested Newt Species Complex in Europe) for its conservation and protection [32]. Triturus macedonicus is currently designated as “Vulnerable” (VU), as its populations are decreasing due to the loss of its aquatic habitats through a combination of factors such as agricultural and livestock activities and the introduction of predatory fish [33].
Our study sought to assess the conservation status of the Macedonian crested newt inhabiting a protected area in the southern limit of its distribution. Our findings are expected to advance the current knowledge on the conservation requirements of the species in the area and may inform future conservation strategies. We employed two types of molecular markers, mtDNA haplotypes and multilocus microsatellite genotypes, to (a) complement the phylogeny of the species by resolving the phylogenetic position of the populations inhabiting Northern Pindos National Park; (b) infer the genetic conservation status of populations by assessing levels of genetic diversity, population structure, and connectivity through gene flow; and (c) address management priorities for the conservation of the species and its habitats in the area. We hypothesize that breeding individuals on the opposite sides of the major geographic barriers (Aoos River valley and Vikos Gorge) might constitute distinct and highly differentiated gene pools.
2. Materials and Methods
2.1. Laboratory Procedures
Tissue samples (toe clips and/or tail tips) from 309 individuals were collected from 38 water bodies in Northern Pindos National Park during April–July 2021. Additionally, seven samples collected in earlier samplings from sites outside of the national park were included in the subsequent genetic analyses (Table S1), resulting in a total of 316 samples. Total genomic DNA was extracted using the Macherey-Nagel NucleoSpin DNA Extraction kit following the manufacturer’s protocol.
For the mtDNA analysis, a partial fragment of the mitochondrial subunit 4 of the NADH dehydrogenase gene complex gene (ND4) was amplified for 86 samples using the primer combination KARF4 (AGC-GCC-TGT-CGC-CGG-GTC-AAT-A) and DOBR2 (GTG-TTT-CAT-AAC-TCT-TCT-TGG-T) [34]. Reactions were performed in a final volume of 20 µL containing 1X Taq buffer contained 1.5 mM of MgCl2, 0.4 µL of 10 mM of dNTPs, 2U Taq polymerase, 1 µL of each primer (10 µM), and 30 ng of DNA template, and the reactions occurred under the following conditions: an initial denaturation step at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 58 °C for 45 s, extension at 72 °C for 60 s, and a final extension step at 72 °C for 5 min. The PCR products were purified according to the NucleoSpin PCR clean-up kit following the manufacturer’s protocol. A Sanger sequencing of amplicons (~750 bp) was performed on an ABI 3730XL sequencer (Applied Biosystems) by CeMIA (Cellular and Molecular Immunological Applications, Larissa, Greece).
Sequences were edited in MEGA X software [35] and aligned using the ClustalW algorithm [36]. A dataset was created that consisted of 158 available homologous sequences from GenBank for the ND4 fragment analysis (244 mtDNA sequences, 658 bp) (Table S1), with five additional outgroups from related newt species: Triturus carnifex (KT364610 [37]), Triturus ivanbureschi (KT364629 [38]), Triturus marmoratus (KY762259 [39]), Pleurodeles waltl (KY938490 [40]), and Pleurodeles poireti (KX577053 [41]).
For the microsatellite analysis, 304 samples from 39 breeding sites were genotyped at 7 microsatellite loci, which were developed and tested specifically for the Macedonian crested newt in a previous study [42] (Table S2). Genetic loci were amplified by the PCR technique using forward primers labeled with fluorescent dyes (FAM, ROX, and HEX) in two multiplex reactions. Each reaction had a final volume of 12.5 µL, containing 15–20 ng of genomic DNA, 0.2 µM of each primer, 6.25 µL of KAPA2G Mix (Kapa Biosystems), and ddH2O. The PCR temperature profile consisted of an initial denaturation step at 95 °C for 3 min, 30 cycles of 15 s at 95 °C (denaturation-denaturation), 30 s at 60 °C (hybridization-annealing), 30 s at 72 °C (extension-extension), and a final extension step at 72 °C for 10 min. Genotyping of the seven microsatellite loci (Genescan 500 LIZ, Applied Biosystems) was performed using an ABI 3730XL sequencer (Applied Biosystems, CeMIA-Cellular and Molecular Immunological Applications, Larissa, Greece). Genotype scoring was performed using STRand ver. 2.4.110 [43].
2.2. Mitochondrial DNA Analysis
2.2.1. Phylogeography
Mitochondrial haplotypes were identified in DnaSP ver.6.12 [44]. We constructed an intraspecific phylogeny while following three different approaches: (a) maximum likelihood estimation (ML) [45], (b) Bayesian inference (BI) [46], and (c) neighbor joining (NJ) [47]. The general time-reversible (GTR) model [48] with corrections for invariant sites (+I) was selected as the best-fit nucleotide substitution model based on the Bayesian information criterion (BIC) [49] for both the ML and BI approaches, which was incorporated using the Smart Model Selection software ver.1.8.4 (SMS) [50] implemented in the online version of the PhyML 3.0 program [51]. The ML haplotype tree was constructed in the online version of PhyML 3.0, and the robustness of the topology was evaluated by 1000 bootstrap replications. The BI haplotype tree was constructed in MrBayes ver.3.2.2 [52], where four MCMC chains ran for 2 × 106 generations and were sampled every 100 generations; the first 25% of the generated trees were discarded as the burn-in. For tree visualization, we used FigTree ver.1.4.4 [53]. The NJ haplotype tree was constructed using MEGA X based on the p-distance matrix using 10,000 bootstrap replications to assess branch statistical support. To visualize the relationships among the detected haplotypes, a median-joining (MJ) network [54] was constructed using PopArt ver.1.7 [55].
2.2.2. Historical Demography
To investigate the demographic history of the species in the national park, a mismatch distribution analysis [56] was performed in DnaSP software for the clades with a sufficient number of sequences. Three parameters were estimated for N0 and N1 (the initial population and the post-expansion population, respectively): τ, which refers to the time estimated from the beginning of the expansion in units of mutational time (τ = 2μt, where μ is the mutation rate of the genetic locus, and t is the number of generations since the beginning of the expansion), and θ0 and θ1, which refer to the size of the population before and after the expansion, respectively (θ0 = 2N0μ, and θ1 = 2N1μ) [57]. Additionally, the goodness-of-fit and sudden expansion models were evaluated by 100,000 bootstrap replications. The sum of squared deviations (SSDs) between the observed and expected distributions was used as a statistical control, and for p > 0.05, the sudden expansion model can be accepted [56]. Furthermore, Tajima’s D [58] and Fu’s Fs [59] tests for departure from neutrality were also estimated in Arlequin ver. 3.5 [57].
2.2.3. Genetic Diversity
For each identified clade, the nucleotide (Pi) and haplotype (Hd) diversity were estimated in DnaSP. Pairwise genetic distances (p-distance) within and between clades, as well as between detected clades, were estimated using MEGA X using 1000 bootstraps.
2.3. Microsatellite Analysis
Individual genotypes were checked for selective amplification due to allele dropout and the presence of null alleles in Micro-Checker ver. 2.2.3 (CI = 99%, 4000 iterations of MCMC–Monte Carlo Markov Chains); we only performed this check for sites with a sample size of greater than six individuals as Micro-Checker only considers the sites with a sample size of greater than six individuals in order to achieve an accurate dataset that is representative of the entire study area [60].
The deviation from the Hardy–Weinberg equilibrium was tested through the inbreeding coefficient (FIS) per genetic locus and per breeding site (120,000 randomizations) using FSTAT ver. 2.9.4 [61]. Linkage disequilibrium tests were performed using GenePop on the Web (10,000 iterations of MCMC) [62].
