Abstract
The present study describes three new species of monogeneans parasitizing the gills of anostomid fishes from the Upper Paraná River basin, southeastern Brazil: Jainus beccus n. sp. and Jainus radixelongatus n. sp. on Leporinus friderici, Leporinus octofasciatus, Leporinus striatus, and Megaleporinus elongatus; and Jainus ornatus n. sp. on L. friderici. The new species differ from other congeners by the morphology of the accessory piece. There is a semicircular distal portion resembling a “bird’s beak” in Jainus beccus n. sp. It composed of two subunits—one ventral and more sclerotized, sickle-shaped, and another dorsal with three projections—in Jainus radixelongatus n. sp. There are two elongated and sclerotized subunits, both of which have a sickle-shaped distal portion, in Jainus ornatus n. sp. Supplementary observations not reported in the original descriptions of the type-species Jainus piava Karling, Bellay, Takemoto & Pavanelli, 2011 are proposed, as follows: the presence of a thin and delicate ventral bar, which can vary greatly in shape; an accessory piece not articulated with the MCO’s base. This paper provides the first phylogenetic study based on LSU rDNA and COI mtDNA gene sequences for Jainus, improving and clarifying the understanding of host–parasite relationships in neotropical characiforms.
Keywords:
Jainus; new species; Ancyrocephalinae; ectoparasites; taxonomy; neotropical region; phylogeny; COI mtDNA; LSU rDNA 1. Introduction
In Brazil until 2011, 1034 species of parasites were registered in 451 species of freshwater fishes [1]. However, these data show that only 10% of known fish species had their parasitic fauna investigated. Among the taxonomic groups of fish parasites, monogeneans can be considered one of the most diverse [2,3]. After that, Cohen et al. [4] in 2013 registered 651 species of monogeneans that parasitized fishes, amphibians, and reptiles in South America, and 67% parasitized only fishes from Brazil. The majority of these occurrences were concentrated in freshwater fishes from the basins of the Amazon and Paraná Rivers.
In most of the taxonomic studies on monogeneans, descriptions are based on morphological data and do not provide the molecular characterization of the species described [5]. Considering molecular data as a key tool for the delimitation of new species, recent studies have used molecular techniques to support morphological and taxonomic descriptions of monogenean species, providing ways of performing phylogenetic analysis based on different markers [5,6,7,8,9], improving the knowledge about this diverse group and elucidating the relationship between species and their hosts [5,10,11].
Jainus Mizelle, Kritsky & Crane, 1968 comprises gill parasites of neotropical characiforms. Currently, the genus includes seven species, and six of them have been described in fishes from Brazilian ecosystems: Jainus jainus Mizelle, Kritsky & Crane, 1968 (type species) parasitizing Chalceus macrolepidotus Cuvier, 1818 (Chalceidae); Jainus robustus Mizelle, Kritsky & Crane, 1968 from Bryconops affinis (Günther, 1864) (Creatochanes affinis) (Iguanodectidae); Jainus iocensis Cohen, Kohn & Boeger, 2012 from Salminus brasiliensis (Cuvier, 1816) (Bryconidae); Jainus piava Karling, Bellay, Takemoto & Pavanelli, 2011 from Schizodon borellii (Boulenger, 1900) (Anostomidae); Jainus leporini Abdallah, Azevedo & Luque, 2012 from Hypomasticus copelandii (Steindachner, 1875) (Leporinus copelandii) (Anostomidae); and Jainus amazonensis Kritsky, Thatcher & Kayton, 1980 from Brycon melanopterus (Cope, 1872) (Bryconidae) [12,13,14,15,16].
As part of our ongoing studies about the biodiversity of helminth parasites of freshwater fishes from tributaries of the Upper Paraná River basin, southeastern Brazil, three new species of Jainus are described from the gills of four anostomid fishes (Leporinus friderici (Block, 1794), Leporinus striatus Kner, 1858, Leporinus octofasciatus Steindachner, 1915, and Megaleporinus elongatus (Valenciennes, 1850)) supported by morphological and molecular data. We also present new morphological features that are added to the diagnosis of J. piava and its relationship with congeners and other dactylogyrids parasites of neotropical characiforms, using sequences of the partial LSU rDNA gene.
2. Materials and Methods
2.1. Host Sampling and Parasitological Procedures
Fish specimens were collected in two localities from the Upper Paraná River basin, São Paulo state, Brazil, from 2011 to 2013: Jurumirim Reservoir (23°12′17″ S, 49°13′19″ W) and Sapucaí-Mirim River (20°29′38.38″ S, 47°51′33.11″ W). Data on host species; locality; number of sampled hosts; and mean standard length (SL) in centimeters (cm) ± standard deviation (SD), followed by the range (minimum–maximum values), are in Table 1.

Table 1.
List of host species sampled in the present study in the Upper Paraná River basin, São Paulo, Brazil (SL—mean standard length; SD—standard deviation).
Hosts were collected under the license number SP/538/88 DEFOP (Department of the Development of Fishery and Inspection) for the Jurimirim Reservoir, and SISBio 13794-1 (Permanent License for the Collection of Zoological Material) for the Sapucaí-Mirim River. All animal procedures were performed in full compliance with the Ethics Committee for Animal Experimentation (CEUA/protocols n. 120 and 942) of the Universidade Estadual Paulista (São Paulo State University—UNESP). Fishes were collected using a nylon monofilament gillnet. After capture, some specimens were individually stored in plastic bags and frozen for later necropsy at the laboratory. Some freshly killed hosts were examined in situ, and some specimens of monogeneans were placed into 96% molecular-grade ethanol for molecular analyses. In the laboratory, gills were removed, placed in Petri dishes with water, and checked for monogeneans under a stereomicroscope. Monogeneans were mounted on slides with Hoyer’s and Gray and Wess’s medium [17].
The morphology of the parasites was analyzed using V3 Leica Application Suite (LAS) computerized system for image analysis adapted in a microscope with differential interference contrast. Illustrations were performed with the aid of a drawing tube (camera lucida) mounted on a Leica DMLS microscope equipped with phase-contrast optics. Measurements of the sclerotized structures of the haptor (bar, anchors, and hooks) and copulatory complex were performed according to the scheme shown in Figure 1a–l. For the copulatory complex comprising a sclerotized accessory piece and a male copulatory organ (MCO), or cirrus, the direction of the MCO’s rings (counterclockwise vs. clockwise) follows Kritsky et al. [18]. The numbering and distribution of hook pairs and the specific terminology of the genus Jainus follow Mizelle [19] and Mizelle et al. [12], respectively. All measurements are represented in micrometers (μm) and expressed as the means followed by the respective ranges and the numbers of specimens measured in parentheses. The prevalence and mean intensity of infection were calculated following Bush et al. [20].
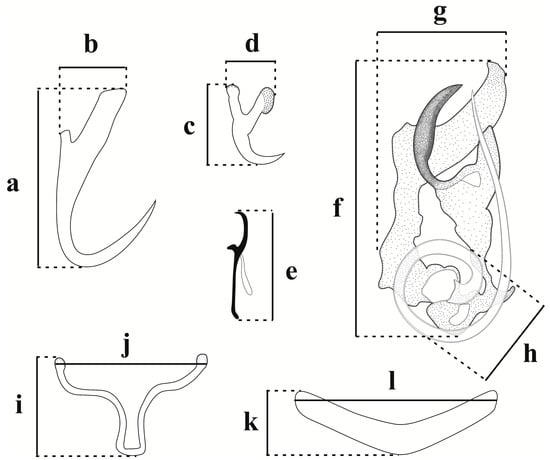
Figure 1.
Scheme of measurements of the sclerotized structures of the haptor and copulatory com-plex of Jainus spp. Dorsal anchor: (a) total length, (b) width; ventral anchor: (c) total length, (d) width; hook: (e) total length; accessory piece: (f) total length, (g) total width; MCO: (h) diameter of the ring; ventral bar: (i) total length, (j) total width; dorsal bar: (k) total length, (l) total width.
Type and voucher specimens of the monogeneans were deposited in the Helminthological Collection of the Instituto Oswaldo Cruz (CHIOC), Rio de Janeiro (state), Brazil. Other vouchers were deposited in the Helminthological Collection of the Institute of Biosciences (CHIBB), Botucatu, São Paulo (state), Brazil. According to Brazilian laws, species registration for scientific research purposes was carried out at SisGen (A8F53FE). Paratypes and vouchers examined for comparative purposes: J. leporini (paratypes 535a–c) and J. amazonensis (paratypes MZUSP, USNM, UNSM) deposited in the zoological collection of the Instituto Nacional de Pesquisas da Amazônia (INPA), Amazonas, Brazil; J. iocensis (paratypes 37635, 37700, 37712) and J. piava (paratypes 37236, 37235B, 37235C, and photomicrographs of holotype 37235A) deposited in CHIOC.
