Abstract
Dicephalospora is a genus of Helotiaceae (Helotiales) that presently comprises 14 species, all of which were collected from Asia. The current study describes three additional species and a collection from Chiang Rai and Chiang Mai Provinces, Thailand. The new fungi were identified based on morphological characteristics coupled with phylogenetic analyses of combined LSU and ITS nrDNA loci. Dicephalospora chiangraiensis is characterized by small asci (90–110 × 5–10 µm) and ascospores of 20–25 × 2–4 µm, featuring a non-mucilaginous cap. Dicephalospora irregularis is characterized by sessile apothecia, non-amyloid asci, branched, filiform paraphyses, and fusoid-clavate to ellipsoid ascospores with a mucilaginous cap, while D. inthanonensis is characterized by unbranched and aseptate paraphyses, a partly globose blue reaction with Melzers reagent at ascal apices, and fusoid ascospores in the range of 24–32 × 3–5 µm with a non-mucilaginous cap. With the present study, the number of species of Dicephalospora known from Thailand has now increased to three. A dichotomous key to the species of the genus is also provided.
1. Introduction
Helotiaceae Whetzel is one of the largest families of Helotiales (Leotiomycetes), typified by Helotium Pers. (1801). Their genera have been traditionally divided based on their excipular structure, hairs, and the morphology of their asci and paraphyses [1]. The family has been subjected to several revisions [2,3,4], and many molecular studies have been conducted to resolve the phylogeny [2,3,4,5,6,7].
Dicephalospora Spooner is a genus in Helotiaceae typified by D. calochroa (Syd. and P. Syd.) Spooner [8]. The genus was established to accommodate two inoperculate cup fungi in Sclerotiniaceae found on stromatized plant tissues and characterized by an ectal excipulum of textura prismatica, asci with amyloid tips in Melzer’s reagent, and mucilaginous-capped ascospores [8]. The sclerotiniaceous fungi were divided into two clades [9]. One clade contains Botryotinia, Ciboria, Ciborinia, Lambertella, Lanzia, Monilinia, Rutstroemia, and Sclerotinia. The other clade includes Dicephalospora and Lanzia, i.e., two genera that stromatize plant tissues and do not produce sclerotia [9]. Dicephalospora has historically been included in Rutstroemiaceae [10], Helotiaceae, and Sclerotiniaceae [4]. However, recent phylogenetic studies clearly showed the affinities of Dicephalospora to Helotiaceae [2,6,11,12].
Dicephalospora species are usually found on rotten wood, twigs and leaf petioles [13]. The distinguishing characteristics of this genus are erumpent or superficial, stipitate, yellow, orange, red to blackish apothecia, an ectal excipulum composed of cells of textura prismatica, and medullary excipulum composed of cells of textura intricata, filiform, paraphyses straight or slightly curved at the apex, and hyaline, subellipsoid to fusoid, guttulate ascospores with a mucilaginous cap at the poles [14]. Ascospore shape and size are criteria for identifying Dicephalospora species [9]. Dicephalospora was reported to produce an interesting secondary metabolite named dicephalosterol. This triterpenoid from D. rufocornea (Berk. and Broome) Spooner inhibits testosterone 5α-reductase, a key enzyme that is regarded as a valid target for the development of drugs against prostatic hypertrophy [14]. There are 14 Dicephalospora species listed in the Index Fungorum [15], out of which 13 were reported from China (D. rufocornea, D. sessilis, D. dentate, D. calochroa, D. huangshanica, D. aurantiaca, D. yunnanica, D. damingshanica, D. pinglongshanica, D. albolutea, D. shennongjiana, D. phaeoparaphysis, and D. contracta [2,9,16]). In this study, we introduce three new species in Thailand, D. chiangraiensis, D. irregularis, and D. inthanonensis, based on morphological characteristics and phylogenetic data.
2. Materials and Methods
2.1. Sample Collection and Examination of Specimens
Dicephalospora specimens were collected from northern Thailand. Specimens were examined using a Motic SMZ-168 stereomicroscope (Motic Instruments, Richmond, BC, Canada). Sections of the fruiting bodies were mounted in water and preserved in lacto-glycerol to examine the morphological characteristics. Photographs were taken by using a Canon (Ota, Japan) EOS 600D digital camera connected to a Nikon Ni compound microscope (Nikon Corporation, Tokyo, Japan). The photo-plate was made using Adobe Photoshop CS6 Extended version 13.0 × 64 (Adobe Systems, San José, CA, USA). Measurements were taken using the Tarosoft (R) Image Frame Work program v. 0.9.7 (Tarosoft, Nontha Buri, Thailand). Specimens were deposited at the Mae Fah Luang University (MFLU) herbarium, Chiang Rai, Thailand.
2.2. DNA Extraction, PCR Amplification, Sequencing and Phylogenetic Analysis
Genomic DNA was extracted directly from the apothecia using the Forensic DNA Kit-D3591-01 (OMEGA bio-tek). The internal transcribed spacer (ITS) and the 28S large subunit rRNA (LSU) gene regions were amplified using primer pairs ITS5/ITS4 [17] and LR0R/LR5 [18], respectively. The total volume of PCR mixtures was 25 μL, containing 8.5 μL ddH2O, 12.5 μL 2× Easy Taq PCR SuperMix (Beijing Trans Gen Biotech Co., Chaoyang District, Beijing, China), 2 μL of DNA template, and 1 μL of each forward and reverse primers (10 pM). PCR amplification conditions for all regions were as follows: an initial denaturation step of 5 min at 94 °C, 35 cycles of denaturation at 94 °C for 1 min, annealing at 53 °C for 50 s, elongation at 72 °C for 3 min, and a final extension step at 72 °C for 10 min. The quality of PCR products was checked on 1% agarose gel stained with ethidium bromide. The PCR products were sent for sequencing at Sangon Biotech (Shanghai, China).
Sequences of available Dicephalospora species were analyzed (Table 1) using nucleotide BLAST searches in GenBank and also from recent publications [2]. Pleuroascus nicholsonii (CBS 345.73) and Connersia rilstonii (CBS 537.74) from the Bulgariella clade (Helotiales) were used as outgroup taxa. The LSU and ITS sequence datasets were selected to construct the phylogenetic tree. The sequences were assembled with Geneious Prime v2019.1.1. The datasets were compiled with BioEdit v.7.2.5 [19], manually trimmed, and aligned with MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html, accessed on 30 June 2022) using default settings [20]. The individual gene alignments were analyzed separately for any conflicts in the tree. The phylogeny website tool “ALTER” [21] was used to convert the alignment file for maximum likelihood (ML) analysis. For Bayesian analysis, Readseq (2.2) on CIPRES was used to change to nexus file format. All the newly generated sequence data were deposited in GenBank (Table 1), and the alignment was deposited in the Supplementary Materials.