2.3.1. Population Structure and Gene Flow
The genetic population structure of all breeding populations in the study area was investigated using a Bayesian inference method in Structure ver. 2.3.4. [63]. Breeding sites with a small sample size (fewer than six individuals) were included to increase the representability of the area. Initially, to achieve unbiasedness in the model, parameter λ was calculated assuming that all sampled genotypes belong to one gene pool [64]; setting K = 1 with ten iterations; and using the “admixture model”, “correlated allele frequencies”, and LOCPRIOR (500,000 MCMC iterations, burn-in period of 250,000 iterations). The estimated λ was fixed for the following analysis using the same priors as before. To assess the optimal number of gene pools, ΔK was calculated using Structure Harvester [65] following the method of [66]. An analysis of molecular variance (AMOVA) [67] was conducted using Arlequin to examine the distribution of genetic variation at three hierarchical levels: among the two geographic areas (eastern park—E.P.; western park—W.P.), among sites within areas, and within sites. The significance of the variance components was tested using 30,000 permutations.
To infer the gene flow rates among breeding sites, the rate of recent migration rates (i.e., during the last three generations) was calculated using BayesAss ver. 1.3 [68] with 3 × 106 MCMC iterations, a burn-in period of 106 iterations, and a sampling frequency of 2000 iterations. The mixing parameters were adjusted to achieve an acceptable ratio ranging between 0.4 and 0.6 for the average number of changes per total iterations [69]. The delta (Δ) values were set to 0.3, 0.2, and 0.65 for allele frequency (deltaP), migration rate (deltam), and inbreeding (deltaF), respectively. The 95% confidence intervals of the results obtained from the dataset were compared with the 95% confidence intervals for cases with insufficient data [70].
2.3.2. Genetic Diversity, Differentiation, and Isolation by Distance
Genetic diversity indices based on microsatellite data (average number of alleles per locus (A), number of private alleles (P), expected heterozygosity (He), and observed heterozygosity (Ho) were calculated in GenAlEx ver. 6.5 [71]. The Allelic richness (Ar) and Global FST or θ index (CI = 95%, 15,000 permutations) [72] were calculated using FSTAT. The pairwise FST was calculated in FSTAT, and its significance was tested by 10,000 permutations in Arlequin. The effective population size (Ne) of the gene pools was estimated using the program NeEstimator version 2.1 [73] using the molecular coancestry method [74].
For the entire park, and for each of the two geographic groups of breeding sites (W.P. and E.P.), the correlation between two matrices, a genetic distance matrix (FST/1 − FST), and a Euclidean geographic distance matrix (log km), generated by the dist() command in R software [75], was calculated using the Mantel test [76] (CI = 95%, 10,000 permutations) in GenAlEx.
3. Results
3.1. Mitochondrial DNA
3.1.1. Phylogeography and Historical Demography
Within the entire species distribution, the phylogenetic analysis identified 34 haplotypes. Four new haplotypes (Tmac31, Tmac32, Tmac33, and Tmac34) were added to the existing knowledge [27,31]. Out of 658 nucleotide positions aligned, 583 were conserved and 75 were variable, all of which were coding for ND4.
Identical tree topologies were estimated by all of the different approaches (ML, BI, and NJ). However, some of the nodes are not highly supported for the ML and NJ methods (Figure 1b). Our phylogenetic analysis identified three main clades that differed by a substantial number of mutations (13–17 steps) while converging on an ancestral haplotype.
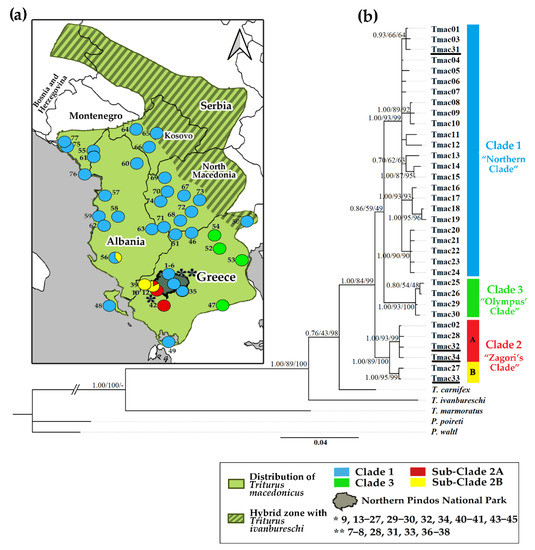
Figure 1.
(a) Geographic distribution of Triturus macedonicus (based on [31]). Location numbers correspond to Table S1. (b) Bayesian phylogeny of T. macedonicus haplotypes and five outgroups based on a 658 bp fragment of the mtDNA gene ND4. Numbers along internal branches represent posterior probability of Bayesian inference (left) and bootstrap support for maximum likelihood (middle) and neighbor joining (right). The coloration of clades follows [31]. Newfound haplotypes (Tmac31, Tmac32, Tmac33, and Tmac34) are underlined.
Clade 1 (“Northern Clade”) consists of the individuals found mainly in the Northern distribution of the species, Corfu Island (Tmac16, 17, 18, 19) and Amvrakikos Gulf (Tmac13) (Figure 1 and Figure A1). It includes 24 haplotypes that constitute an extensive complex mutational network, which originate from a central ancestral haplotype (Tmac04) (Figure A1). Clade 2 (“Zagori’s Clade”) includes six haplotypes and is further divided into two subclades. Subclade 2A includes individuals from Central Zagori and Ioannina Basin and shows a simpler typical star-like network, where some private haplotypes derive from a high-frequency ancestral haplotype (Tmac02) with one mutational step (Figure 1 and Figure A1). Sub-Clade 2B includes individuals from the northwestern border of the national park (Tmac33) up to southern Albania (Tmac27) (Figure 1 and Figure A1). Finally, Clade 3 (“Olympus’ Clade”) includes individuals from the eastern distribution of the species around Mount Olympus (Figure 1 and Figure A1). The clade includes four haplotypes and shows a reticulation structure in which haplotypes are equidistant by one mutation step (Figure A1).
The examined T. macedonicus individuals in the national park fall within Clades 1 and 2. All individuals from the eastern part of the park belong to Clade 1 (“Northern Clade”), while all individuals from the western part belong to Clade 2 (“Zagori’s Clade”).
The sudden expansion model appears to be supported for both Clades 1, 2, and Subclade 2A (Figure S1 and Table S3). The expansion time, represented by τ, was longer for Clade 1 than for the others. The θ0 value was approximately zero for all clades, indicating the possible existence of a bottleneck event prior to expansion. Regarding the neutrality tests, the D value was negative and statistically significant only for Clade 1 and Sub-Clade 2A (p = 0.047 and p = 0.010, respectively), indicating population growth after a bottleneck event [58]. The Fs value was negative and statistically significant only for Sub-Clade 2A (p = 0.000), indicating an excess number of haplotypes due to recent growth [59] or genetic hitchhiking [77]. Clade 2 showed a negative D value, whereas a positive Fs value reflected a deficiency in haplotypes due to a recent bottleneck event [59]. Finally, the available data (nine sequences, two haplotypes) were insufficient for conducting a demographic analysis of Sub-Clade 2B.
3.1.2. Genetic Diversity
Clade 1 had the highest haplotype (Hd = 0.820) and nucleotide diversity (Pi = 0.00622). Between the two Sub-Clades 2A and 2B, Subclade 2B showed a higher haplotype (Hd = 0.389) and nucleotide diversity (Pi = 0.00059) than Subclade 2A (Hd = 0.103, Pi = 0.00016) (Table S4).
The genetic distance (p-distance) between and within the clades and sub-clades of T. macedonicus and the related species T. carnifex and T. ivanbureschi is shown in Table 1. The greatest distance was observed between Subclade 2A and T. ivanbureschi. Within the T. macedonicus lineages, the largest distance was observed between Subclade 2A and Clade 3 (3.3%), whereas the smallest distance was observed between Subclades 2A and 2B (1%). The genetic distance between Clades 1 and 2 was 2.8%.