2.2. Molecular and Phylogenetic Analyses
Monogenean specimens were placed into 96% molecular-grade ethanol and subsequently used for molecular characterization. Conspecific specimens (paragenophores; see Pleijel et al. [21]) were mounted in Hoyer’s or Gray and Wess’s medium and deposited in the CHIBB. The total genomic DNA of the parasites was extracted using 200 μL of a 5% suspension of Chelex in deionized water and 2 μL of proteinase K, followed by overnight incubation at 56 °C, boiling at 90 °C for 8 min, and centrifugation at 14,000 rpm for 10 min, producing a final volume of 20 μL [8]. Partial fragments of the LSU rDNA and COI mtDNA genes were amplified by conventional polymerase chain reaction (PCR) performed with a total volume of 20 μL for each reaction, containing 3 μL of extracted DNA, 10 μL of 2xMyFiTM Mix (Bioline, Taunton, MA, USA), 3.8 μL of pure water, and 1.6 μL of each PCR primer. For the LSU rDNA gene and COI mtDNA gene, the primers used and the thermocycling profile are expressed in Table 2.

Table 2.
Primers used in this study. New primers obtained for the present study are in bold.
PCR products (2 µL) were run on an agarose gel (1%) using GelRed™ fluorescent nucleic acid dye and loading buffer to confirm amplicon size and yield. Amplicons were purified with the QIAquick PCR Purification Kit (Qiagen) and sequenced using the BigDye v.3.1 Terminator Cycle Sequencing kit (Applied Biosystems). Sequences were run on an Applied Biosystems ABI 3500 DNA genetic analyzer. Forward and reverse sequences were assembled and edited using Sequencher version 5.2.4 (Gene Codes, Ann Arbor, MI, USA). Datasets were separately aligned using the default parameters of the MUSCLE algorithm implemented on Geneious version 7.1.3 [24].
In total, five newly sequences for three species were submitted to the GenBank database: LSU rDNA gene—two partial sequences, (GenBank accession numbers OQ843018, OQ843019); and COI—three partial sequences, (accession numbers OQ833545, OQ833544, OQ833543) (see Table 3).
Phylogenetic relationships were reconstructed for each alignment under Bayesian inference (BI) and maximum likelihood (ML), applying the model GTR+I+G, which was selected as the best-fitting model of nucleotide evolution by the JModel Test v.2.1 using the Akaike information criterion [25] for all data sets. The BI analyses were run using MrBayes [26]. The Markov chain Monte Carlo (MCMC) chains were run with 106 generations and sampling tree topologies every 100th generation. The first 25% of trees were discarded as burn-in, and the remaining trees were used for calculating the Bayesian posterior probabilities. The ML analyses were estimated using RaxML [27], with 1000 bootstrap replications. Both ML and BI analyses were performed on the computational resource CIPRES [28]. Genetic divergence was calculated with the uncorrected p-distances model in MEGA7 [29,30]. Phylogenetic trees were edited in the software FigTree v1.4 [31].

Table 3.
List of monogeneans included in the phylogenetic analyses, with details of the hosts, localities, and GenBank accession numbers of sequences from the LSU rDNA and COI mtDNA genes. New sequences obtained in the present study are in bold.
Table 3.
List of monogeneans included in the phylogenetic analyses, with details of the hosts, localities, and GenBank accession numbers of sequences from the LSU rDNA and COI mtDNA genes. New sequences obtained in the present study are in bold.
| Monogenea Species | Host Species | Locality | Accession Numbers | References | |
|---|---|---|---|---|---|
| LSU rDNA | COI | ||||
| Dactylogyridae | |||||
| Ameloblastella chavarriai (Price, 1938) | Rhamdia quelen | Mexico | KP056251 | [6] | |
| Ameloblastella edentensis Mendoza-Franco, Mendoza-Palmero & Scholz, 2020 | Hypophthalmus edentatus | Peru | KP056255 | [6] | |
| Ameloblastella unapioides Mendoza-Franco, Mendoza-Palmero & Scholz, 2020 | Sorubim lima | Peru | KP056254 | [6] | |
| Aphanoblastella aurorae Mendoza-Palmero, Scholz, Mendoza-Franco & Kuchta, 2012 | Goeldiella eques | Peru | KP056239 | [6] | |
| Aphanoblastella magna Yamada, Acosta, Yamada, Scholz & Silva, 2018 | Pimelodella avanhandavae | Brazil | MH688484 | [32] | |
| Aphanoblastella travassosi (Price, 1938) | Rhamdia guatemalensis | Mexico | MK358458 | [9] | |
| Cacatuocotyle papilionis Zago, Franceschini, Müller & Silva, 2018 | Astyanax lacustris | Brazil | MG832889 | [33] | |
| Characithecium paranapanemense Zago, Franceschini, Abdallah, Müller, Azevedo & Silva, 2021 | Psalidodon paranae | Brazil | MZ408907 | [34] | |
| Cosmetocleithrum bifurcum Mendoza-Franco, Mendoza-Palmero & Scholz, 2000 | Hassar sp. | Peru | KP056217 | [6] | |
| Cosmetocleithrum bulbocirrus Kritsky, Thatcher & Boeger, 1986 | Pterodoras granulosus | Brazil | MG001326 | [9] | |
| Demidospermus prolixus Franceschini, Zago, Müller, Francisco, Takemoto & Silva, 2017 | Loricaria prolixa | Brazil | KY766955 | [5] | |
| Demidospermus anus Suriano, 1983 | Loricariichthys platymetopon | Brazil | KY766957 | [5] | |
| Demidospermus mortenthaleri Mendoza-Palmero, Scholz, Mendoza-Franco & Kuchta, 2012 | Brachyplatystoma juruense | Peru | KP056245 | [6] | |
| Demidospermus rhinelepisi Acosta, Scholz, Blasco-Costa, Alves & Silva, 2019 | Rhinelepis aspera | Brazil | MG001324 | [9] | |
| Demidospermus spirophallus Franceschini, Zago, Müller, Francisco, Takemoto & Silva, 2017 | Loricaria prolixa | Brazil | KY766954 | [5] | |
| Diaphorocleidus magnus Zago, Franceschini, Abdallah, Müller, Azevedo & Silva, 2021 | Astyanax lacustris | Brazil | MZ408903 | MZ408253 | [34] |
| Diaphorocleidus neotropicalis Zago, Franceschini, Abdallah, Müller, Azevedo & Silva, 2021 | Astyanax lacustris | Brazil | MZ408906 | MZ408254 | [34] |
| Diaphorocleidus petrosusi Mendoza-Franco, Aguirre-Macedo & Vidal-Martínez, 2007 | - | Panama | MF939878 | [35] | |
| Heteropriapulus anchoradiatus Acosta, Franceschini, Zago, Scholz & Silva, 2017 | Pterygoplichthys ambrosettii | Brazil | MF116371 | [7] | |
| Heteropriapulus heterotylus Acosta, Franceschini, Zago, Scholz & Silva, 2017 | Pterygoplichthys ambrosettii | Brazil | MF116370 | [7] | |
| Heteropriapulus simplex Acosta, Franceschini, Zago, Scholz & Silva, 2017 | Pterygoplichthys ambrosettii | Brazil | MF116372 | [7] | |