Reconstruction of the maximum likelihood (ML) analysis was performed via RAxMLv.9 on the CIPRES web portal as part of the “RAxML-HPC2 on XSEDE 8.2.10 on TG tool” (http://www.phylo.org/potal2/) [22]. RAxML rapid bootstrapping and subsequent ML search used distinct model/data partitions with joint branch length optimization executing 1000 rapid bootstrap inferences and thereafter a thorough ML search. All free model parameters were estimated by RAxML and ML estimate of 25 per site rate categories. The likelihood of the final tree was evaluated and optimized under GTR-GAMMA. The GAMMA model parameters were estimated to an accuracy of 0.1000000000 log-likelihood units. Every 100th tree was saved. Phylogenetic trees were illustrated using the FigTree v1.4.0 program [23]. The Bayesian command was generated using Fabox 1.41 [24]. The best-fit model was determined using MrModeltest v.2.3 and selected the TrN+I+G model for the LSU gene and TIM2+I+G for the ITS gene [25].
The Bayesian posterior probability analysis was performed using MrBayes 3.2.6 run on XSEDE at the CIPRES web portal [26], using the parameter settings of two parallel runs, four chains, run for 2,000,000 generations, and sample frequency at every 100th generation. The first 8000 trees representing the burn-in phase of the analyses were discarded, and the remaining 32,000 trees were used to calculate posterior probabilities (BYPP) in the majority rule consensus tree (critical value for the topological convergence diagnostic set to 0.01).
2.3. Pairwise Homoplasy Index (PHI) Analysis
The genealogical concordance phylogenetic species recognition (GCPSP) model with a pairwise homoplasy index (PHI) test was used to analyze new species and the most closely related species [27]. The recombination level within phylogenetically closely related species was determined by the PHI test performed in SplitsTree4 [28,29]. The PHI analysis was conducted separately for each single loci and the combined LSU and ITS dataset. Two separate analyses were performed for Dicephalospora chiangraiensis and Dicephalospora inthanonensis, along with their most closely related taxa. The relationships of these two species were visualized in split graphs with both the LogDet transformation and splits decomposition options. A pairwise homoplasy index below the 0.05 threshold (ϕw < 0.05) indicated the presence of significant recombination in the dataset.


Table 1.
Taxa used in the phylogenetic analysis and their GenBank accession numbers. New taxa are printed in bold.
Table 1.
Taxa used in the phylogenetic analysis and their GenBank accession numbers. New taxa are printed in bold.
| Species | Isolate 1 | GenBank Accession No. 2 | Reference | |
|---|---|---|---|---|
| LSU | ITS | |||
| Amylocarpus encephaloides Curr. 1859 | CBS 129.60 | MH869464 | MH857920 | [30] |
| Amylocarpus encephaloides | 017cN | KM272361 | KM272369 | [31] |
| Bryoscyphus dicrani (Ade and Höhn.) Spooner 1984 | M141 | EU940107 | EU940183 | [32] |
| g Connersia rilstonei (C. Booth) Malloch 1974 | CBS 537.74 | AF096189 | KJ755499 | [33] |
| Crocicreas amenti (Batsch) S.E. Carp. 1980 | F-147481 | FJ005124 | FJ005093 | [34] |
| Crocicreas cacaliae (Pers.) S.E. Carp. 1980 | F-148706 | FJ005126 | FJ005107 | [34] |
| Crocicreas tomentosum (Dennis) S.E. Carp. 1980 | MFLU 17-0082 | MK592008 | MK584988 | [2] |
| Crocicreas cyathoideum (Bull.) S.E. Carp. 1980 | MFLU 18-0698 | MK591970 | MK584943 | [2] |
| Cudoniella clavus (Alb. and Schwein.) Dennis 1964 | AFTOL-ID 166 | DQ470944 | DQ491502 | [35] |
| Cyathicula microspora Velen. 1934 | M267 | EU940088 | EU940165 | [36] |
| Dicephalospora albolutea H.D. Zheng and W.Y. Zhuang 2019 | HMAS 279693 | - | MK425601 | [16] |
| Dicephalospora aurantiaca (W.Y. Zhuang) W.Y. Zhuang and Z.Q. Zeng 2016 | MFLU 16-0591a | MK591988 | MK584962 | [2] |
| Dicephalospora aurantiaca | MFLU 16-0591b | - | MK584958 | [2] |
| Dicephalospora chiangraiensis K. Phutthacharoen and K.D. Hyde 2022 | MFLU 21-0020 * | MZ241828 | MZ241819 | In this study |
| Dicephalospora chiangraiensis | MFLU 21-0019 * | MZ241827 | MZ241818 | In this study |
| Dicephalospora chiangraiensis | MFLU 21-0018 * | MZ241826 | MZ241817 | In this study |
| Dicephalospora chrysotricha (Berk.) Verkley 2004 | PDD:93932 | - | MH578487 | Unpublished |
| Dicephalospora chrysotricha | PDD:91762 | - | KF727411 | Unpublished |
| Dicephalospora chrysotricha | PDD:81537 | - | KF727410 | Unpublished |
| Dicephalospora chrysotricha | PDD:58197 | - | KF727409 | Unpublished |
| Dicephalospora dentata Xiao X. Liu and W.Y. Zhuang 2015 | 3093 | - | KP204263 | [9] |
| Dicephalospora huangshanica (W.Y. Zhuang) W.Y. Zhuang and Z.Q. Zeng 2016 | MFLU 18-1828 | MK591979 | MK584979 | [2] |
| Dicephalospora huangshanica | HMAS 279694 | - | MK425602 | [16] |
| Dicephalospora huangshanica | HMAS 74836 | - | DQ986485 | [9] |
| Dicephalospora huangshanica | HMAS 81363 | - | DQ986483 | [9] |
| Dicephalospora huangshanica | HMAS 81364 | - | DQ986484 | [9] |
| Dicephalospora huangshanica | KUS-F52405 | JN086711 | JN033408 | [6] |
| Dicephalospora inthanonensis K. Phutthacharoen, Chethana and K.D. Hyde 2022 | MFLU 22-0050 * | ON606312 | ON604634 | In this study |
| Dicephalospora inthanonensis | MFLU 22-0053 * | ON606313 | ON604635 | In this study |
| Dicephalospora irregularis Lestari, Pasouvang and K.D Hyde 2022 | MFLU 22-0054 * | ON514038 | ON511117 | In this study |
| Dicephalospora rufocornea (Berk. and Broome) Spooner 1987 | 10106 | - | KU668565 | [14] |
| Dicephalospora rufocornea | HMAS 275559 | MH729336 | - | [16] |
| Dicephalospora rufocornea | HMAS 279695 | - | MK425603 | [16] |
| Dicephalospora rufocornea | HMAS 279696 | - | MK425604 | [16] |
| Dicephalospora rufocornea | HMAS 279697 | - | MK425605 | [16] |
| Dicephalospora rufocornea | HMAS 75518 | - | DQ986480 | [9] |