Table 1.
p-distance (%) within and between clades and sub-clades of Triturus macedonicus and two sister Triturus species. Below diagonal: mean % p-distance between clades and sub-clades; Above diagonal: standard error estimates (after 1000 bootstrap replicates); Diagonal: % p-distance within clades and sub-clades with respective S.E. in parenthesis.
Within the national park, Clade 1 was represented by only three haplotypes. Similarly, Clade 2 was represented by three haplotypes. One dominant haplotype for each clade was identified: Tmac01 for Clade 1 and Tmac02 for Clade 2 (Figure 2).
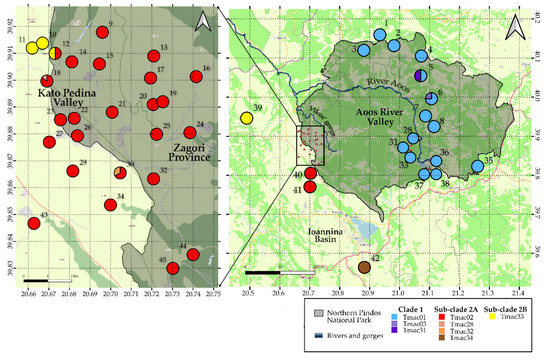
Figure 2.
Spatial distribution of haplotype diversity in Northern Pindos National Park and the surrounding area. Location numbers correspond to Table S1.
Out of the new haplotypes found, Tmac31 was a private haplotype in site 5 and Tmac32 in site 30, whereas Tmac03 within the park was only detected once in site 6. The Tmac28 haplotype, which is a private haplotype of site 18 [27], was not recorded.
3.2. Microsatellite Analysis
All microsatellite loci were polymorphic in all studied breeding sites. The Micro-Checker results indicated the possible presence of null alleles at the TCM-83 locus in sites 27 and 30. For the remaining sites, this locus showed no evidence of null alleles, and the data were included in further analyses. No linkage disequilibrium (LD) was detected (p > 0.05) for all pairs of loci in all sites, while the inbreeding coefficient FIS ranged from −0.139 (site 24) to 0.198 (site 11) (Table S5); however, heterozygote deficiency and excess were not statistically significant (p < 0.00036). Specifically, the number of alleles per locus ranged from 2 (for TCM-83) to 18 (for TCM-517) (total of 47 alleles) (Table S2).
3.2.1. Population Structure and Gene Flow
The results obtained by the Evanno method showed the highest ΔK value (2672.3), which corresponded to K = 2; the second highest ΔK value (71.1) for K = 3 was considerably lower, suggesting that the optimal number of gene pools is two. The Bayesian clustering analysis of all individual genotypes indicates a clear population genetic structure, with extremely low admixture between the two distinct gene pools. Gene pool 1 (blue) consists of individuals from sites located east of the Aoos River valley (E.P.), while gene pool 2 (red) consists of individuals from sites located west of the valley (W.P.) (Figure 3). The AMOVA analysis revealed significant genetic differentiation between the two geographic areas (% variation = 17.82, FST = 0.23, p = 0.0000) (Table S6). The genetic differentiation among sites within areas (% variation= 5.16, FSC = 0.62, p = 0.0000) and within sites (% variation= 77.02, FCT = 0.18, p = 0.0000) was also significant (Table S6).
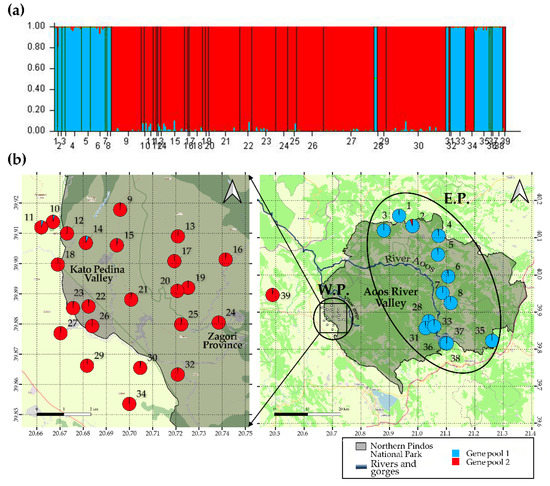
Figure 3.
Genetic population structure of Triturus macedonicus in Northern Pindos National Park: (a) STRUCTURE barplot showing two gene pools, comprising breeding sites from the eastern and western parts of the national park, respectively, with extremely low admixture between them. Vertical bars represent single individuals with their site of origin on the x-axis and the respective coefficients of membership on y-axis. (b) Geographic location and membership of each site in the two identified gene pools. Pie diagrams represent the average coefficient of membership of each site. E.P.: “Eastern Park” group; W.P.: “Western Park” group.
When simulating the effect of having insufficient data for migration rates estimation, we obtained values of 0.675–0.992 (CI = 95%) for the non-migration rates (proportions of individuals originating from the “source” population of each generation) and 1.43 × 10−19–0.0933 (CI = 95%) for the immigration rates. For the non-migration rates, the confidence intervals that were recovered from the full dataset were significantly smaller than those obtained from the null hypothesis (insufficient data), suggesting that the dataset contained useful information to support the results [68]. For the entire dataset, only seven sites exhibited significant migration rates (Table S7). Sites 27, 9, and 4 appear to be “source” populations, with site 27 being the most prominent, showing migration rates to four other sites (18, 21, 23, 26). The highest migration rate (0.172) was found between “source” population 27 and site 26. Sites 4 and 9 showed significant migration rates to sites 5, 35, and 21, respectively (Table S7).
3.2.2. Genetic Diversity and Isolation by Distance
The mean number of alleles per locus (A) was estimated to be 4.9 in gene pool 1 and 4.4 in gene pool 2 (Table 2). In total, 25 private alleles were identified, 14 of which were private for gene pool 1 and 11 of which were private for gene pool 2 (Table 2 and Table S8). The allelic richness (Ar) significantly differed, with a mean value of 3.0 for gene pool 1 and 1.9 for gene pool 2 (Table 2). The expected (He) and observed (Ho) heterozygosity was estimated to be 51.2% and 46.4% for gene pool 1 and 56.8% and 50.8% for gGene pool 2, respectively (Table 2). On the breeding site level, the mean number of alleles per locus ranged from 2.6 in site 25 to 4.0 in site 30 (Table S9). Two private alleles were identified: a 264 bp allele at the TCM-517 locus, private to site 4, and a 184 bp allele at the TCM-496 locus, private to site 38 (Table S9). The expected heterozygosity ranged from 40.4% to 56.3%, with a mean value of 48.35%. Accordingly, the observed heterozygosity ranged from 39% to 61.2%, with a mean value of 50.1%. The intra-population genetic diversity significantly differed between the breeding sites; the highest was observed in site 22 (Ar = 3.4, He = 0.537), and the lowest was observed in site 6 (Ar = 2.6, He = 0.404) (Table S9). Furthermore, the sites with the lowest genetic diversity were 6, 9, 25, 35, and 38 (Table S9).

Table 2.
Indices of genetic diversity per detected gene pool. Listed in order are: the sample size (n); mean number of alleles per locus (A); corrected allelic richness (Ar); number of private alleles (Ap); expected (He) and observed (Ho) heterozygosity, with corresponding ± SE in parentheses; and effective population size (Ne), with the 95% CI in parenthesis.
The overall (global) FST value in the national park was 0.131 (95% CI = 0.092–0.159), and the pairwise FST values ranged from 0 to 0.332 (76% of the comparisons were statistically significant).