| Heteropriapulus sp. | Pterygoplichthys ambrosettii | Brazil | MF116373 | [7] | |
| Jainus beccus n. sp. | Leporinus friderici | Brazil | OQ833545 | Present study | |
| Jainus piava Karling, Bellay, Takemoto & Pavanelli, 2011 | Schizodon nasutus | Brazil | OQ843019 | OQ833543 | Present study |
| Jainus radixelongatus n. sp. | Leporinus striatus | Brazil | OQ843018 | OQ833544 | Present study |
| Nanayella aculeatrium Acosta, Mendoza-Palmero, Silva & Scholz, 2019 | Sorubim lima | Peru | KP056228 | [6] | |
| Nanayella amplofalcis Acosta, Mendoza-Palmero, Silva & Scholz, 2019 | Hemisorubim platyrhynchos | Brazil | MG001325 | [9] | |
| Nanayella fluctuatrium Acosta, Mendoza-Palmero, Silva & Scholz, 2019 | Sorubim lima | Brazil | MG001327 | [9] | |
| Nanayella megorchis Acosta, Mendoza-Palmero, Silva & Scholz, 2019 | Sorubim lima | Peru | MK367407 | [9] | |
| Nanayella processusclavis Acosta, Mendoza-Palmero, Silva & Scholz, 2019 | Hemisorubim platyrhynchos | Brazil | MG001328 | [9] | |
| Parasciadicleithrum octofasciatum Mendoza-Palmero, Blasco-Costa, Hernández-Mena & Pérez-Ponce de León, 2017 | Rocio octofasciata | Mexico | KY305885 | [36] | |
| Sciadicleithrum meekii Mendoza-Palmero, Blasco-Costa, Hernández-Mena & Pérez-Ponce de León, 2017 | Thorichthys meeki | Mexico | KY305889 | [36] | |
| Sciadicleithrum mexicanum Mendoza-Palmero, Blasco-Costa, Hernández-Mena & Pérez-Ponce de León, 2017 | Rocio octofasciata | Mexico | KY305886 | [36] | |
| Sciadicleithrum panamensis Mendoza-Franco, Aguirre-Macedo & Vidal-Martínez, 2007 | - | Panama | MF939864 | [35] | |
| Sciadicleithrum splendidae Kritsky, Vidal-Martínez & Rodriguez-Canul, 1994 | Parachromis friedrichsthalii | Mexico | KY305890 | [36] | |
| Trinigyrus anthus Franceschini, Acosta, Zago, Müller & Silva, 2020 | Hypostomus regani | Brazil | MN947622 | MN916719 | [37] |
| Trinigyrus carvalhoi Franceschini, Acosta, Zago, Müller & Silva, 2020 | Hypostomus ancistroides | Brazil | MN947608 | MN922321 | [37] |
| Trinigyrus peregrinus Nitta & Nagasawa, 2016 | Pterygoplychthys ambrosettii | Brazil | MN944890 | MN913212 | [37] |
| Trinigyrus peregrinus Nitta & Nagasawa, 2016 | Pterygoplichthys disjunctivus | Japan | LC104308 | [38] | |
| Unibarra paranoplatensis Suriano & Incorvaia, 1995 | Aguarunichthys torosus | Peru | KP056219 | [6] | |
| Unilatus unilatus Mizelle & Kritsky, 1967 | Pterygoplichthys ambrosettii | Brazil | MF102106 | [7] | |
| Urocleidoides cultellus Mendoza-Franco & Reina, 2008 | - | Panama | MF939848 | [35] | |
| Urocleidoides digitabulum Zago, Yamada, Yamada, Franceschini, Bongiovani & Silva, 2020 | Megaleporinus elongatus | Brazil | MT556796 | [39] | |
| Urocleidoides indianensis Oliveira, Silva, Vieira & Acosta, 2020 | Parodon nasus | Brazil | OK482868 | [40] | |
| Urocleidoides paradoni Oliveira, Silva, Vieira & Acosta, 2020 | Parodon nasus | Brazil | OK482867 | [40] | |
| Urocleidoides paradoxus Kritsky, Thatcher & Boeger, 1986 | Leporinus friderici | Brazil | MT556795 | [39] | |
| Urocleidoides sinus Zago, Yamada, Yamada, Franceschini, Bongiovani & Silva, 2020 | Schizodon nasutus | Brazil | MT556799 | [39] | |
| Urocleidoides tenuis Zago, Yamada, Yamada, Franceschini, Bongiovani & Silva, 2020 | Parodon nasus | Brazil | OK465455 | [40] | |
| Urocleidoides uncinus Zago, Yamada, Yamada, Franceschini, Bongiovani & Silva, 2020 | Gymnotus inaequilabiatus | Brazil | MT556798 | MT594473 | [39] |
| Vancleaveus janauacaensis Kritsky, Thatcher & Boeger, 1986 | Pterodoras granulosus | Peru | KP056247 | [6] | |
| Walteriella conica Mendoza-Palmero, Mendoza-Franco, Acosta & Scholz, 2019 | Brachyplatystoma juruense | Peru | MK834513 | [41] | |
| Walteriella ophiocirrus Mendoza-Palmero, Mendoza-Franco, Acosta & Scholz, 2019 | Platystomatichthys sturio | Peru | MK834515 | [41] | |
| Diplectanidae | |||||
| Murraytrema pricei Caballero, Bravo & Grocott, 1955 | Nibea albiflora | China | DQ157672 | [42] | |
| Pseudorhabdosynochus epinepheli (Yamaguti, 1938) | Epinephelus brunneus | China | AY553622 | [43] | |
| Pseudorhabdosynochus lantauensis (Beverley-Burton & Suriano, 1981) | Epinephelus brunneus | China | AY553624 | [43] |
3. Results
3.1. Description of Jainus beccus n. sp.
(Figure 2).
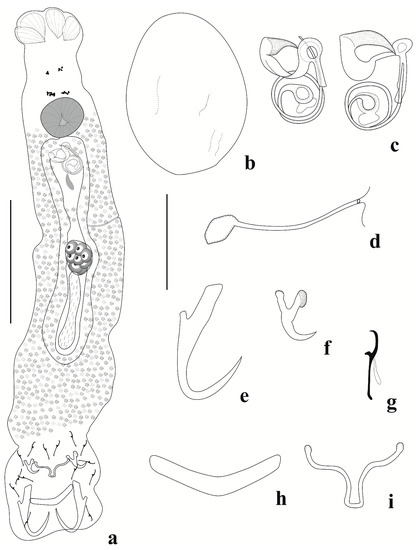
Figure 2.
Jainus beccus n. sp. from Leporinus friderici (Characiformes, Anostomidae). (a) Whole mount (composite, ventral view); (b) egg; (c) copulatory complex (ventral view); (d) vagina; (e) dorsal anchor; (f) ventral anchor; (g) hook; (h) dorsal bar; (i) ventral bar (Scale bars: (a) = 50 µm; (b–i) = 20 µm).
(urn:lsid:zoobank.org:act:A6A834C5-E70E-45B3-8B05-08C3AC3332CD).
3.1.1. Taxonomic Summary
Type host: Leporinus friderici (Bloch, 1794) (Characiformes, Anostomidae).
Type locality: Sapucaí-Mirim River (20°29′38.38″ S, 47°51′33.11″ W), Upper Paraná River basin, São Paulo, Brazil.
Other hosts: Leporinus octofasciatus Steindachner, 1915; Leporinus striatus Kner, 1858 and Megaleporinus elongatus (Valenciennes, 1850) (Characiformes: Anostomidae).
Other locality: Jurumirim Reservoir, Paranapanema River (23°12′17″ S, 49°13′19″ W), Upper Paraná River basin, São Paulo, Brazil.
Site of infection: Gill filaments.
Prevalence, mean intensity of infection. and range of intensity: Sapucaí-Mirim River: L. friderici: 31 infected fish of 40 analyzed (77.5%), 17.38 ± 3.07 (1–55); L. octofasciatus: 1 infected fish of 17 analyzed (5.88%), 1. Jurumirim Reservoir: L. friderici: 34 infected fish of 116 analyzed (29.31%), 8.06 ± 1.97 (1–51); L. octofasciatus: 8 infected fish of 15 analyzed (53.3%), 3.2 ± 0.7 (1–6); L. striatus: 1 infected fish of 25 analyzed (4%); M. elongatus: 12 infected fish of 30 analyzed (40%), 2.7 ± 0.5 (1–7).
Type-material: Holotype CHIOC (39959a); paratypes CHIOC (39959b, 39960–39964, 39965a, 39965b, 39966, 39967), vouchers CHIBB (703L–710L).
Molecular sequence data: A COI mtDNA (903 bp) partial sequence obtained from one monogenean specimen from L. friderici was deposited in GenBank under accession numberOQ833545.
Etymology: The specific epithet is derived from Latin (beccus = bird’s beak) and refers to the shape of the distal portion of the accessory piece.