| Dicephalospora rufocornea | JS140813-06 | KP161277 | - | [37] |
| Dicephalospora rufocornea | KUS F52274 | JN086704 | JN033401 | [6] |
| Dicephalospora rufocornea | MFLU 16-0585 | MK591984 | MK584955 | [2] |
| Dicephalospora rufocornea | MFLU 16-1858 | MK584991 | MK592010 | [2] |
| Dicephalospora rufocornea | MFLU 16-1860 | MK592011 | MK584989 | [2] |
| Dicephalospora rufocornea | MFLU 18-0674a | MK584959 | - | [2] |
| Dicephalospora rufocornea | MFLU 18-0674b | MK591989 | MK584960 | [2] |
| Dicephalospora rufocornea | MFLU 18-0675 | MK591987 | MK584961 | [2] |
| Dicephalospora rufocornea | MFLU 18-1825 | MK591976 | MK584949 | [2] |
| Dicephalospora rufocornea | MFLU 18-1827 | MK591978 | MK584978 | [2] |
| Dicephalospora rufocornea | MHHNU 8663 | - | MK253761 | Unpublished |
| Dicephalospora rufocornea | TNS:F-40024 | AB926123 | AB926055 | [11] |
| Dicephalospora rufocornea | MFLU 19-2085 * | MZ241829 | MZ241820 | In this study |
| Dicephalospora rufocornea | MFLU 19-2073 * | - | MZ241814 | In this study |
| Dicephalospora rufocornea | MFLU 19-2082 * | MZ241824 | MZ241815 | In this study |
| Dicephalospora rufocornea | MFLU 19-2083 * | MZ241825 | MZ241816 | In this study |
| Dicephalospora rufocornea | MFLU 19-2087 * | MZ241830 | MZ241821 | In this study |
| Dicephalospora rufocornea | MFLU 19-2089 * | MZ241831 | MZ241822 | In this study |
| Dicephalospora rufocornea | MFLU 19-2071 * | - | MZ241813 | In this study |
| Dicephalospora sessilis Ekanayaka and K.D. Hyde 2019 | MFLU 18-1823 | MK591974 | NR_163779 | [2] |
| Dicephalospora shennongjiana H.D. Zheng and W.Y. Zhuang 2019 | HMAS 279698 | - | MK425606 | [16] |
| Dicephalospora yunnanica H.D. Zheng and W.Y. Zhuang 2019 | HMAS 279701 | - | MK425609 | [16] |
| Dicephalospora yunnanica | HMAS 279700 | - | MK425608 | [16] |
| Dicephalospora yunnanica | HMAS 279699 | - | MK425607 | [16] |
| Dicephalospora yunnanica | HMAS 61850 | - | DQ986486 | [16] |
| Endoscypha perforans Syd. 1924 | PDD:102231 | MK039717 | KF727424 | Unpublished |
| Glarea lozoyensis Bills and Peláez 1999 | ATCC 20868 | - | NR_137138 | [38] |
| Glarea sp. | C2B | - | KX610435 | [39] |
| Gloeotinia granigena (Quél.) T. Schumach. 1979 | CBS 417.50 | MH868212 | - | [30] |
| Hymenoscyphus fructigenus (Bull.) Gray 1821 | CBS 186.47 | MH867741 | MH856211 | [30] |
| Hymenoscyphus occultus Andr. Gross and J.G. Han 2015 | KUS_F52847 | - | KP068064 | [40] |
| Hymenoscyphus pseudoalbidus Queloz, Grünig, Berndt, T. Kowalski, T.N. Sieber and Holdenr. 2011 | Hokk_14 | - | KJ511191 | [41] |
| Hymenotorrendiella eucalypti (Berk.) P.R. Johnst., Baral and R. Galán 2014 | PDD:70105 | - | MH578483 | Unpublished |
| Lanzia berggrenii (Cooke and W. Phillips) Spooner 1987 | ICMP:19614 | KC164640 | KC164645 | [42] |
| Ombrophila violacea Fr. 1849 | WZ0024 | AY789365 | AY789366 | [43] |
| Phaeohelotium epiphyllum (Pers.) Hengstm. 2009 | TNS:F_40042 | AB926130 | AB926061 | [44] |
| Pirottaea palmicola P.R. Johnst. 1998 | PDD:60282 | - | KM677208 | Unpublished |
| Pirottaea palmicola | PDD:65971 | - | KM677206 | Unpublished |
| Pleuroascus nicholsonii Massee and E.S. Salmon 1901 | CBS 345.73 | AF096196 | KJ755519 | [45] |
| Roesleria subterranea (Weinm.) Redhead 1985 | CBS 339.96 | EF608074 | EF060308 | [46] |
| Roesleria subterranea | CBS 407.51 | - | MH856922 | [30] |
| Torrendiella eucalypti (Berk.) Spooner 1987 | CPC 11050 | DQ195800 | DQ195788 | [44] |
| Torrendiella madsenii (G.W. Beaton and Weste) Spooner 1987 | PRJ D672 | KJ606676 | AY755336 | [42] |
1 CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. MFLU: Mae Fah Luang University, Thailand. M: HMAS: JS: KUS: MHHNU: TNS: National Museum of Nature and Science, Tsukuba, Japan. PDD: New Zealand Fungal and Plant Disease Herbarium. ICMP: International Collection of Microorganisms from Plants. 2 LSU: 28S large subunit rRNA. ITS: Internal transcribed spacer. Ex-type strains are indicated in bold. The new strains are indicated by an asterisk (*).
3. Results
3.1. Molecular Phylogeny
A phylogram (Figure 1) was generated from maximum likelihood (ML) analysis based on combined LSU and ITS sequence data for the family Helotiaceae. Seventy-nine strains were included in the combined analyses and the dataset comprised 1287 characteristics after alignment (832 characteristics for LSU and 455 characteristics for ITS), including gaps. Single gene analyses were performed to compare the topology and clade stability with combined gene analyses. The tree topology of the ML analysis is similar to the BI analysis. The best RaxML tree with a final likelihood value of −8372.349462 is presented in Figure 1. The ML analysis resulted in 489 distinct alignment patterns, with 31.10% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.235011, C = 0.232700, G = 0.281575, T = 0.250714; substitution rates AC = 1.529216, AG = 2.397210, AT = 1.365721, CG = 0.605708, CT = 6.021470, GT = 1.000000; gamma distribution shape parameter α = 0.577342. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with the final average standard deviation of split frequencies = 0.009064. A separate analysis of ITS sequence data was performed to confirm our new species. The same dataset was used for this analysis with seventy-six strains. The best RaxML tree with a final likelihood value of −5024.994 is presented in Figure 2. The ML analysis resulted in 264 distinct alignment patterns, with 30.02% undetermined characteristics or gaps. Estimated base frequencies were as follows: A = 0.250, C = 0.250, G = 0.250, T = 0.250; substitution rates AC = 1.91925, AG = 3.16738, AT = 1.91925, CG = 1.00000, CT = 6.94378, GT = 1.000000; gamma distribution shape parameter α = 0.638. Bayesian posterior probabilities (BYPP) from MCMC were evaluated with the final average standard deviation of split frequencies = 0.00293.