The mean FST between the two genetic groups was equal to 0.184 (95% CI = 0.092–0.257) while the within genetic group FST was 0.091 (0.064–0.126) and 0.055 (0.038–0.075) for the eastern and the western group, respectively. There were 50% (18/36) and 59.2% (116/196) of the pairwise FSTs in the eastern and western genetic groups, respectively, that were found to be statistically significant and relatively high (0.05–0.23 and 0.02–0.19, respectively) (Figure S2).
The effective population sizes were estimated using the molecular coancestry method to be 290.2 (95% CI= 0.3–1456.7) for gene pool 1 (E.P.) and 37.9 (95% CI = 0.0–190.0) for gene pool 2 (W.P.) (Table 2).
No indication of IBD was found in each of the two population groups: W.P.: r = 0.185, p = 0.23, R2 = 0.034; E.P.: r = 0.209, p = 0.117, R2 = 0.044. In contrast, the entirety of breeding sites in the study area follows the pattern of IBD (r = 0.81, p = 0.0001, R2 = 0.655).
4. Discussion
Our multimarker approach on the phylogeography and population genetic structure of the Macedonian crested newt within the biodiversity hot spot of Northern Pindos National Park adds new insights to the current phylogeny of the species as well as its conservation status in the area.
4.1. Identification and Characterization of ESUs
The phylogenetic reconstruction coincides with the one proposed by [31], which confirms the existence of the three major clades (Clade 1 or “Northern Clade”, Clade 2 or “Zagori’s Clade”, and Clade 3 or “Olympus’ Clade”) but differs in the basal topology. Contrary to [31], our analysis seems to support the basal bifurcation between Clade 2 and the ancestor of the Clade 1 and Clade 3 branches (Figure 1b). However, the distinction between Clade 1 and Clade 3, although suggested by all three methods, is not as clear. The same basal node is also low supported in [31]. Moreover, our analysis revealed the existence of two subclades within Clade 2 (Figure 1b). Our dense sampling in the study area and the four new haplotypes that were identified, three of which were recovered in the Zagori’s Clade (Tmac32, 33, and 34; Figure 1b), might explain the finer resolution of species phylogeny. The degree of genetic differentiation between the three clades is considerably high (Table 1). Similar levels of genetic differentiation have been observed among recognized subspecies of the alpine newt (Ichthyosaura alpestris) [78], raising the question of whether the three Macedonian crested newt lineages might deserve a subspecific designation despite the cytonuclear discordance that has been observed for the species [31]. In any case, sequencing of larger genome fragments and the analysis of additional genomic markers is expected to reconstruct the phylogenetic history of lineages with greater accuracy and fidelity (e.g., [79]).
The combination of high haplotype and low nucleotide diversity that was observed for both the entire species and the individual mitochondrial clades (Table S4) might reflect rapid demographic growth from an initial small source population [80]. However, the recent expansion model is not supported for all the clades. Specifically, Tajima’s D and Fu’s Fs tests did not support the recent expansion model for Clade 2, whereas Tajima’s D test did support the recent expansion of Clade 1. Clade 1 seems to have been expanded from an already identified refugium in the western coasts of Albania [27,28,31]. In contrast, Clade 2 shows long demographic stability, and the populations probably survived in an independent refugium during the Last Glacial Maximum. Central Zagori was a suitable area that was absent of glaciers compared with the rest of the Northern Pindos area [81,82]. However, the recent expansion model is supported for Sub-Clade 2A. This suggests that Sub-clade 2A individuals might have recently expanded across the Zagori Province and Ioannina Basin (Figure 1 and Figure 2) after a bottleneck, which was probably caused by human intervention in the water table of the area [83]. Such an expansion could have been facilitated by the existence of a dense network of permanent artificial ponds in the area, as has been the case in the close relative Triturus cristatus [84,85].
All Triturus macedonicus populations from the Northern Pindos National Park belong to two previously described phylogenetic clades. Sites from the eastern and northern parts of the national park constitute marginal populations at the southern periphery of the geographically extended northern lineage (Clade 1), while sites from the western parts of the national park belong to the geographically more localized Zagori lineage (Clade 2) (Figure 2).
These two population groups are geographically separated by physical features such as the Aoos River valley and Vikos Gorge (Figure 2 and Figure 3), which appear to have been insurmountable barriers for the gene flow. Such a long-lasting physical separation could explain the observed high differentiation in both mitochondrial and nuclear markers (2.8% p-distance and 0.184 FST) and the formation of two independent gene pools with complete or extremely low admixture between them (Figure 2 and Figure 3). Moreover, the detection of several private alleles in each gene pool indicates separate evolutionary histories in isolation [86].
Therefore, as these two population groups were found to be reciprocally monophyletic at the mtDNA level, differ considerably in their allele frequencies, and show unique genetic characteristics and diagnostic genetic diversity, they meet the criteria to be characterized as separate evolutionary significant units (ESUs), with particular importance for targeted conservation [4,5,6].
4.2. Genetic Variation and Population Connectivity
The overall genetic diversity of the Macedonian crested newt in the study area was found to be lower than that observed in other studies of closely related species at various spatial scales [87,88,89,90].
Although the studied breeding sites comprise two homogeneous gene pools, significant migration within each of them was detected on a few occasions. Only seven cases were observed: two in the eastern group and five in the western one. This can be indicative of reduced functional connectivity between breeding sites or might reflect a poor performance of the Bayesian approach that was used due to uneven sample sizes; thus, this observation should be treated with caution. However, 50% (18/36) and 59.2% (116/196) of the pairwise FSTs in the eastern and western genetic group, respectively, were found to be statistically significant and relatively high (0.05–0.23 and 0.02–0.19, respectively), having been higher than those found in similar studies [87,89,90,91]. Such levels of differentiation, independent of the geographic distance between sites (absence of IBD), implies local isolation and differentiation due to drift [1,20]. The quite low Ne estimates in both geographic areas, being particularly low in the western one in spite of the larger number of sampled individuals, can be considered as indicative of a strong drift [92].
The significant differentiation among breeding sites of the eastern population group, the most prominent being between the sites residing in the northern and southern parts of the E.P., could be attributed to their geographic isolation, as the dispersal ability of Triturus newts is much lower than the between breeding sites geographic distance [85,93].
Although individuals from the western group occupy a smaller geographic area and breed in a dense network of artificial ponds, significant genetic differentiation and low dispersal between sites have been detected, which was probably due to the denser network of paved and dirt roads that might act as barriers to gene flow [94], promoting local differentiation even on a small scale (e.g., sites 15 and 34; Figure 3 and Figure S2). The negative effects of road networks on amphibian dispersal have been addressed, such as the impact of road traffic on annual migration routes [95] and the high heat storage on paved surfaces, which poses a high risk of dehydration for amphibians [96]. The negative impact of the road network on the population structure of Lissotriton graecus in the study area has been previously addressed [89]. However, a more detailed landscape genetic analysis in the area could locate these barriers to gene flow more concretely.
4.3. Conservation Status and Management Priorities
Our results on the levels of differentiation (Figure S2), AMOVA (Table S6), and effective population size estimates (Table 2) indicate the local isolation of breeding sites and genetic differentiation due to drift in both geographic areas. Apart from the negative impact of drift on the genetic variation levels [1], the lack of functional connectivity makes local populations susceptible to alterations of their local habitat conditions [20].
The individuals of the two genetic groups exhibit contrasting habitat preferences (Table S1). Individuals from the eastern population mostly occupy natural water bodies, while individuals from the western population inhabit almost exclusively artificial cattle ponds (Table S1). Artificial ponds provide stable habitats, and their construction has been proved successful in boosting amphibian populations in the past [97,98,99,100,101]; their potential for conservation of crested newts has been demonstrated in situ [85]. In the neighboring area of the Ioannina Basin, agricultural and livestock ponds have been identified as ideal habitats for newts to thrive [102].