3.1.2. Description (Based on 30 Specimens: 15 Mounted in Gray and Wess’s Medium, and 15 Mounted in Hoyer’s Medium)
Body elongate, fusiform 279 (207–400, n = 28) long; 58 (27–105, n = 23) wide near midlength. Cephalic lobes moderately developed, two pairs of head organs lying in cephalic lobes. Cephalic glands not visible. Four eyespots present; anterior pair smaller than posterior one; distance between eyespots of anterior pair smaller than that between posterior one; one member of each pair sometimes absent. Accessory granules are small, generally elongate, and frequently in the anterior trunk. Pharynx subspherical 16 (12–21, n = 13) wide; esophagus short. Peduncle short. Haptor sub-square, 41 (30–54, n = 22) long, 48 (36–55, n = 24) wide. Anchors dissimilar in shape and size. Ventral anchor 10 (8–11, n = 25) long, 6 (5–7, n = 25) wide, curved point and shaft, superficial root dilatated in distal portion and with grooves and elongated deep root. Dorsal anchor 27 (22–33, n = 27) long, 9 (7–11, n = 27) wide, with elongated curved point, straight and short shaft, deep root poorly developed, superficial root elongated and rectangular. Ventral bar 10 (7–14, n = 22) long, 22 (17–30, n = 24) wide, delicate and filamentous. Dorsal bar 8 (5–11, n = 24) long, 25 (20–31, n = 24) wide, robust, V-shaped. Seven pairs of similar and marginal hooks, 12 (10–15, n = 34) long, with erect thumb, curved point, erect shank; filament hook (FH) loop half of shank length. Copulatory complex comprising a male copulatory organ (MCO) with two counterclockwise rings 10 (9–14, n = 20) in diameter, a sclerotized expanded and ornamented base, and an accessory piece 16 (13–19, n = 18) wide, well developed, with proximal portion formed by a concave rod that guides the MCO and a semicircular distal portion resembling a “bird’s beak”, not articulated to the MCO. Vagina simple, sinistral, comprising a delicate tube. Gonads intercaecal, in tandem. Germarium 17 (14–21, n = 4) long, 13 (9–17, n = 4) wide. Testis 42 (30–63, n = 4) long, 10 (6–14, n = 4) wide, elongated and posterior to germarium. Seminal vesicle a dilation of vas deferens. Seminal receptacle anterior to germarium. Prostatic reservoir present. Oviduct, ootype, and uterus not observed. Vitellaria densely dispersed throughout trunk, absent in reproductive organ regions. Egg 77 (n = 1) long, 47 (n = 1) wide, without filament.
3.1.3. Remarks
Jainus beccus n. sp. can be distinguished from other congeners by the morphology of the accessory piece (well developed with a proximal portion formed by a concave rod that guides the MCO, and the semicircular distal portion resembling a “bird’s beak”) and the ventral anchor (a superficial root dilatated in the distal portion, with grooves). The new species closely resembles J. piava and J. leporini by having a delicate ventral bar and dorsal bar curved or V-shaped. However, Jainus beccus n. sp. differs from these congeners by having a simple vagina (vagina associated with a plate-like sclerotized structure in J. leporini; vagina with terminal bulbous expansion in J. piava). Analyses of the paratypes of J. leporini described by Abdallah et al. [16] and holotypes and paratypes of J. piava described by Karling et al. [14] confirmed these resemblances and differences reported in the current study.
3.2. Description of Jainus ornatus n. sp.
(Figure 3).
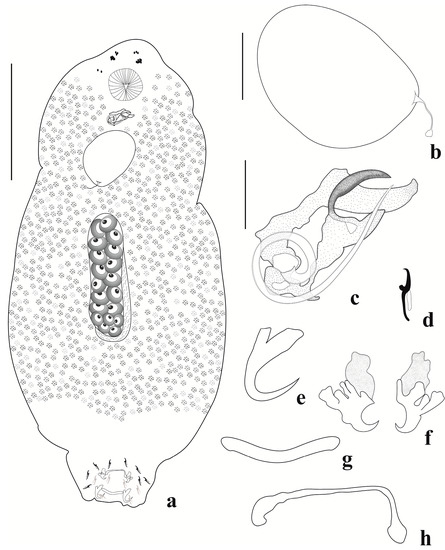
Figure 3.
Jainus ornatus n. sp. from Leporinus friderici (Characiformes, Anostomidae). (a) Whole mount (composite, ventral view); (b) egg; (c) copulatory complex (ventral view); (d) hook; (e) dorsal anchor; (f) ventral anchors; (g) dorsal bar; (h) ventral bar (scale bars: (a) = 200 µm; (b) = 50 µm; (c–h) = 20 µm).
(urn:lsid:zoobank.org:act:AE22AE7F-7474-43C0-8292-C8DB1B943081).
3.2.1. Taxonomic Summary
Type host: Leporinus friderici (Bloch, 1794) (Characiformes, Anostomidae).
Type locality: Jurumirim Reservoir, Paranapanema River (23°12′17″ S, 49°13′19″ W), Upper Paraná River basin, São Paulo, Brazil.
Site of infection: Gill filaments.
Prevalence, mean intensity of infection, and range of intensity: 8 infected fish of 116 analyzed (6.89%), 3.2 ± 0.79 (1–8).
Type-material: Holotype CHIOC (39968a); paratypes CHIOC (39968b–39968d, 39969); vouchers CHIBB (711L–719L).
Etymology: The specific epithet is derived from Latin (ornatus = adorned) and refers to the presence of an adorned root of the ventral anchor.
3.2.2. Description (Based on 23 Specimens Mounted in Hoyer’s Medium)
Body robust, fusiform, 1041 (774–1586, n = 15) long, and 429 (325–620, n = 14) wide near midlength. Cephalic lobes poorly developed; head organs and cephalic glands not observed. Four eyespots. Anterior pair is smaller than posterior one; one member of each pair sometimes absent. Accessory granules small, generally elongate, and frequently in anterior trunk. Pharynx subspherical 54 (41–64, n = 16) wide. Peduncle short.
Haptor sub-square, 76 (61–93, n = 9) long, 136 (118–157, n = 9) wide. Anchors dissimilar in shape and size. Ventral anchor 25 (23–27, n = 8) long, 19 (16–22, n = 8) wide, with deep and superficial roots variable in shape and bearing irregular sclerotization (like an ornament), blade-like point and shaft. Dorsal anchor 23 (14–25, n = 12) long, 13 (11–14, n = 12) wide, with curved point and shaft, well-developed deep and superficial roots. Ventral bar 12 (7–19, n = 8) long, 42 (26–56, n = 8) wide, delicate, curved, and with enlarged ends. Dorsal bar 7 (6–7, n = 5) long, 36 (31–42, n = 12) wide, V-shaped. Seven pairs of similar and marginal hooks, 15 (14–16, n = 26) long, with erect thumb, curved point, arcuated shank; FH loop ¾ shank length. Copulatory complex comprising a MCO with one counterclockwise ring 15 (15–16, n = 7) in diameter, and accessory piece 59 (54–63, n = 5) long, 21 (20–24, n = 6) wide, robust, comprising two elongated and sclerotized subunits, both with sickle-shaped distal portions, articulated to the MCO. Gonads intercaecal, overlapping. Germarium 225 (136–313, n = 3) long, 59 (42–71, n = 3) wide. Testis 205 (155–315, n = 5) long, 65 (52–91, n = 5) wide, elongated, and dorsal to germarium. Vitellaria densely dispersed throughout trunk, absent in reproductive organ regions. Vagina, seminal vesicle, seminal receptacle, prostatic reservoir, oviduct, ootype, and uterus not observed. Egg 116 (74–154, n = 7) long, 89 (69–104, n = 5) wide, with filament.
3.2.3. Remarks
Jainus ornatus n. sp. can be distinguished from most of its congeners by the morphology of the accessory piece (comprising two elongated and sclerotized subunits, both with sickle-shaped distal portions) and ventral anchor (with deep and superficial roots variable in shape and bearing irregular sclerotization, blade-like point, and shaft). Based on the shape of the ventral anchor, Jainus ornatus n. sp. closely resembles J. iocensis by possessing roots with irregular sclerotization. However, the new species differs from J. iocensis by having a delicate ventral bar with enlarged ends (V-shaped and with a posteromedian projection in J. iocensis), and an accessory piece with two elongated and sclerotized subunits, both with a sickle-shaped distal portion (hook-shaped in J. iocensis). Examination of paratypes of J. iocensis described by Cohen et al. [15] confirmed these resemblances and differences reported in the current study.
3.3. Description of Jainus radixelongatus n. sp.
(Figure 4).