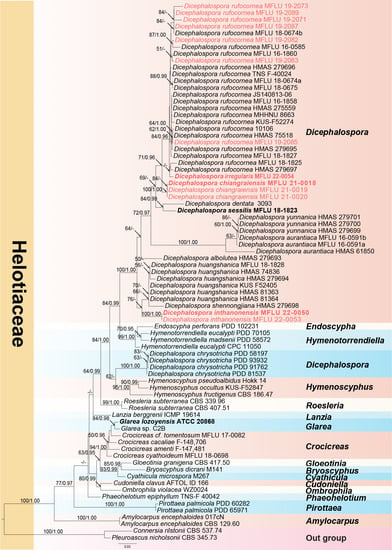
Figure 1.
Phylogram of RAxML analysis based on combined LSU and ITS sequence dataset. Bootstrap support values for maximum likelihood equal to or greater than 50%, and Bayesian posterior probabilities equal to or greater than 0.95 BYPP are indicated at the nodes as ML/BYPP. The tree is rooted to Connersia rilstonii (CBS 537.74) and Pleuroascus nicholsonii (CBS 345.73). Ex-type strains are in bold. The newly generated sequences are in red font.
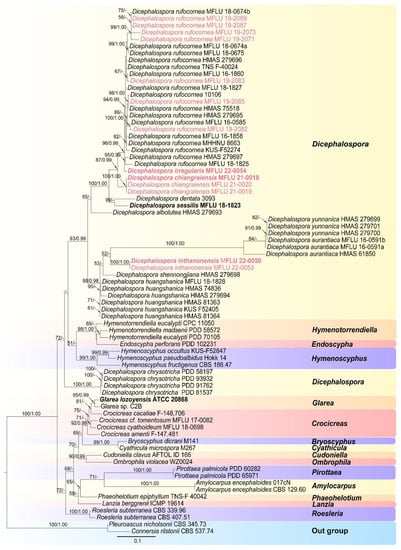
Figure 2.
Phylogram of RAxML analysis based on ITS sequence dataset. Bootstrap support values for maximum likelihood equal to or greater than 50%, and Bayesian posterior probabilities equal to or greater than 0.95 BYPP are indicated at the nodes as ML/BYPP. The tree is rooted to Connersia rilstonii (CBS 537.74) and Pleuroascus nicholsonii (CBS 345.73). Ex-type strains are in bold. The newly generated sequences are in red font.
As shown in Figure 1, Dicephalospora yunnanica grouped together with strains MFLU 16-0591a and MFLU 16-0591b. This evidence suggests that MFLU 16-0591a and MFLU 16-0591b are conspecific with D. yunnanica. The morphology of MFLU 16-0591a and MFLU 16-0591b is also similar to that of D. yunnanica except for the amyloid ascal apex, which is lacking in D. yunnanica. This evidence is in our opinion insufficient to segregate the Thai specimens from Dicephalospora yunnanica. Sequence data of additional DNA loci and a study of additional specimens will be needed to resolve this species complex.
3.2. Pairwise Homoplasy Index (PHI) Analysis
In the phylogenetic analyses, our novel taxa (D. chiangraiensis, D. irregularis, and D. inthanonensis) clustered separately from other known taxa. To further confirm their evolutionary independence, a pairwise homoplasy index was determined for our novel species. Dicephalospora chiangraiensis shares similar morphologies with D. rufocornea and D. irregularis. Similarly, D. inthanonensis shares similarities with D. huangshanica. A pairwise homoplasy index (PHI or ϕw) below 0.05 provides evidence for the presence of significant recombination within a dataset. According to our analysis, our dataset showed a PHI of 0.99, indicating no significant genetic recombination between D. chiangraiensis and its sister taxa, D. rufocornea and D. irregularis (Figure 3). Hence, it is concluded that these taxa were significantly different. The analysis conducted for D. inthanonensis and D. huangshanica resulted in a PHI of 1.0, indicating no significant genetic recombination between D. inthanonensis and its sister taxon, D. huangshanica (Figure 4). Hence, it is concluded that these two taxa are significantly different.
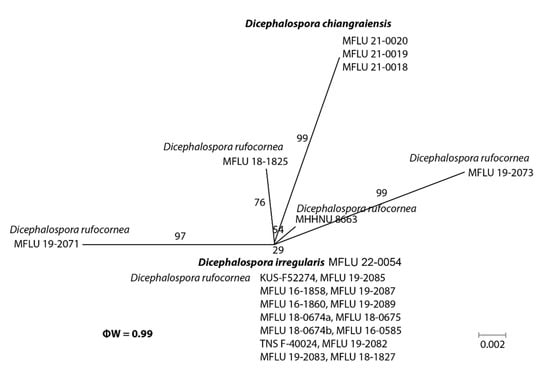
Figure 3.
The results of the pairwise homoplasy index (PHI) test for the closely related species of Dicephalospora chiangraiensis using both LogDet transformation and splits decomposition. PHI test results (ϕw) < 0.05 indicate significant recombination within the dataset.
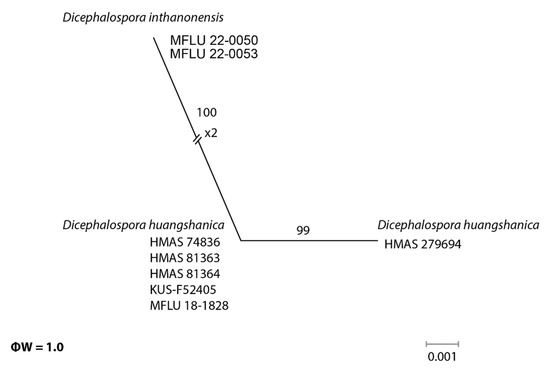
Figure 4.
The results of the pairwise homoplasy index (PHI) test for the closely related species of Dicephalospora inthanonensis using both LogDet transformation and splits decomposition. PHI test results (ϕw) < 0.05 indicate significant recombination within the dataset.
3.3. Taxonomy
Dicephalospora chiangraiensis K. Phutthacharoen & K.D. Hyde, sp. nov. (Figure 5).

Figure 5.
Dicephalospora chiangraiensis (MFLU 21-0018, holotype). (a,b) D. chiangraiensis on the stem; (c) cross section of the apothecium; (d) close up of the excipulum; (e) margin of the apothecium; (f) filiform paraphyses; (g) cylindrical asci; (h) apical part of asci in Melzer’s reagent; (i) fusiform ascospores. Scale bars: (a,b) = 500 µm; (c) = 100 µm; (d) = 50 µm; (e–g) = 20 µm; (h,i) = 5 µm.
- Index Fungorum number:—IF558614, Facesoffungi number:—FOF09715
- Etymology:—“chiangraiensis” refers to the locality where the fungus was collected.