Although artificial cattle ponds might seem stable enough (stable hydroperiod) and could potentially serve as effective breeding sites for numerous taxa, they simultaneously proved vulnerable to significant pressures locally. For example, fish introductions (releases) as well as high mechanical and chemical disturbances from cattle activity have been confirmed in the study area [25]. Such pressures negatively affect artificial ponds by altering and downgrading habitat quality, with subsequent negative effects on aquatic communities [103]. Introduced fish often prey on eggs or disrupt the spawning sites of newts and other aquatic breeders, reducing their evolutionary potential [103,104,105,106]. In the neighboring Ioannina Basin, the presence of invasive fish in suburban ponds has been found to be responsible for the absence of the species from these sites [102]. At the same time, the transition of traditional livestock practices (i.e., sheep farming) to the more “commercial” or financially profitable cattle farming has caused high mechanical and chemical disturbance of breeding sites in the study area [25]. Other newt species, such as Triturus dobrogicus in Romania [107] and Lissotriton lantzi in Georgia [108], have been found to systematically avoid such problematic breeding environments. Within the study area, two robust and diverse breeding sites of the western genetic group, which served as a source to many others in a metapopulation context some 15 years ago [89], were extensively pressured to extinction due to the severe degradation of their habitat by both practices [25].
Certain management measures that would preserve and secure the quality and stability of the aquatic habitat of T. macedonicus, particularly in the western part of the national park, should implement fish removals, fencing of ponds, and pond restoration [109,110,111,112]. Additionally, pond construction could facilitate migration and dispersal movements, increasing the functional connectivity of local breeding sites [85]. The maintenance of old artificial ponds and the establishment of new ones in the study area is expected to facilitate the metapopulation networks of Clade 2 and reinforce the presence of Clade 1 populations through natural colonization by individuals over a few years [85]. At the same time, the aforementioned measures are anticipated to have a positive impact on agriculture and livestock farming [113].
Management actions should also be aimed at peripheral populations, such as sites 27 and 9 in the western group and site 4 in the eastern group, as these are found to be the most important sources of gene flow (Table S7). Peripheral populations are often characterized as genetic diversity hotspots [114,115,116,117] and have been the focus of numerous conservation programs [118,119,120].
Because T. macedonicus is considered as an “umbrella” species [121], the preservation and management of its habitats, apart from being beneficial for the breeding populations of the species, is expected to benefit a wide spectrum of aquatic breeders as well, which would thus preserve biodiversity in the hotspot that is Northern Pindos National Park. The coordination and cooperation of several public agencies are required to conserve each ESU and maintain the evolutionary continuum of the species [122,123].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d15050671/s1: Figure S1. Mismatch distributions and neutrality tests for Clades 1, 2, and Sub-Clade 2A; Figure S2. Heatmap of pairwise FST values among the analyzed sites (n ≥ 6, Table S9) within the (a) Eastern and (b) Western genetic group; Table S1. Sample localities of Triturus macedonicus used in this study; Table S2. Primer pairs used in this study for the amplification of seven microsatellite loci; Table S3. Mismatch analysis and neutrality tests for clades and sub-clades with a sufficient number of sequences (Clades 1 and 2, Sub-Clade 2A); Table S4. Distribution of haplotypes and diversity measures of each clade and sub-clade identified in the phylogenetic analysis; Table S5. Inbreeding coefficient (FIS) and percentage of randomizations that gave smaller (FIS 95% CI+) or larger (FIS 95% CI−) values than observed; Table S6. Hierarchical analysis of molecular variance (AMOVA) among the two geographic areas in the park; Table S7. Posterior distributions of recent migration rates among breeding sites; Table S8. List of private alleles detected in each of the two gene pools; Table S9. Indices of genetic diversity per analyzed breeding site [124,125].
Author Contributions
Conceptualization, K.S. and A.B.; methodology, K.S. and A.B.; software, T.D. and A.T.; validation, K.S.; formal analysis, T.D., A.T. and K.S.; investigation, T.D., A.T., E.-A.T., A.B., K.S. and A.K.; resources, K.S. and A.K.; data curation, T.D. and A.T.; writing—original draft preparation, T.D. and A.T.; writing—review and editing, K.S., E.-A.T., A.B., A.K., T.D. and A.T.; visualization, T.D. and A.T.; supervision, K.S.; project administration, K.S.; funding acquisition, K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Management Agency of Northern Pindos National Park/ΥΜΕΠΕΡAA Operational Programme Transport Infrastructure, Environment and Sustainable Development MIS 5033216.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of the Ministry of Environment and Energy of Greece (28162/962, 25 April 2021).
Data Availability Statement
The novel sequences obtained in the present study are openly available in GenBank (Accession numbers: OQ877047—OQ877050; Table S1). The multilocus genotypic data produced in this study are available on request from the corresponding author. The genotypic data are not publicly available due to the restrictions imposed by the project execution contract.
Acknowledgments
We thank the personnel of the Management Unit of Northern Pindos National Park for assisting in the field; Ben Wielstra and three anonymous reviewers provided insightful comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had a role in the decision to publish the results.
Appendix A
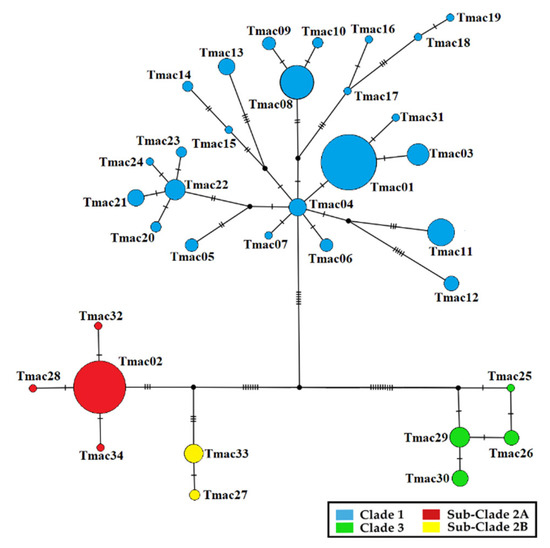
Figure A1.
Median-joining haplotype network for T. macedonicus based on a 658 bp fragment of the mtDNA gene ND4. The coloration of clades follows [31] and Figure 1.