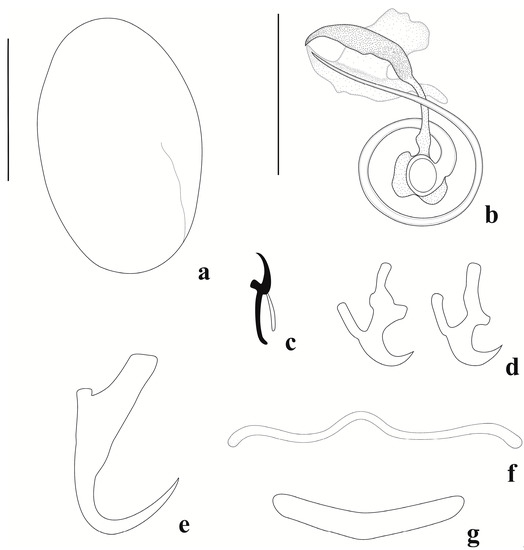
Figure 4.
Jainus radixelongatus n. sp. from Leporinus striatus (Characiformes, Anostomidae). (a) Egg; (b) copulatory complex (ventral view); (c) hook; (d) ventral anchor; (e) dorsal anchor; (f) ventral bar; (g) dorsal bar (scale bars: (a) = 50 µm; (b–g) = 20 µm).
(urn:lsid:zoobank.org:act:7B431077-6265-4DA9-9D04-20CA031BD673).
3.3.1. Taxonomic Summary
Type host: Leporinus striatus Kner, 1858 (Characiformes: Anostomidae).
Type locality: Jurumirim Reservoir, Paranapanema River (23°12′17″ S, 49°13′19″ W), Upper Paraná River basin, São Paulo, Brazil.
Other hosts: Leporinus friderici (Bloch, 1794), Leporinus octofasciatus Steindachner, 1915, and Megaleporinus elongatus (Valenciennes, 1850) (Characiformes: Anostomidae).
Other locality: Sapucaí-Mirim River (20°29′38.38″ S, 47°51′33.11″ W), Upper Paraná River basin, São Paulo, Brazil.
Site of infection: Gill filaments.
Prevalence, mean intensity of infection, and range of intensity: Jurumirim Reservoir: L. striatus: 6 infected fish of 25 analyzed (24%), 2 ± 0.5 (1–4); L. octofasciatus: 6 infected fish of 15 analyzed (40%), 2.1 ± 0.4 (1–4). Sapucaí-Mirim River: L. friderici: 27 infected fish of 40 analyzed (67.5%), 5.53 ± 1.31 (1–30); L. octofasciatus: 7 infected fish of 17 analyzed (41.18%), 2 ± 0.54 (1–4); M. elongatus: 1 infected fish of 22 analyzed (4.55%), 4.
Type-material: Holotype CHIOC (39970a); paratypes CHIOC (39970b, 39971, 39972); vouchers CHIBB (720L, 721L).
Molecular sequence data: LSU rDNA (1369 bp) and COI mtDNA (895 bp) partial sequences obtained of the same monogenean specimen from L. striatus were deposited in GenBank under accession numbers OQ843018 andOQ833544, respectively.
Etymology: The specific name derives from Latin (radix= root) and refers to the large size of the superficial and deep roots of the ventral anchor.
3.3.2. Description (Based on Nine Specimens Mounted in Gray and Wess’s Medium)
Body elongate, fusiform, 547 (415–651, n = 4) long, and 73 (34–100, n = 3) wide near midlength. Cephalic lobes poorly developed. Cephalic glands not visible. Four eyespots present. Anterior pair is smaller than posterior pair; distance between eyespots of anterior pair smaller than posterior one; one member of each pair sometimes absent. Accessory granules elongate, frequently in anterior trunk. Pharynx subspherical 19 (13–24, n = 7) wide. Peduncle short. Haptor sub-square, 50 (34–69, n = 4) long, 48 (40–54, n = 4) wide. Anchors dissimilar in shape and size. Ventral anchor 14 (13–14, n = 8) long, 8 (6–9, n = 8) wide, short point and shaft, superficial and deep root elongated, with small protuberance on the inner surface of base. Dorsal anchor 24 (23–25, n = 7) long, 9 (7–12, n = 7) wide, with elongate curved point, short and curved shaft, poorly developed deep root, elongated and square-like superficial root. Ventral bar 10 (6–12, n = 4) long, 25 (18–32, n = 6) wide, delicate, filamentous, and enlarged ends. Dorsal bar 7 (5–11, n = 7) long, 24 (17–27, n = 7) wide, broadly V-shaped. Seven pairs of marginal and similar hooks, 12 (10–13, n = 23) long, with erect thumb, slightly curved point, arcuated shank, and FH loop half of shank length. Copulatory complex comprising a MCO with 1½ counterclockwise rings 12 (11–13, n = 9) in diameter; and an accessory piece 29 (26–32, n = 9) long, 12 (9–16, n = 9) wide, robust, composed of two subunits: one ventral and more sclerotized, sickle-shaped, and another dorsal with three projections (proximal projection elongated, middle projection enlarged, and lateral, distal projection, spine-shaped), articulated with base of the MCO. Vagina simple, sinistral. Prostatic reservoir present. Oviduct, ootype, and uterus not observed. Vitellaria densely dispersed throughout trunk. Egg 95 (n = 1) long, 61 (n = 1) wide, without filament.
3.3.3. Remarks
Jainus radixelongatus n. sp. can be distinguished from its congeners mainly by the morphology of the accessory piece, which is composed of two subunits: one ventral and more sclerotized and sickle-shaped, and another dorsal with three projections (proximal projection, elongated; middle projection, enlarged and lateral; distal projection, spine-shaped). The morphologies of these subunits of the accessory piece are markedly different from those observed in Jainus ornatus n. sp. The new species resembles J. amazonensis by possessing a ventral anchor with superficial and deep roots with elongated termination; however, these species differ by the morphology of the dorsal bar (V-shaped in Jainus radixelongatus n. sp. and simple and arcuate in J. amazonensis) and the delicate ventral bar, being filamentous with enlarged ends in the new species and broadly V-shaped with small medial projection in J. amazonensis). Examination of paratypes of J. amazonensis described by Kritsky et al. [13] and deposited in the zoological collection of the Instituto Nacional de Pesquisas da Amazônia (INPA), Amazonas, Brazil, confirmed these resemblances and differences reported in the current study. There were not appropriate specimens for a whole-mount illustration.
3.4. Supplementary Observations of Jainus piava Karling, Bellay, Takemoto & Pavanelli, 2011
(Figure 5).
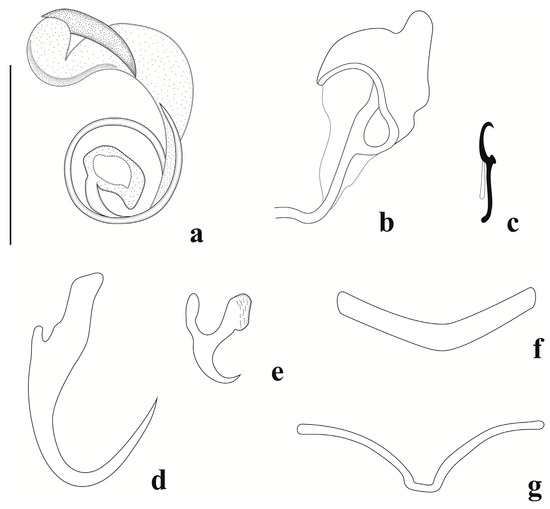
Figure 5.
Jainus piava from Schizodon nasutus (Characiformes, Anostomidae). (a) Copulatory complex (ventral view); (b) vagina; (c) hook; (d) dorsal anchor; (e) ventral anchor; (f) dorsal bar; (g) ventral bar (Scale bars: (a–g) = 20 µm).
3.4.1. Taxonomic Summary
Type host: Schizodon borellii (Boulenger, 1900) (Characiformes, Anostomidae).
Type-locality: Upper Paraná River floodplain, Brazil (22°50′–22°70′ S, 53°15′–53°40′ W).
Current records: Schizodon nasutus Kner, 1858 (Characiformes, Anostomidae) from Jurumirim Reservoir (23°12′17″ S, 49°13′19″ W) and Sapucaí-Mirim River (20°29′38.38″ S, 47°51′33.11″ W), Upper Paraná River basin, São Paulo, Brazil.
Site of infection: Gill filaments.
Prevalence, mean intensity of infection, and range of intensity: Jurumirim Reservoir: 10 infected fish of 30 analyzed (33.3%), 3.4 ± 0.9 (1–9). Sapucaí-Mirim River: 12 infected fish of 40 analyzed (30%), 2.86 ± 0.63 (1–7).
Specimens deposited: vouchers CHIOC (39973a, 39973b, 39974) and CHIBB (722L–724L).