- Holotype:—MFLU 21-0018
Saprobic on dead stems. Sexual morph: Apothecia 290–850 × 250–500 µm (x = 420 × 360 µm, n = 10), when dry arising solitary or gregarious, scattered on wood, central short stipitate, superficial, orange when fresh, flat cupulate. The stipe is 250–350 µm (x = 300 µm, n = 10). The receptacle is flat. The disc is flat and orange. The margins are orange and smooth. The ectal excipulum is 70–150 µm (x = 80 µm, n = 10), multi-layered, and thin-walled, with hyaline to light yellow cells of textura porrecta, small cells condensed, and small globose at the tips. The medullary excipulum is 30–60 µm (x = 55 µm, n = 10) and multi-layered, with hyaline to light orange cells of textura intricate and small cells condensed. The hymenium is yellow and intensive with asci and paraphyses inside. The paraphyses are 1.5–2.5 µm wide (x = 2 µm, n = 40), thin-walled, numerous, filiform, aseptate, and swollen at the apex. The asci are 90–110 × 5–10 µm (x = 100 × 7.5 µm, n = 20), eight-spored, unitunicate, inoperculate, long and cylindrical, and thin-walled, with a tapered long stipitate base, blunt apices, and an apical apparatus bluing in Melzer’s reagent. Ascospores are 20–25 × 2–4 µm (x = 22 × 3 µm, n = 20), biseriate, hyaline, fusiform, pseudo-septate, and thin-walled, with a non-mucilaginous cap.
The details of the material examined are as follows: Thailand, Chiang Rai Province, Wiang Chiang Rung District, on dead stem, 22 August 2018, Kunthida Phuttacharoen, HMS03 (MFLU 21-0018, holotype); ibid., HMS04 (MFLU 21-0019, paratype), ibid., HMS05 (MFLU 21-0020, paratype).
The GenBank accession information is as follows: MFLU 21-0018: LSU-MZ241826, ITS-MZ241817; MFLU 21-0019: LSU-MZ241827, ITS-MZ241818; MFLU 21-0020: LSU-MZ241828, ITS-MZ241819.
Notes: Our collection grouped sister to Dicephalospora rufocornea with 71% ML and 0.70 BYPP support (Figure 1). Furthermore, the pairwise homoplasy index (PHI) showed no significant genetic recombination between D. chiangraiensis and its sister taxa D. rufocornea and D. irregularis (Figure 2). Dicephalospora chiangraiensis differs from D. rufocornea in having smaller, orange apothecia (290–850 × 250–500 vs. 500–4000 × 300–3000 µm). The ectal excipulum of D. chiangraiensis is composed of textura porrecta cells, while the cells are textura prismatica to textura epidermoidea in D. rufocornea. Paraphyses of our species were more swollen at the apex than D. rufocornea. Asci (90–110 × 5–10 vs. 135–170 × 10–15 µm) and ascospores (20–25 × 2–4 vs. 20–30 × 4–8 µm) of new species are smaller when compared to D. rufocornea.
Dicephalospora inthanonensis K. Phutthacharoen, Chethana and K.D. Hyde, sp. nov. (Figure 6).

Figure 6.
Dicephalospora inthanonensis (MFLU 22-0050, holotype). (a,b) Dicephalospora species on a leaf; (c,d) cross section of the apothecium; (e) close up of the excipulum; (f) filiform paraphyses and asci; (g) fusiform ascospores; (h) apical part of ascus in Melzer’s reagent; (i) cylindrical asci. Scale bars (a) = 1 mm; (b) = 400 µm; (c) = 200 µm; (d) = 100 µm; (f,i) = 20 µm; (g) = 10 µm; (h) = 5 µm.
- Index Fungorum number: IF559758; Facesoffungi number: FOF12543.
- Etymology: “inthanonensis” refers to the locality where the fungus was collected.
- Holotype: MFLU 22-0050.
Saprobic on dead leaf. Sexual morph: Apothecia 0.3–0.5 diameter mm, when dry arising solitary or gregarious in a small group, scattered on wood, centrally stipitate, superficial, yellow to orange when fresh, become red when dry. The stipe is 0.3–0.7 mm and dark at the base. The receptacle is orange and cupulate. The margins are smooth and orange to dark orange. The disc is slightly convex and orange or red wine. The ectal excipulum is composed of thick-walled, hyaline to light orange, gelatinized yellowish cells of textura globulosa to textura angularis. The medullary excipulum is composed of thin-walled, hyaline to yellowish, gelatinized cells of textura porrecta to textura intricata. The hymenium is hyaline to yellowish and inner mixed with asci and paraphyses. Paraphyses are 2.0–3.0 µm wide (x = 2.5 µm, n = 25) at the terminal cell, filiform, and numerous, and have lengths exceeding the asci; they are unbranched, aseptate, and are swollen, small, and globose at the apex. Asci are 95–115 × 8–11 µm (x = 105 × 9 µm, n = 20), eight-spored, unitunicate, cylindrical, and clavate, with the amyloid (J+) having a rounded apex in Melzer’s reagent. Ascospores are 24–32 × 3–5 µm (x = 27 × 4 µm, n = 15), uniseriate to biseriate, and fusiform, with a non-mucilaginous cap. Asexual morphology: undetermined.
The details of the material examined are as follows: Thailand, Chiang Mai Province, Chom Thong District, Doi Inthanon National Park, Kew Mae Pan nature trail, on a dead leaf, 20 October 2021, Kunthida Phuttacharoen, KMPD3.6 (MFLU 22-0050, holotype); ibid., KMPD3.6a (MFLU 22-0051, isotype); ibid., KMPD3.6b (MFLU 22-0052, isotype); ibid., Ang Ka nature trail, on dead leaf, 18 October 2021, Kunthida Phuttacharoen, AG2.15 (MFLU 22-0053, paratype).
The GenBank accession information is as follows: MFLU 22-0050: LSU-ON604634, ITS-ON606312; MFLU 22-0053: LSU-ON604635, ITS-ON606313.
Notes: Our specimens MFLU 22-0050 and MFLU 22-0053 grouped as a sister clade to Dicephalospora shennongjiana HMAS 279698 and Dicephalospora huangshanica clade (Figure 1), with 76% ML bootstrap support and 0.94 Bayesian posterior probabilities. Furthermore, the pairwise homoplasy index (PHI) showed no significant genetic recombination between D. inthanonensis and its sister taxa D. huangshanica (Figure 4).
Dicephalospora inthanonensis differs from D. shennongjiana and D. huangshanica by the shape of its ascopores, paraphyses, and the Melzer reaction at the apex of asci. Our new species has fusiform ascospores, unbranched and aseptate paraphyses, and a partly globose, blue reaction observed with Melzer’s reagent at ascal apices. In contrast, D. shennongjiana has elliptical-subfusoid ascospores, branched and septate paraphyses, and two blue lines observed with Melzer’s reagent at ascal apices [15]. Hence, these two species can be distinguished by morphology. Our species differs from D. huangshanica by having textura globulosa to textura angularis cells in the ectal excipulum and aseptate paraphyses, whereas D. huangshanica has textura prismatica cells in the ectal excipulum and septate paraphyses [13]. Comparison of ITS sequences showed 2.5% base pair differences between D. inthanonensis and D. huangshanica. Dicephalospora inthanonensis also shares a similar ascospore morphology with D. chiangraiensis, but D. inthanonensis differs by the presence of small yellow guttules in the paraphyses. However, they are distantly related in the phylogenetic tree. A comparison of LSU and ITS sequences of D. inthanonensis with D. chiangraiensis shows 2.43% and 6.5% base pair differences, respectively.