References
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics, 2nd ed.; Cambridge University Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Byrne, M. Phylogenetics and the Conservation of a Diverse and Ancient Flora. Comptes Rendus Biol. 2003, 326, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Canestrelli, D.; Bisconti, R.; Sacco, F.; Nascetti, G. What Triggers the Rising of an Intraspecific Biodiversity Hotspot? Hints from the Agile Frog. Sci. Rep. 2014, 4, 5042. [Google Scholar] [CrossRef] [PubMed]
- Ryder, O.A. Species Conservation and Systematics: The Dilemma of Subspecies. Trends Ecol. Evol. 1986, 1, 9–10. [Google Scholar] [CrossRef]
- Waples, R.S. Pacific Salmon, Oncorhynchus Spp. & the Definition of ‘Species’ under the Endangered Species Act. Mar. Fish. Rev. 1991, 53, 11–12. [Google Scholar]
- Moritz, C. Defining ‘Evolutionarily Significant Units’ for Conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Houlahan, J.E.; Findlay, C.S.; Schmidt, B.R.; Meyer, A.H.; Kuzmin, S.L. Quantitative Evidence for Global Amphibian Population Declines. Nature 2000, 404, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Wake, D.B.; Vredenburg, V.T. Are We in the Midst of the Sixth Mass Extinction? A View from the World of Amphibians. Proc. Natl. Acad. Sci. USA 2008, 105, 11466–11473. [Google Scholar] [CrossRef] [PubMed]
- IUCN. The IUCN Red List of Threatened Species. Version 2022-2; IUCN: Gland, Switzerland, 2023. [Google Scholar]
- Erwin, K.L. Wetlands and Global Climate Change: The Role of Wetland Restoration in a Changing World. Wetl. Ecol. Manag. 2009, 17, 71–84. [Google Scholar] [CrossRef]
- Cayuela, H.; Arsovski, D.; Thirion, J.-M.; Bonnaire, E.; Pichenot, J.; Boitaud, S.; Miaud, C.; Joly, P.; Besnard, A. Demographic Responses to Weather Fluctuations Are Context Dependent in a Long-Lived Amphibian. Glob. Chang. Biol. 2016, 22, 2676–2687. [Google Scholar] [CrossRef]
- Weinbach, A.; Cayuela, H.; Grolet, O.; Besnard, A.; Joly, P. Resilience to Climate Variation in a Spatially Structured Amphibian Population. Sci. Rep. 2018, 8, 14607. [Google Scholar] [CrossRef]
- Blondel, J.; Aronson, J. Biology and Wildlife of the Mediterranean Region; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Préau, C.; Bertrand, R.; Sellier, Y.; Grandjean, F.; Isselin-Nondedeu, F. Climate Change Would Prevail over Land Use Change in Shaping the Future Distribution of Triturus marmoratus in France. Anim. Conserv. 2022, 25, 221–232. [Google Scholar] [CrossRef]
- Bowler, D.E.; Benton, T.G. Causes and Consequences of Animal Dispersal Strategies: Relating Individual Behaviour to Spatial Dynamics. Biol. Rev. 2005, 80, 205–225. [Google Scholar] [CrossRef] [PubMed]
- Ronce, O. How Does It Feel to Be Like a Rolling Stone? Ten Questions About Dispersal Evolution. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 231–253. [Google Scholar] [CrossRef]
- Tromeur, E.; Rudolf, L.; Gross, T. Impact of Dispersal on the Stability of Metapopulations. J. Theor. Biol. 2016, 392, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hanski, I.; Gaggiotti, O. Metapopulation Biology. In Ecology, Genetics and Evolution of Metapopulations; Elsevier: Amsterdam, The Netherlands, 2004; pp. 3–22. [Google Scholar]
- Wright, S. Evolution and the Genetics of Populations. Vol. 4. Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Lande, R. Risk of Population Extinction from Fixation of New Deleterious Mutations. Evolution 1994, 48, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.; Bürger, R. Mutational Meltdowns In Sexual Populations. Evolution 1995, 49, 1067–1080. [Google Scholar] [CrossRef]
- Slatkin, M. Gene Flow and Genetic Drift in a Species Subject to Frequent Local Extinctions. Theor. Popul. Biol. 1977, 12, 253–262. [Google Scholar] [CrossRef]
- Semlitsch, R.D. Differentiating Migration and Dispersal Processes for Pond-Breeding Amphibians. J. Wildl. Manag. 2008, 72, 260–267. [Google Scholar] [CrossRef]
- Griffiths, I.; Kryštufek, B.; Reed, J. Balkan Biodiversity; Springer Netherlands: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Sotiropoulos, K.; Danelis, T.; Theodoropoulos, A.; Toli, E.-A.; Bounas, A. Investigation of the Distribution and Conservation Status of the Priority Species of the 92/43 Macedonian Crested Newt (Triturus macedonicus) and Its Habitats, in the Protected Area of the Northern Pindos National Park. Deliverable B1: Project Final Report; University of Ioannina: Ioannina, Greece, 2022. (In Greek) [Google Scholar]
- Valakos, E.; Pafilis, P.; Sotiropoulos, K.; Lymberakis, P.; Maragou, P.; Foufopoulos, J. The Amphibians and Reptiles of Greece; Chimaira Editions: Frankfurt am Main, Germany, 2008. [Google Scholar]
- Wielstra, B.; Crnobrnja-Isailović, J.; Litvinchuk, S.N.; Reijnen, B.T.; Skidmore, A.K.; Sotiropoulos, K.; Toxopeus, A.G.; Tzankov, N.; Vukov, T.; Arntzen, J.W. Tracing Glacial Refugia of Triturus Newts Based on Mitochondrial DNA Phylogeography and Species Distribution Modeling. Front. Zool. Vol. 2013, 10, 13. [Google Scholar] [CrossRef]
- Wielstra, B.; Arntzen, J.W. Postglacial Species Displacement in Triturus Newts Deduced from Asymmetrically Introgressed Mitochondrial DNA and Ecological Niche Models. BMC Evol. Biol. 2012, 12, 161. [Google Scholar] [CrossRef]
- Wielstra, B.; Arntzen, J.W. Exploring the Effect of Asymmetric Mitochondrial DNA Introgression on Estimating Niche Divergence in Morphologically Cryptic Species. PLoS ONE 2014, 9, e95504. [Google Scholar] [CrossRef] [PubMed]
- Wielstra, B.; Arntzen, J.W. Unraveling the Rapid Radiation of Crested Newts (Triturus cristatus Superspecies) Using Complete Mitogenomic Sequences. BMC Evol. Biol. 2011, 11, 162. [Google Scholar] [CrossRef]
- Wielstra, B.; Arntzen, J.W. Extensive Cytonuclear Discordance in a Crested Newt from the Balkan Peninsula Glacial Refugium. Biol. J. Linn. Soc. 2020, 130, 578–585. [Google Scholar] [CrossRef]
- Edgar, P.; Bird, D.R. Action Plan for the Conservation of the Crested Newt Triturus cristatus Species Complex in Europe. In Proceedings of the Council of the European Union; Council of Europe: Strasbourg, France, 2006. [Google Scholar]
- IUCN SSC Amphibian Specialist Group. Triturus macedonicus. The IUCN Red List of Threatened Species 2022; IUCN: Gland, Switzerland, 2022. [Google Scholar]
- Arntzen, J.W.; Espregueira Themudo, G.; Wielstra, B. The Phylogeny of Crested Newts (Triturus cristatus Superspecies): Nuclear and Mitochondrial Genetic Characters Suggest a Hard Polytomy, in Line with the Paleogeography of the Centre of Origin. Contrib. Zool. 2007, 76, 261–278. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Wielstra, B.; Canestrelli, D. Next-Generation Phylogeography of the Italian Crested Newt Triturus carnifex Confirms Allopatric Glacial Refugia Positioned in Two Mediterranean Peninsulas; GenBank: KT364610 Direct Submission; Terrestrial Zoology, Naturalis Biodiversity Center, Darwinweg 2: Leiden, The Netherlands, 2015. [Google Scholar]
- Wielstra, B.; Burke, T.; Butlin, R.K.; Avcı, A.; Üzüm, N.; Bozkurt, E.; Olgun, K.; Arntzen, J.W. A Genomic Footprint Confirms Hybrid Zone Movement in Crested Newts. Evol. Lett. 2017, 1, 93–101. [Google Scholar] [CrossRef]
- Carvalho, S.B.; Velo-Antón, G.; Tarroso, P.; Portela, A.P.; Barata, M.; Carranza, S.; Moritz, C.; Possingham, H.P. Spatial Conservation Prioritization of Biodiversity Spanning the Evolutionary Continuum. Nat. Ecol. Evol. 2017, 1, 0151. [Google Scholar] [CrossRef]
- Gutiérrez-Rodríguez, J.; Barbosa, A.M.; Martínez-Solano, Í. Integrative Inference of Population History in the Ibero-Maghrebian Endemic Pleurodeles waltl (Salamandridae). Mol. Phylogenet Evol. 2017, 112, 122–137. [Google Scholar] [CrossRef]
- Escoriza, D.; Gutiérrez-Rodríguez, J.; Ben Hassine, J.; Martínez-Solano, I. Genetic Assessment of the Threatened Microendemic Pleurodeles poireti (Caudata, Salamandridae), with Molecular Evidence for Hybridization with Pleurodeles nebulosus. Conserv. Genet. 2016, 17, 1445–1458. [Google Scholar] [CrossRef]
- Sotiropoulos, K.; Tsaparis, D.; Eleftherakos, K.; Kotoulas, G.; Legakis, A.; Kasapidis, P. New Polymorphic Microsatellite Loci for the Macedonian Crested Newt, Triturus macedonicus, and Cross-Priming Testing in Four Other Crested Newt Species. Mol. Ecol. Resour. 2008, 8, 1402–1404. [Google Scholar] [CrossRef] [PubMed]
- Toonen, R.J.; Hughes, S.S. Increased Throughput for Fragment Analysis on an ABI PRISM 377 Automated Sequencer Using a Membrane Comb and STRand Software. Biotechniques 2001, 31, 1320–1324. [Google Scholar] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F.; Nielsen, R.; Bollback, J.P. Bayesian Inference of Phylogeny and Its Impact on Evolutionary Biology. Science 2001, 294, 2310–2314. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tavaré, S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. In Some Mathematical Questions in Biology—DNA Sequence Analysis; Miura, R.M., Ed.; American Mathematical Society: Providence, RI, USA, 1986; Volume 17, pp. 57–86. [Google Scholar]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. 2007. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 9 April 2023).