Molecular sequence data: LSU rDNA (1377 bp) and COI mtDNA (651 bp) partial sequences, obtained from the same specimen, were deposited in GenBank under accession numbers OQ843019 and OQ833543, respectively.
3.4.2. Supplementary Observations (Based on 30 Specimens Mounted in Gray and Wess’s Medium)
Body elongate, fusiform 390 (221–642, n = 29) long; 85 (40–170, n = 26) wide near midlength. Two pairs of cephalic lobes; two pairs of head organs; cephalic glands not visible. Four eyespots; anterior pair smaller than the posterior one; distance between the eyespots of the anterior pair smaller than the posterior one. Accessory granules small, generally elongate, present in some specimens in the cephalic area. Pharynx subspherical, 19 (13–27, n = 25) wide; esophagus short. Peduncle short. Haptor sub-square, 52 (34–68, n = 25) long, and 47 (34–72, n = 25) wide. Anchors dissimilar in shape and size. Ventral anchor 10 (8–11, n = 28) long and 7 (6–9, n = 28) wide. It had a short point and shaft, an elongate deep root, a superficial root with a striate, and expanded terminal extension. Dorsal anchor 24 (22–26, n = 30) long and 9 (7–10, n = 30) wide, with an elongate curved point, encurved shaft, poorly developed deep root, and elongate superficial root. Ventral bar 9 (6–13, n = 22) long and 21 (12–28, n = 24) wide, delicate, and variable in shape. Dorsal bar 6 (4–8, n = 27) long, 23 (20–28, n = 30) wide, and arcuated. Seven pairs of similar and marginal hooks, 11 (11–12, n = 24) long, with erect thumb, curved point, and erect shank with slightly curved distal portion; FH loop ¾ shank length. Copulatory complex comprising a MCO with 1½ counterclockwise rings 12 (9–13, n = 26) in diameter, a sclerotized expanded base, and an accessory piece 21 (16–24, n = 28) long, well developed, variable in shape, not articulated to the MCO. Vagina sinistral, marginal, sclerotized, adorned, and connected to the seminal receptacle. Gonads intercaecal and overlapping. Germarium elongate, 59 (n = 1) long, and 12 (n = 1) wide. Testis elongate, dorsal to germarium, 86 (n = 1) long, and 46 (n = 1) wide. Seminal vesicle as a dilation of the vas deferens. Seminal receptacle anterior to the germarium. Prostatic reservoir present. Oviduct, ootype, and uterus not observed. Vitellaria densely dispersed throughout trunk, absent in reproductive organ regions.
3.4.3. Remarks
Jainus piava was described by Karling et al. [14] in the gills of S. borellii. Since then, its occurrence in several other host species has been reported. Analyzing the holotype and paratypes (CHIOC 37235A and CHIOC 37236, 37235B-C, respectively) we can confirm the identity of the specimens found in S. nasutus as J. piava. Nonetheless, comparative analyses of the morphology of the specimens analyzed for the original description and that found in the present study revealed differences not reported in the original description, such as the presence of a thin and delicate ventral bar, with great variation in its shape; a ventral anchor presenting a striated region at the end of the superficial root; an accessory piece not articulated with MCO base; and the ends of the accessory piece being smooth and not present as many undulations, as in the drawing of the original description.
3.5. Molecular Analyses
We successfully obtained two LSU rDNA partial sequences from specimens of Jainus radixelongatus n. sp. (GenBank accession number OQ843018; length of 1369 bp) and of Jainus piava (GenBank accession numberOQ843019; length of 1377 bp) and three COI mtDNA partial sequences from specimens of Jainus radixelongatus n. sp. (GenBank accession numberOQ833544; length of 895 bp), Jainus beccus n. sp. (GenBank accession number OQ833545; length of 903 bp), and J. piava (GenBank accession number OQ833543; length of 651 bp).
For the LSU rDNA gene, a dataset was constructed including the newly generated sequences of Jainus spp., 46 sequences of the Dactylogyridae retrieved from GenBank and 3 sequences of the Diplectanidae (used as outgroup) (Table 3). The final alignment was 1472 bp long considering the longest newly generated sequence (J. piavaOQ843019). The BI and ML phylogenetic analyses of the LSU rDNA produced phylograms with similar topologies to most clades supported (Figure 6). In both analyses, Jainus was recovered as monophyletic; Jainus radixelongatus n. sp. and J. piava were grouped together in a monophyletic clade (pp = 1; bootstrap = 100), closely related to Trinigyrus spp., Unilatus unilatus Mizelle & Kritsky, 1967 (MF 1021106), and Heteropriapulus spp., which are described as parasites of fishes belonging to Loricariidae (Siluriformes).
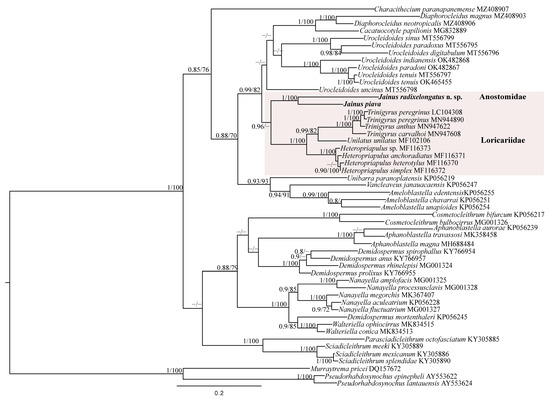
Figure 6.
Bayesian topology based on partial LSU rDNA sequences of selected monogenean species. Murraytrema pricei and Pseudorhabdosynochus spp. were used as the outgroup. GenBank accession numbers are after species names. Newly sequenced species are in bold. The support values are included above the nodes as follows: posterior probabilities for BI analyses, followed by bootstrapping for the ML analyses (posterior probabilities < 0.70 and bootstrap scores < 70 are not shown and are represented by dashes). The branch-length scale bar indicates the number of substitutions per site.
For the COI mtDNA gene, we constructed a dataset including the newly generated sequences of Jainus spp. and nine sequences of the Dactylogyridae retrieved from GenBank, and Sciadicleithrum panamensis Mendoza-Franco, Aguirre-Macedo & Vidal-Martínez, 2007 (MF939864) was used as the outgroup (Table 3). The BI and ML analyses of the partial COI mtDNA alignment also produced phylograms with similar topologies and most supported clades (Figure 7). The newly generated sequences of Jainus radixelongatus n. sp., Jainus beccus n. sp., and J. piava clustered in a monophyletic clade (pp = 0.88; bootstrap = 78), with Urocleidoides spp. and Diaphorocleidus spp. as sister groups (not supported by any of the analyses). The position of Trinigyrus clade appeared unresolved.
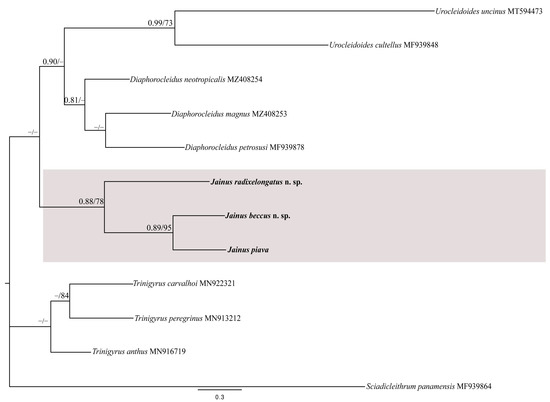
Figure 7.
Bayesian topology based on partial COI mtDNA sequences of dactylogyrids from the neotropical region. Sciadicleithrum panamensis was used as the outgroup. GenBank accession numbers are after species names. Newly sequenced species are in bold. The support values are included above the nodes as follows: posterior probabilities for BI, followed by bootstraps for the ML analyses (posterior probabilities < 0.70 and bootstrap scores < 70 are not shown and are represented by dashes). The branch-length scale bar indicates the number of substitutions per site.
The genetic divergence found in the LSU rDNA partial sequences between Jainus radixelongatus n. sp. and J. piava was 14.3% (213 bp), and the genetic divergence in the COI mtRNA partial sequences ranged from 14.6% (54 bp—J. piava versus J. beccus n. sp.) to 21.8% (80 bp—J. piava versus J. radixelongatus n. sp.).