Dicephalospora irregularis Lestari, Pasouvang and K.D Hyde, sp. nov. (Figure 7).
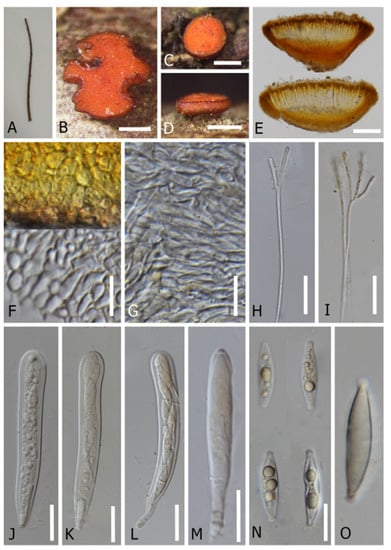
Figure 7.
Dicephalospora irregularis (MFLU 22-0054, holotype). (A) Dead twig substrate; (B–D) top and side view of apothecia; (E) a cross section of an apothecium; (F) cells of ectal excipulum; (G) cells of medullary excipulum; (H,I) a branched paraphyses; (J–M) asci (mounted in Melzer’s reagent; (N,O) ascospores (mounted in KOH 5%). Scale bars: (B–D) = 500 µm; (E) = 200 µm; (F,G) = 13 µm; (H–L) = 25 µm; (M) = 30 µm; (N) = 20 µm.
Index Fungorum number: IF555157; Facesoffungi number: FOF11034.
Etymology: “irregularis” refers to irregular shape of apothecia.
Holotype: MFLU 22-0054.
It is saprobic on dead twigs of an unidentified tree. Sexual morph: Apothecia 0.5–1.3 × 0.3–1.2 mm, arising singly or in small groups, sessile, erumpent, and irregular in shape. The receptacle is cupulate and orange to yellow-brown. The margins are smooth and brown to dark orange. The disc is slightly convex and orange. The ectal excipulum is 30–45 µm (x = 40 µm, n = 15) in lower flanks, and composed of thick-walled, yellowish cells of textura globulosa to textura angularis. The medullary excipulum is 42–63 µm (x = 48.8 µm, n = 15) in lower flanks, and composed of thin-walled, yellowish to hyaline cells of textura porrecta to textura intricata. The hymenium is hyaline to yellowish. Paraphyses are 2.5–4.0 µm wide (x = 3 µm, n = 15) at the terminal cell, filiform, and numerous, with length exceeding the asci, unbranched to branched at the top with gelatinous matters, and are apically rounded, aseptate, and guttulate. Asci are 125–150 × 15–20 µm (x = 135 × 17 µm, n = 15), eight-spored, unitunicate, cylindrical clavate, and non-amyloid (J- when mounted in Melzer agent with or without KOH treatment), with a rounded apex, arising from simple septa without basal protuberance. Ascospores are 25–35 × 5.5–7.5 µm (x = 33 × 7.5 µm, n = 15), uniseriate to biseriate, and fusoid-clavate to ellipsoid, with rounded to sub-acute ends, capped with gelatinous obconical collar, and are hyaline and guttulate. Asexual morph: Undetermined.
The details of the material examined are as follows: Thailand, Chiang Mai Province, Mae Taeng District, Pa Pae, on dead stem and twig of an unknown tree, 09 September 2020, Pahua Pasouvang, CM 31 (MFLU 22-0054, holotype).
The GenBank accession information is as follows: ITS-ON511117, LSU-ON514038.
Notes: Our collection MFLU 22-0054 grouped sister to Dicephalospora rufocornea MFLU 18-1825 and HMAS 279697 (Figure 1). The ITS sequence of our specimen shows 96.7% and 98% similarity to the D. rufocornea (MFLU 18-1825) and D. rufocornea (HMAS 279697), respectively, while the LSU data of our specimen show 99% similarity to D. rufocornea (MFLU 18-1825) across 829 bp.
Phylogenetically, our species clusters basal to the Dicephalospora rufocornea clade with 84% ML bootstrap support and 0.71 posterior probability. Our specimen, D. irregularis, is morphologically distinct from D. rufocornea by having sessile apothecia, ectal excipulum of textura globulosa to textura angularis, non-amyloid asci, and fusoid-clavate to ellipsoid ascospores, while D. rufocornea is characterized by having stipitate apothecia, ectal excipulum of textura porrecta to prismatica, hymenoscyphus-type apical ring, and fusoid or fusoid-clavate ascospores [2,8]. Dicephalospora irregularis differs from D. sessilis in apothecial shape and branched, filiform paraphyses [2], and our species is more distantly related to D. sessilis than D. rufocornea in the phylogenetic tree (Figure 8, Figure 9 and Figure 10).

Figure 8.
Apothecial diversity in Dicephalospora rufocornea. (a,b,f,h) Dry apothecia on wood; (a) MFLU 19-2071; (b) MFLU 19-2073; (f) MFLU 19-2085; (h) MFLU 19-2082; (c–e,g,i,j) fresh apothecia on the substrate; (c,d) MFLU 19-2083; (e) MFLU 19-2082; (g) MFLU 19-2085; (i) MFLU 19-2087; (j) MFLU 19-2089. Scale bars: (a,e–g,i) = 0.5 mm; (b–d,j) = 1 mm; (h) = 0.2 mm.
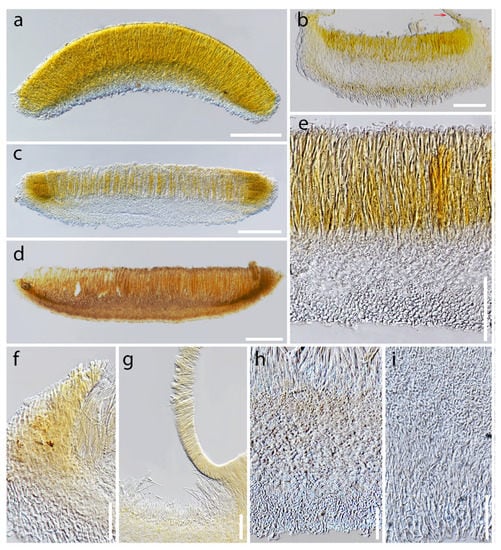
Figure 9.
Apothecial cross sections and excipulum characteristics of Dicephalospora rufocornea. (a–d) Cross section of apothecia; (a) MFLU 19-2085; (b) MFLU 19-2071 arrow pointed out the covered receptacle part; (c) MFLU 19-2089; (d) MFLU 19-2073; (e) close up of the excipulum (MFLU 19-2083); (f) margin of the apothecium (MFLU 19-2082); (g) receptacle covered by apothecium (MFLU 19-2071); (h) ectal excipulum textura epidermoidea (MFLU 19-2073); (i) ectal excipulum textura prismatica (MFLU 19-2071). Scale bars: (a,c,d) = 200 µm; (b,e) = 100 µm; (f–i) = 50 µm.