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT. 2005. Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 9 April 2023).
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.A.; Rannala, B. Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef]
- Faubet, P.; Waples, R.S.; Gaggiotti, O.E. Evaluating the Performance of a Multilocus Bayesian Method for the Estimation of Migration Rates. Mol. Ecol. 2007, 16, 1149–1166. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G. Nonconvergence in Bayesian Estimation of Migration Rates. Mol. Ecol. Resour. 2014, 14, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-Implementation of Software for the Estimation of Contemporary Effective Population Size (Ne) from Genetic Data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Álvarez, I.; Gutiérrez, J.P.; Royo, L.J.; Fernández, I.; Gómez, E.; Arranz, J.J.; Goyache, F. Testing the Usefulness of the Molecular Coancestry Information to Assess Genetic Relationships in Livestock Using a Set of Spanish Sheep Breeds1. J. Anim. Sci. 2005, 83, 737–744. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 9 April 2023).
- Mantel, N.; Valand, R.S. A Technique of Nonparametric Multivariate Analysis. Biometrics 1970, 26, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Fay, J.C.; Wu, C.-I. Hitchhiking Under Positive Darwinian Selection. Genetics 2000, 155, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulos, K.; Eleftherakos, K.; Džukić, G.; Kalezić, M.L.; Legakis, A.; Polymeni, R.M. Phylogeny and Biogeography of the Alpine Newt Mesotriton alpestris (Salamandridae, Caudata), Inferred from MtDNA Sequences. Mol. Phylogenet. Evol. 2007, 45, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Wielstra, B.; Salvi, D.; Canestrelli, D. Genetic Divergence across Glacial Refugia Despite Interglacial Gene Flow in a Crested Newt. Evol. Biol. 2021, 48, 17–26. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hughes, P.D.; Woodward, J.C.; Gibbard, P.L.; Macklin, M.G.; Gilmour, M.A.; Smith, G.R. The Glacial History of the Pindus Mountains, Greece. J. Geol. 2006, 114, 413–434. [Google Scholar] [CrossRef]
- Telbisz, T.; Stergiou, C.L.; Mindszenty, A.; Chatzipetros, A. Geological and Geomorphological Characteristics of Vikos Gorge and Tymphi Mountain (Northern Pindos National Park, Greece) and Karst-Related Social Processes of the Region. Acta Carsolog. 2019, 48, 29–42. [Google Scholar] [CrossRef]
- Kolettas, S.X. The Lakes of Ioannina & Lapsista (Historical, Geographical-Ecological Study); Prefecture & Union of Municipalities of Ioannina: Ioannina, Greece, 2000. (In Greek) [Google Scholar]
- Arntzen, J.W.; Teunis, S.F.M. A Six Year Study on the Populations Dynamics of the Crested Newt (Triturus cristatus) Following the Colonisation of a Newly Created Pond. Herpetol. J. 1993, 3, 99–110. [Google Scholar]
- Kupfer, A.; Kneitz, S. Population Ecology of the Great Crested Newt (Triturus cristatus) in an Agricultural Landscape Dynamics, Pond Fidelity and Dispersal. Herpetol. J. 2000, 10, 165–171. [Google Scholar]
- Kalinowski, S.T. Counting Alleles with Rarefaction: Private Alleles and Hierarchical Sampling Designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Jehle, R.; Wilson, G.A.; Arntzen, J.W.; Burke, T. Contemporary Gene Flow and the Spatio-Temporal Genetic Structure of Subdivided Newt Populations (Triturus cristatus, T. carmoratus). J. Evol. Biol. 2005, 18, 619–628. [Google Scholar] [CrossRef]
- Maletzky, A.; Kaiser, R.; Mikulíček, P. Conservation Genetics of Crested Newt Species Triturus cristatus and T. carnifex within a Contact Zone in Central Europe: Impact of Interspecific Introgression and Gene Flow. Diversity 2009, 2, 28–46. [Google Scholar] [CrossRef]
- Sotiropoulos, K.; Eleftherakos, K.; Tsaparis, D.; Kasapidis, P.; Giokas, S.; Legakis, A.; Kotoulas, G. Fine Scale Spatial Genetic Structure of Two Syntopic Newts across a Network of Ponds: Implications for Conservation. Conserv. Genet. 2013, 14, 385–400. [Google Scholar] [CrossRef]
- Unglaub, B.; Cayuela, H.; Schmidt, B.R.; Preißler, K.; Glos, J.; Steinfartz, S. Context-dependent Dispersal Determines Relatedness and Genetic Structure in a Patchy Amphibian Population. Mol. Ecol. 2021, 30, 5009–5028. [Google Scholar] [CrossRef] [PubMed]
- Schön, I.; Raepsaet, A.; Goddeeris, B.; Bauwens, D.; Mergeay, J.; Vanoverbeke, J.; Martens, K. High Genetic Diversity but Limited Gene Flow in Flemish Populations of the Crested Newt, Triturus cristatus. Belg. J. Zool. 2011, 141, 3–13. [Google Scholar]
- Nei, M.; Tajima, F. Genetic Drift And Estimation Of Effective Population Size. Genetics 1981, 98, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Preisser, E.L.; Kefer, J.Y.; Lawrence, J.D.; Clark, T.W. Vernal Pool Conservation in Connecticut: An Assessment and Recommendations. Environ. Manag. 2000, 26, 503–513. [Google Scholar] [CrossRef]
- Vos, C.C.; Stumpel, A.H.P. Comparison of Habitat-Isolation Parameters in Relation to Fragmented Distribution Patterns in the Tree Frog (Hyla arborea). Landsc. Ecol. 1996, 11, 203–214. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, C.; Campo, D.; Pola, I.G.; Garcia-Vazquez, E. Rural Road Networks as Barriers to Gene Flow for Amphibians: Species-Dependent Mitigation by Traffic Calming. Landsc. Urban. Plan. 2012, 104, 171–180. [Google Scholar] [CrossRef]
- Asaeda, T.; Ca, V.T.; Wake, A. Heat Storage of Pavement and Its Effect on the Lower Atmosphere. Atmos. Environ. 1996, 30, 413–427. [Google Scholar] [CrossRef]
- Bray, R. Case Study: A Programme of Habitat Creation and Great Crested Newt Introduction to Restored Opencast Land for British Coal Opencast. In Proceedings of the Conservation and Management of Great Crested Newts: Proceedings of a Symposium, Richmond, UK, 11 January 1994; English Nature (ENS20): Peterborough, UK, 1994; pp. 113–125. [Google Scholar]
- Kinne, O. Successful Re-Introduction of the Newts Triturus cristatus and T. vulgaris. Endanger. Species Res. 2004, 1, 25–40. [Google Scholar] [CrossRef]
- McNeill, D.C. Translocation of a Population of Great Crested Newts (Triturus cristatus): A Scottish Case Study. PhD Thesis, University of Glasgow, Glasgow, UK, 2010. [Google Scholar]