4. Discussion
The new species proposed here were erected with support by a combination of the differences observed in the morphological and molecular data among Jainus spp. To date, nine species of Jainus (including the three new species here proposed) were described in Brazil. Only one, Jainus hexops Kritsky & Leiby, 1972, was reported to parasitize Astyanax fasciatus (Cuvier, 1819) (Characidae) from Puntarenas Province, Costa Rica. Species of this genus were reported to parasitize mainly characiforms belonging to Chalceidae, Anostomidae, Iguanodectidae, and Bryconidae [12,13,14,15,16,44].
In the present study, the monogenean species recovered from the gills of the anostomid fishes in the southeast of Brazil are allocated to Jainus due to their possession of a robust body with a poorly developed peduncle; one testis and one ovary in tandem or overlapping; a modified ventral anchor; a vaginal sinistral (not observed in Jainus ornatus n. sp.); and a male copulatory organ that is nonarticulated proximally (observed in Jainus beccus n. sp.) (see [12]). In addition, as emended by Karling et al. [14] they include: a male copulatory organ and accessory piece articulated proximally (observed in Jainus ornatus n. sp. and Jainus radixelongatus n. sp.); and a male copulatory organ with coils (observed in all new species described herein) or not. In general, Jainus spp. presents a robust accessory piece that is variable in shape; a coiled MCO; a delicate or filamentous ventral bar; a V-shaped dorsal bar; a delicate ventral anchor with modified deep and superficial roots; similar hooks; and a simple vagina. However, J. hexops runs outside of these morphological patterns by presenting a different haptoral hook and a sinuous MCO.
To date, there is only one sequence of Jainus spp. available in GenBank—that of J. hexops (OM397908)—which was not used in the final alignment because it was too short (299 bp) for the chosen alignment criteria. Nevertheless, we included it and tested it in previous alignments (not shown here); however, we obtained inconclusive results, since this sequence was not placed together with the other sequences of Jainus. The sequence identified as that of J. hexops (OM397908) might not belong to the genus Jainus; therefore, we recommend a revision of this species and an improvement in the sequence length to a better fit for future alignment and analyses. According to the phylogeny of the LSU, Jainus spp. (recovered from fishes belonging to Anostomidae, Characidae, and Bryconidae) are sister taxa to Trinigyrus spp., Unilatus unilatus (MF 1021106), and Heteropriapulus spp. (recovered from fishes belonging to Loricariidae). This relationship is not well defined, since there is no synapomorphy in the morphology. Therefore, more studies should be carried out in the future to confirm its relationship.
Our results described three new species of Jainus based on morphological and molecular data (partial LSU rDNA and COI mtDNA genes) and supplementary data on the morphology of J. piava. We provide the first phylogenetic study based on LSU rDNA and COI mtDNA gene sequences for species of this genus, improving and clarifying the understanding of host–parasite relationships in neotropical characiforms.
Author Contributions
Conceptualization, P.d.O.F.Y., A.C.Z. and F.H.Y.; Methodology, P.d.O.F.Y., A.C.Z., F.H.Y., L.F., M.I.M. and M.B.E.; Software, M.I.M. and M.B.E.; Validation, R.J.d.S.; Formal Analysis, P.d.O.F.Y., A.C.Z. and F.H.Y.; Investigation, P.d.O.F.Y., A.C.Z. and F.H.Y.; Data Curation, P.d.O.F.Y. and A.C.Z.; Writing—Original Draft Preparation, P.d.O.F.Y. and F.H.Y.; Writing—Review and Editing, P.d.O.F.Y., A.C.Z., F.H.Y., L.F., M.I.M., M.B.E. and R.J.d.S.; Visualization, M.I.M. and M.B.E.; Supervision, R.J.d.S.; Funding Acquisition, R.J.d.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by CNPq—National Council for Scientific and Technological Development (grant numbers—P.d.O.F.Y.: 140872/2017-5; F.H.Y.: 304502/2022-7; R.J.d.S. 311635/2021-0); FAPESP—São Paulo Research Foundation (grant numbers—M.I.M.: 2017/16546-3; A.C.Z.: 2011/23588-8, 2015/11542-4, 2016/07829-9; F.H.Y.: 2011/22603-3, 2014/14298-4; M.B.E: 2021/12779-9; L.F.: 2012/07850-7, 2015/11543-0; R.J.d.S.: 2020/05412-9); CAPES—Coordination for the Improvement of Higher Education Personnel (grant numbers—M.I.M.: 3005/2010); and Pro-Rectory of Research (PROPe—UNESP) (grant numbers—M.I.M.: 02/2016; M.B.E.: 04/2022). We would also like to thank Duke Energy and CELAN (Central Elétrica Anhanguera) for the financial and logistical support for the sampling expeditions to Sapucaí-Mirim River.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Marcela Fontes Bongiovani for assistance with parasite identifications; and Edmir Daniel Carvalho (in memoriam), Sandro Geraldo de Castro Britto (in memoriam), Diogo Freitas Souza, and Marcos Gomes Nogueira for their logistical support during the collections of fish hosts in the Sapucaí-Mirim River. We are also thankful to the curators of the parasitological collections of the museums CHIOC and INPA for lending us the monogenean paratypes and vouchers and providing some photomicrographs of the deposited specimens.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Eiras, J.C.; Takemoto, R.M.; Pavanelli, G.C.; Adriano, E.A. About the biodiversity of parasites of freshwater fish from Brazil. Fish Pathol. 2011, 31, 161–168. [Google Scholar]
- Kritsky, D.C.; Boeger, W.A.; Jegu, M. Neotropical Monogenoidea. 28. Ancyrocephalinae (Dactylogyridae) of Piranha and Their Relatives (Teleostei, Serrasalmidae) from Brazil and French Guiana: Species of Notozothecium Boeger and Kritsky, 1988, and Mymarothecium gen. n. J. Helminthol. 1996, 63, 153–175. [Google Scholar]
- Almeida, K.S.S.; Cohen, S.C. Diversidade de Monogenea (Platyhelminthes) parasitos de Astyanax altiparanae do reservatório da Usina Hidrelétrica de Itaipu. Saúde Ambiente em Revista. 2011, 6, 31–41. [Google Scholar]
- Cohen, S.C.; Kohn, A.; Justo, M.C.N. South American Monogenoidea Parasites of Fishes, Amphibians and Reptiles; Oficina de Livros: Rio de Janeiro, Brazil, 2013; 663p. [Google Scholar]
- Franceschini, L.; Zago, A.C.; Müller, M.I.; Francisco, C.J.; Takemoto, R.M.; Silva, R.J. Morphology and molecular characterization of Demidospermus spirophallus n. sp., D. prolixus n. sp. (Monogenea: Dactylogyridae) and a redescription of D. anus in siluriform catfish from Brazil. J. Helminthol. 2017, 92, 228–243. [Google Scholar] [CrossRef]
- Mendoza-Palmero, C.A.; Blasco-Costa, I.; Scholz, T. Molecular phylogeny of Neotropical monogeneans (Platyhelminthes: Monogenea) from catfishes (Siluriformes). Parasites Vectors 2015, 8, 1–11. [Google Scholar] [CrossRef]
- Acosta, A.A.; Franceschini, L.; Zago, A.C.; Scholz, T.; Silva, R.J. Six new species of Heteropriapulus (Monogenea: Dactylogyridae) from South American fishes with an amended diagnosis to the genus. Zootaxa 2017, 4290, 459–482. [Google Scholar] [CrossRef]
- Acosta, A.A.; Scholz, T.; Blasco-Costa, I.; Alves, P.V.; Silva, R.J. A new genus and two new species of dactylogyrid monogeneans from gills of Neotropical catfishes (Siluriformes: Doradidae and Loricariidae). Parasitol. Int. 2018, 67, 4–12. [Google Scholar] [CrossRef]
- Acosta, A.A.; Mendoza-Palmero, C.A.; Silva, R.J.; Scholz, T. A new genus and four new species of dactylogyrids (Monogenea), gill parasites of pimelodid catfishes (Siluriformes: Pimelodidae) in South America and the reassignment of Urocleidoides megorchis Mizelle et Kritsky, 1969. Folia Parasitol. 2019, 66, 1–12. [Google Scholar] [CrossRef]