Figure 10.
Asci and ascospore characteristics of Dicephalospora rufocornea. (a–c) Filiform paraphyses; (a) MFLU 19-2083 in Melzer’s reagent; (b) MFLU 19-2085; (c) MFLU 19-2083; (d,e) ascospores; (d) MFLU 19-2083; (e) MFLU 19-2082; (f) amyloid ascal apex in Melzer’s reagent (MFLU 19-2089); (g,h) ellipsoidal to fusiform ascospores; (g) MFLU 19-2071; (h) MFLU 19-2085; (i) fusiform ascospores (MFLU 19-2073); (j) amyloid ascal apex in Melzer’s reagent (MFLU 19-2071); (k,l) fusiform ascospores; (k) MFLU 19-2083; (l) MFLU 19-2087. Scale bars (a–c) = 20 µm; (d,e) = 30 µm; (f–l) = 10 µm.
Index Fungorum number: IF130937; Facesoffungi number:—FOF05900.
Saprobic on dead stems. Sexual morph: Apothecia 0.5–4 × 0.3–3 µm (x = 1 × 0.7 µm, n = 60), when dry arise solitary or gregarious in a small group, scattered on wood, superficial, are yellow to orange when fresh, and become red when dry, with centrally short stipitate. The stipe is 400–700 µm (x = 500 µm, n = 60) and dark at the base. The receptacle is cupulate. The disc is flat to concave and orange or red wine. Margins are orange or red more than the disc, and smooth. The ectal excipulum is 65–140 µm (x = 95 µm, n = 50) and multi-layered, with hyaline to light orange cells of textura prismatica or textura epidermoidea. The medullary excipulum is 60–100 µm (x = 75 µm, n = 50), multi-layered, and hyaline to light orange, with cells of textura intricata. The hymenium is yellow rather than light orange and inter mixed with asci and paraphyses. Paraphyses are 2–3 µm wide (x = 2.5 µm, n = 100), numerous, filiform, thin-walled, aseptate, and branched at the lower part. Asci are 135–170 × 10–15 µm (x = 150 × 10 µm, n = 100), eight-spored, unitunicate, inoperculate, long cylindrical, and thin-walled, with a tapered short stipitate base, blunt apices, and the amyloid ascal apex bluing in Melzer’s reagent. Ascospores are 20–30 × 4–8 µm (x = 25 × 6 µm, n = 100), partially biseriate, hyaline, ellipsoidal to fusiform, thick-walled, and guttulate, with a mucilaginous cap.
The details of the material examined are as follows: Thailand, Chiang Mai, Mae On District, on dead stem, 16 May 2018, Kunthida Phuttacharoen, CM023 (MFLU 19-2071); ibid., CM025 (MFLU 19-2073); ibid., Mueang Chiang Mai District, 8 August 2018, Kunthida Phuttacharoen, DCM10 (MFLU 19-2082); ibid., DCM11 (MFLU 19-2083); ibid., PD03 (MFLU 19-2089); ibid., Mae Taeng District, 7 August 2018, Kunthida Phuttacharoen, MRC02 (MFLU 19-2085); ibid., MRC05 (MFLU 19-2087) (reference specimens).
The GenBank accession information is as follows: MFLU 19-2085: LSU-MZ241829, ITS-MZ241820; MFLU 19-2073: ITS-MZ241814; MFLU 19-2082: LSU-MZ241824, ITS-MZ241815; MFLU 19-2083: LSU-MZ241825, ITS-MZ241816; MFLU 19-2087: LSU-MZ241830, ITS-MZ241821; MFLU 19-2089: LSU-MZ241831, ITS-MZ241822; MFLU 19-2071: ITS-MZ241813.
Notes: Our collection from northern Thailand clustered with Dicephalospora rufocornea, a species with worldwide distribution. Most have been reported from different provinces (Yunnan, Honghe, Jinghong, Xishuangbanna, Zeng Ming, etc.) in China. In Thailand, D. rufocornea has been reported in Chiang Rai and Chiang Mai provinces. Dicephalospora rufocornea is characterized by yellowish or red apothecia, a central stipe, and ascospores ranging from 27–39 × 4–6 µm, following the taxonomic key of Zheng and Zhuang [16]. ITS and LSU sequences of our isolates are 99% similar to other D. rufocornea specimens in the dataset. However, their morphologies are slightly different from our collection. For example, the apothecial color varies between yellow, orange, and red. In other collections, the receptacle part is usually bare, but our collection (MFLU 19-2071) had receptacle cells covered by the apothecia. The ectal excipulum is composed of textura prismatica and epidermoidea. The ascospores in our collection are 25 × 6 µm, within the range indicated in the taxonomic key [16].
A dichotomous key to the species of Dicephalospora
- 1.
- Sessile apothecia…………………………………………………………….………………………2
- -
- Stipitate apothecia……………………………………………………………………………………5
- 2.
- Receptacle with surface hairs.…………………………………………………...…D. chrycotricha
- -
- Receptacle without hairs……………………………………………………………………………3
- 3.
- Asci J+……………………………………………………………………………………D. calochroa
- -
- Asci J-.……………………………………………………………………………..……………..……4
- 4.
- Disc concave with unbranched paraphyses..……………………….…….……………D. sessilis
- -
- Disc slightly convex with branched paraphyses…………………..….……………D. irregularis
- 5.
- Margin dentate..……………………………….…………………………………………D. dentata
- -
- Margin not dentate…………………………………………………………………………………6
- 6.
- Disc cream to yellowish, white apothecia.…………………………………….……D. albolutea
- -
- Disc concolorous……………………………………….……………………………………..……7
- 7.
- Paraphyses with dark pigment contents…………………………….………D. phaeoparaphysis
- -
- Paraphyses without dark pigment contents ……………………………….……………………8
- 8.
- Asci J-…………………………………………………………………………….D. pinglongshanica
- -
- Asci J+……………………………………………………….………………………………………9
- 9.
- Ascospore cap mucilaginous.………………………………………………………………….10
- -
- Ascospore cap non-mucilaginous...……………………………………………………………11
- 10.
- Ascospore lemon-shaped, 9−12.7 μm wide..………………………………D. damingshanica
- -
- Ascospore fusoid, 27–39 × 4–6 µm wide………………………………….………D. rufocornea
- 11.
- Ascospores constricted in the middle……………………………………………D. contracta
- -
- Ascospores not constricted in the middle……………………………………………………12
- 12.
- Ascospores elliptical-subfusoid………………………………………...….D. shennongjiana
- -
- Ascospores fusoid ………………………………………………………………………………13
- 13.
- Disc convex……….……………………….…………………………………D. inthanonensis
- -
- Disc flat …………………………………………………………………………………………14
- 14.
- Paraphyses septate ………………………………….…………………………...…………15
- -
- Paraphyses aseptate ……………………………………….…………….……………………16
- 15.
- Ascospores multiseriate, ectal excipulum globose at the tips.…………D. huangshanica
- -
- Ascospores biseriate, 16.5−25.3 × 3.3−3.5 μm...…………………………………D. yunnanica
- 16.