- Furnborough, P.; Kirby, P.; Lambert, S.; Pankhurst, T.; Parker, P.; Piec, D. The Effectiveness and Cost Efficiency of Different Pond Restoration Techniques for Bearded Stonewort and Other Aquatic Taxa. Report on the Second Life for Ponds Project at Hampton Nature Reserve in Peterborough; Froglife: Cambridgeshire, UK, 2011. [Google Scholar]
- Lewis, B. An Evaluation of Mitigation Actions for Great Crested Newts at Development Sites. Ph.D. Thesis, The Durrell Institute of Conservation and Ecology, Canterbury, UK, 2012. [Google Scholar]
- Bounas, A.; Keroglidou, M.; Toli, E.; Chousidis, I.; Tsaparis, D.; Leonardos, I.; Sotiropoulos, K. Constrained by Aliens, Shifting Landscape, or Poor Water Quality? Factors Affecting the Persistence of Amphibians in an Urban Pond Network. Aquat. Conserv. 2020, 30, 1037–1049. [Google Scholar] [CrossRef]
- Kats, L.B.; Ferrer, R.P. Alien Predators and Amphibian Declines: Review of Two Decades of Science and the Transition to Conservation. Divers. Distrib. 2003, 9, 99–110. [Google Scholar] [CrossRef]
- Van Buskirk, J.; Schmidt, B.R. Predator-Induced Phenotypic Plasticity in Larval Newts: Trade-Offs, Selection, and Variation in Nature. Ecology 2000, 81, 3009. [Google Scholar] [CrossRef]
- Joly, P.; Miaud, C.; Lehmann, A.; Grolet, O. Habitat Matrix Effects on Pond Occupancy in Newts. Conserv. Biol. 2001, 15, 239–248. [Google Scholar] [CrossRef]
- Denoël, M.; Ficetola, G.F.; Ćirović, R.; Radović, D.; Džukić, G.; Kalezić, M.L.; Vukov, T.D. A Multi-Scale Approach to Facultative Paedomorphosis of European Newts (Salamandridae) in the Montenegrin Karst: Distribution Pattern, Environmental Variables, and Conservation. Biol. Conserv. 2009, 142, 509–517. [Google Scholar] [CrossRef]
- Covaciu-Marcov, S.-D.; Cicort-Lucaciu, A.-S.; Ferenczi, I.; Kovács, É.H.; Ferenti, S.; Sas, I. Which Aquatic Habitat Is Better for the Feeding of a Protected Newt Species (Triturus dobrogicus) in Carei Plain Natural Protected Area? Olten. Stud. Comun. Ştiinţele Nat. 2012, 28, 115–120. [Google Scholar]
- Tarkhnishvili, D.N. Amphibians of the Caucasus: Status of Natural Populations and Perspectives on Conservation. In Amphibian Populations in the Commonwealth of Independent States: Current Status and Declines; Kuzmin, S.L., Dodd, C.K., Jr., Pikulik, M.M., Eds.; Pensoft Publishers: Moscow, Russia, 1995; pp. 106–124. [Google Scholar]
- Phillips, R.A.; Patterson, D.; Shimmings, P. Increased Use of Ponds by Breeding Natterjack Toads, Bufo Calamita, Following Management. Herpetol. J. 2002, 12, 75–78. [Google Scholar]
- Smith, R.K.; Sutherland, W.J. Amphibian Conservation: Global Evidence for the Effects of Interventions; Pelagic Publishing: Exeter, UK, 2014. [Google Scholar]
- Smith, R.K.; Meredith, H.; Sutherland, W.J. Amphibian Conservation. In What Works in Conservation 2020; Sutherland, W.J., Dicks, L.V., Petrovan, S.O., Smith, R.K., Eds.; Open Book Publishers: Cambridge, UK, 2020; pp. 9–64. [Google Scholar]
- Mori, E.; Menchetti, M.; Cantini, M.; Bruni, G.; Santini, G.; Bertolino, S. Twenty Years’ Monitoring of a Population of Italian Crested Newts Triturus Carnifex: Strong Site Fidelity and Shifting Population Structure in Response to Restoration. Ethol. Ecol. Evol. 2017, 29, 460–473. [Google Scholar] [CrossRef]
- Staccione, A.; Broccoli, D.; Mazzoli, P.; Bagli, S.; Mysiak, J. Natural Water Retention Ponds for Water Management in Agriculture: A Potential Scenario in Northern Italy. J. Environ. Manag. 2021, 292, 112849. [Google Scholar] [CrossRef]
- Garner, T.W.J.; Pearman, P.B.; Angelone, S. Genetic Diversity across a Vertebrate Species’ Range: A Test of the Central-Peripheral Hypothesis. Mol. Ecol. 2004, 13, 1047–1053. [Google Scholar] [CrossRef]
- Channell, R.; Lomolino, M.V. Dynamic Biogeography and Conservation of Endangered Species. Nature 2000, 403, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Blank, L.; Sinai, I.; Bar-David, S.; Peleg, N.; Segev, O.; Sadeh, A.; Kopelman, N.M.; Templeton, A.R.; Merilä, J.; Blaustein, L. Genetic Population Structure of the Endangered Fire Salamander (Salamandra infraimmaculata) at the Southernmost Extreme of Its Distribution. Anim. Conserv. 2013, 16, 412–421. [Google Scholar] [CrossRef]
- Peterman, W.E.; Feist, S.M.; Semlitsch, R.D.; Eggert, L.S. Conservation and Management of Peripheral Populations: Spatial and Temporal Influences on the Genetic Structure of Wood Frog (Rana sylvatica) Populations. Biol. Conserv. 2013, 158, 351–358. [Google Scholar] [CrossRef]
- Lesica, P.; Allendorf, F.W. Are Small Populations of Plants Worth Preserving? Conserv. Biol. 1992, 6, 135–139. [Google Scholar] [CrossRef]
- Hunter, M.L.; Hutchinson, A. The Virtues and Shortcomings of Parochialism: Conserving Species That Are Locally Rare, but Globally Common. Conserv. Biol. 1994, 8, 1163–1165. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Eckert, C.G. Population Genetic Consequences of Geographic Disjunction: A Prairie Plant Isolated on Great Lakes Alvars. Mol. Ecol. 2007, 16, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Lambeck, R.J. Focal Species: A Multi-Species Umbrella for Nature Conservation. Especies Focales: Una Sombrilla Multiespecifica Para Conservar La Naturaleza. Conserv. Biol. 1997, 11, 849–856. [Google Scholar] [CrossRef]
- Fraser, D.J.; Bernatchez, L. Adaptive Evolutionary Conservation: Towards a Unified Concept for Defining Conservation Units. Mol. Ecol. 2001, 10, 2741–2752. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing With the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Wielstra, B.; Themudo, G.E.; Güçlü, Ö.; Olgun, K.; Poyarkov, N.A.; Arntzen, J.W. Cryptic Crested Newt Diversity at the Eurasian Transition: The Mitochondrial DNA Phylogeography of Near Eastern Triturus Newts. Mol. Phylogenet. Evol. 2010, 56, 888–896. [Google Scholar] [CrossRef]
- Arntzen, J.W.; Wielstra, B.; Wallis, G.P. The Modality of Nine Triturus Newt Hybrid Zones Assessed with Nuclear, Mitochondrial and Morphological Data. Biol. J. Linn. Soc. 2014, 113, 604–622. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).