- Poulin, R. The evolution of monogenean diversity. Int. J. Parasitol. 2002, 37, 245–254. [Google Scholar] [CrossRef]
- Braga, M.P.; Araújo, S.B.L.; Boeger, W.A. Patterns of interaction between Neotropical freshwater fishes and their gill Monogenoidea (Platyhelminthes). Parasitol. Res. 2014, 113, 481–490. [Google Scholar] [CrossRef]
- Mizelle, J.D.; Kritsky, D.C.; Crane, J.W. Studies on Monogenetic Trematodes. XXXVIII. Ancyrocephalinae from South America with the proposal of Jainus gen. n. Am. Midl. Nat. 1968, 80, 186–198. [Google Scholar] [CrossRef]
- Kritsky, D.C.; Thatcher, V.E.; Kayton, R.J. Neotropical Monogenea. 3. Five new species from South America with the proposal of Tereancistrum gen. n. and Trinibaculum gen. n. (Dactylogyridae: Ancyrocephalinae). Acta Amaz. 1980, 10, 411–417. [Google Scholar] [CrossRef]
- Karling, L.C.; Bellay, S.; Takemoto, R.M.; Pavanelli, G.C. A new species of Jainus (Monogenea), gill parasite of Schizodon borellii (Characiformes, Anostomidae) from the upper Paraná river floodplain, Brazil. Acta Sci. Biol. Sci. 2011, 33, 227–231. [Google Scholar] [CrossRef]
- Cohen, S.C.; Kohn, A.; Boeger, W.A. Neotropical Monogenoidea. 57. Nine new species of Dactylogyridae (Monogenoidea) from the gill of Salminus brasiliensis (Characidae, Characiformes) from the Paraná River, State of Paraná, Brazil. Zootaxa 2012, 3049, 57–68. [Google Scholar] [CrossRef]
- Abdallah, V.D.; Azevedo, R.K.; Luque, L.F. Three new species of Monogenea (Platyhelminthes) parasites of fish in the Guandu river, southeastern Brazil. Acta Sci. Biol. Sci. 2012, 34, 483–490. [Google Scholar] [CrossRef]
- Kritsky, D.C.; Thatcher, V.E.; Boeger, W.A. Neotropical Monogenea. 8. Revision of Urocleidoides (Dactylogyridae, Ancyrocephalinae). Proc. Helminthol. Soc. Wash. 1986, 53, 1–37. [Google Scholar]
- Kritsky, D.C.; Boeger, W.A.; Thatcher, V.E. Neotropical Monogenea. 7. Parasites of the Pirarucu, Arapaima gigas (Cuvier), with descriptions of two new species and redescription of Dawestrema cycloancistrium Price and Nowlin, 1967 (Dactylogyridae: Ancyrocephalinae). Proc. Biol. Soc. Wash. 1985, 98, 321–331. [Google Scholar]
- Mizelle, J.D. New species of trematodes from the gills of Illinois fishes. Am. Midl. Nat. 1936, 17, 785–806. [Google Scholar] [CrossRef]
- Bush, A.O.; Laferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitolgy meets ecology on its own terms: Margolis revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef]
- Pleijel, F.; Jondelius, U.; Norlinder, E.; Nygren, A.; Oxelman, B.; Schander, C.; Sundberg, P.; Thollesson, M. Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Mol. Phylogenet. Evol. 2008, 48, 369–371. [Google Scholar] [CrossRef]
- Plaisance, L.; Rousset, V.; Morand, S.; Littlewood, T.D.J. Colonization of Pacific islands by parasites of low dispersal ability: Phylogeography of two monogenean species parasitizing butterfly fishes in the South Pacific Ocean. J. Biogeogr. 2008, 35, 65–87. [Google Scholar] [CrossRef]
- Lockyer, A.E.; Olson, P.D.; Littlewood, D.T.J. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): Implications and a review of the cercomer theory. Biol. J. Linn. Soc. 2003, 78, 155–171. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Miller, M.A.; Schwartz, T.; Pickett, B.E.; He, S.; Klem, E.B.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol. Bioinform. 2015, 16, 43–48. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4. Molecular Evolution, Phylogenetics and Epidemiology. 2012. Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 8 August 2022).
- Yamada, F.H.; Acosta, A.A.; Yamada, P.O.F.; Scholz, T.; Silva, R.J. A new species of Aphanoblastella Kritsky, Mendoza-Franco and Scholz, 2000 (Monogenea, Dactylogyridae) parasitic on heptapterid catfish (Siluriformes) in the Neotropical region. Acta Parasitol. 2018, 63, 772–780. [Google Scholar] [CrossRef]
- Zago, A.C.; Franceschini, L.; Müller, M.I.; Silva, R.J. A new species of Cacatuocotyle (Monogenea, Dactylogyridae) parasitizing Astyanax spp. (Characiformes, Characidae) from Brazil, including molecular data and a key to species identification. Acta Parasitol. 2018, 63, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Zago, A.C.; Franceschini, L.; Abdallah, V.D.; Müller, M.I.; Azevedo, R.K.; Silva, R.J. Morphological and molecular data of new species of Characithecium and Diaphorocleidus (Monogenea: Dactylogyridae) from Neotropical characid fishes. Parasitol. Int. 2021, 84, 102406. [Google Scholar] [CrossRef] [PubMed]
- Alda, F.; Reina, R.G.; Mendoza, E.; Rios, E.; Torchin, M.E. Freshwater Fish Parasite Barcoding Project (Panama. Museum of Natural Science); Louisiana State University: Baton Rouge, LA, USA, 2017. [Google Scholar]
- Mendoza-Palmero, C.A.; Blasco-Costa, I.; Hernandez-Mena, D.; Perez-Ponce de Leon, G. Parasciadicleithrum octofasciatum n. gen., n. sp. (Monogenoidea: Dactylogyridae), parasite of Rocio octofasciata (Regan) (Cichlidae: Perciformes) from Mexico characterised by morphological and molecular evidence. Parasitol. Int. 2017, 66, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, L.; Acosta, A.A.; Zago, A.C.; Müller, M.I.; Silva, R.J. Trinigyrus spp. (Monogenea: Dactylogyridae) from Brazilian catfishes: New species, molecular data and new morphological contributions to the genus. J. Helminthol. 2020, 94, E126. [Google Scholar] [CrossRef] [PubMed]
- Nitta, M.; Nagasawa, K. Four Alien Monogeneans, Including Trinigyrus peregrinus n. sp., Parasitic on the Invasive Armored Catfish Pterygoplichthys disjunctivus (Siluriformes: Loricariidae) from Okinawa-jima Island, Okinawa Prefecture, Japan. Spec. Div. 2016, 21, 95–104. [Google Scholar] [CrossRef]
- Zago, A.C.; Yamada, F.H.; Yamada, P.O.F.; Franceschini, L.; Bongiovani, M.F.; Silva, R.J. Seven new species of Urocleidoides (Monogenea: Dactylogyridae) from Brazilian fishes supported by morphological and molecular data. Parasitol. Res. 2020, 119, 3255–3283. [Google Scholar] [CrossRef]
- Oliveira, M.S.B.; Santos-Neto, J.F.; Tavares-Dias, M.; Domingues, M.V. New species of Urocleidoides (Monogenoidea: Dactylogyridae) from the gills of two species of Anostomidae from the Brazilian Amazon. Braz. J. Vet. Parasitol. 2020, 29, e007820. [Google Scholar] [CrossRef]
- Mendoza-Palmero, C.A.; Mendoza-Franco, E.F.; Acosta, A.A.; Scholz, T. Walteriella n. g. (Monogenoidea: Dactylogyridae) from the gills of pimelodid catfishes (Siluriformes: Pimelodidae) from the Peruvian Amazonia based on morphological and molecular data. Syst. Parasitol. 2019, 96, 441–452. [Google Scholar] [CrossRef]
- Wu, X.Y.; Zhu, X.Q.; Xie, M.Q.; Li, A.X. The radiation of Haliotrema (Monogenea: Dactylogyridae: Ancyrocephalinae): Molecular evidence and explanation inferred from LSU rDNA sequences. Parasitology 2006, 132, 659–668. [Google Scholar] [CrossRef]
- Wu, X.Y.; Li, A.X.; Zhu, X.Q.; Xie, M.Q. Description of Pseudorhabdosynochus seabassi sp. n. (Monogenea: Diplectanidae) from Lates calcarifer and revision of the phylogenetic position of Diplectanum grouperi (Monogenea: Diplectanidae) based on rDNA sequence data. Folia Parasitol. (Praha). 2005, 52, 231–240. [Google Scholar] [CrossRef]
- Kritsky, D.C.; Leiby, P.D. Dactylogyridae (Monogenea) from the Freshwater Fish, Astyanax fasciatus (Cuvier), in Costa Rica, with Descriptions of Jainus hexops sp. n., Urocleidoides costaricensis, and U. heteroancistrium combs. n. Proc. Helminthol. Soc. Wash. 1972, 39, 227–230. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).