- Ascospore width more than 4 µm………………………….…………………D. aurantiaca
- -
- Ascospore width less than 4 µm, ectal excipulum globose at the tips……D. chiangraiensis
4. Discussion
Dicephalospora now comprises 17 species, including the 3 new species from the current study, and 13 species were reported from China. Among these 17 species, 3 species were reported from Thailand. Only one species, Dicephalospora chrysotricha (Berk.) Verkley, was reported from New Zealand [16]. From the previous studies, the data show Dicephalospora is mostly found in the high humid and cold areas. In this study, three of the new species also found in the northern part Thailand, and one of them, D. inthanonensis, was found at Doi Inthanon, the international park located 2500 m above sea level. The morphology among the species is very similar, however, slight differences are observed in morphology even within the same species. For example, the current study shows the apothecial and ascospore diversity observed within Dicephalospora rufocornea isolates even though the specimens were collected in the same province of Thailand. This study shows that Dicephalospora is very difficult to identify by using only the morphology characteristics, although the same species in the same province also have some differences in characteristics.
Moreover, DNA sequences also play an important role in delineating fungal species in Helotiales [47]. In this study, three new species were introduced based on morphology and phylogeny. The phylogenetic placement of Helotiaceae in our study is similar to that reported by Ekanayaka and coauthors [2]. Eleven species in this genus were grouped with strong 84% ML bootstrap and 0.99 BYPP support (Figure 1). Further, D. chrysotricha grouped with Hymenoscyphus and formed a separate clade with other species of Dicephalospora, with moderate 62% ML bootstrap and 0.95 BYPP support (Figure 1). The salient discriminatory characters of D. chiangraiensis are smaller asci and smaller ascospores compared to the closely related D. rufocornea. They grouped with D. rufocornea with moderate 71% ML bootstrap and 0.96 BYPP support. The base pair comparison for the ITS gene region of D. chiangraiensis with the D. rufocornea revealed 21 base pair differences (4.04%), which is also reflected in the differed morphology. Furthermore, no significant genetic recombination was found between D. chiangraiensis and D. rufocornea. Dicephalospora inthanonensis showed similar morphological characteristics to D. chiangraiensis, but they differ significantly with molecular evidence, both in multi-gene phylogeny and the pairwise homoplasy index. However, the base pair composition between D. irregularis and D. rufocornea were identical. In the multi-gene phylogenetic analysis, these two species show a closer relationship and clustered together in the split tree, but significant differences were observed in their morphology. Thus, base pair comparisons and phylogeny should not be the only criteria for defining new species.
Prior to this study, three Dicephalospora species were identified from Thailand. Hyde and colleagues [48,49] suggested that more than 93% of fungi collected from Thailand are new to science. In the current study, three novel Dicephalospora species were found only in northern Thailand. This shows that there is a need for extensive research throughout Thailand to discover these discomycetes fungal taxa.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14080645/s1, Alignment of the phylogeny in FASTA format.
Author Contributions
Conceptualization, K.D.H.; methodology, K.P., K.W.T.C. and M.S.; validation, K.D.H., K.W.T.C. and M.S.; formal analysis, K.P. and A.S.L.; investigation, K.P.; resources, K.P. and A.S.L.; writing—original draft preparation, K.P.; writing—review and editing, K.D.H., K.W.T.C. and M.S.; supervision, K.D.H., K.W.T.C. and M.S.; funding acquisition, K.D.H., K.P., M.S. and K.W.T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Royal Golden Jubilee PhD Program under the Thailand Research Fund (RGJ), no. PHD/0002/2560, entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Sub region” (grant number RDG6130001) and Mae Fah Luang University entitled “Assessing the taxonomy of Discomycetes and plant pathogenic fungi in Doi Inthanon National Park, Thailand” (grant no. 641A01002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
Shaun Pennycook is thanked for checking and suggesting corrections to the Latin species names. We are immensely grateful to Kriangkrai Chaiphicet, the Director of the Doi Inthanon National Park, and all his staff for their immense help during our collecting trips to explore different areas of the national park in search of discomycetes. K.W. Thilini Chethana would like to thank the National Research Council of Thailand for providing permission to conduct research in the Doi Inthanon National Park, Thailand (no. 0402/2703) and the Mae Fah Luang University (grant no. 641A01002), entitled “Assessing the taxonomy of Discomycetes and plant pathogenic fungi in Doi Inthanon National Park, Thailand”. The authors would like to thank the National Research Council of Thailand for the project entitled “Comparison of diversity and biogeographical distribution of Ascomycetous fungi from two protected areas in Turkey and Thailand” (project no. P-19-52624 and permission no. 0402/2803).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cannon, P.F.; Kirk, P.M. Fungal Families of the World; CABI: Wallingford, UK, 2007. [Google Scholar]
- Ekanayaka, A.H.; Hyde, K.D.; Gentekaki, E.; McKenzie, E.H.C.; Zhao, Q.; Bulgakov, T.S.; Camporesi, E. Preliminary classification of Leotiomycetes. Mycosphere 2019, 10, 310–489. [Google Scholar] [CrossRef]
- Johnston, P.R.; Quijada, L.; Smith, C.A.; Baral, H.-O.; Hosoya, T.; Baschien, C.; Pärtel, K.; Zhuang, W.-Y.; Haelewaters, D.; Park, D.; et al. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. Int. Mycol. Assoc. Fungus 2019, 10, 1. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Crous, P.W.; Quaedvlieg, W.; Hansen, K.; Hawksworth, D.L.; Groenewald, J.Z. Phacidium and Ceuthospora (Phacidiaceae) are congeneric: Taxonomic and nomenclatural implications. Int. Mycol. Assoc. Fungus 2014, 5, 173–193. [Google Scholar] [CrossRef]
- Han, J.-G.; Hosoya, T.; Sung, G.-H.; Shin, H.-D. Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biol. 2014, 118, 150–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Binder, M.; Schoch, C.L.; Johnston, P.R.; Spatafora, J.W.; Hibbett, D.S. evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): A nuclear RDNA phylogeny. Mol. Phylogenetics Evol. 2006, 41, 295–312. [Google Scholar] [CrossRef]
- Spooner, B.M. Helotiales of Australasia: Geoglossaceae, Orbiliaceae, Sclerotiniaceae, Hyaloscyphaceae. Bibl. Mycol. 1987, 116, 1–711. [Google Scholar]
- Liu, X.-X.; Zhuang, W.-Y.; Zeng, Z.-Q.; Zhao, P. Newly discovered sclerotiniaceous fungi from China. Nova Hedwig. 2016, 102, 347–357. [Google Scholar] [CrossRef]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainsworth & Bisby’s Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2008. [Google Scholar]
- Zhao, Y.-J.; Hosaka, K.; Hosoya, T. Taxonomic re-evaluation of the genus Lambertella (Rutstroemiaceae, Helotiales) and allied stroma-forming fungi. Mycol. Prog. 2016, 15, 1215–1228. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Zhuang, W.Y.; Zeng, Z.Q.; Liu, X.X. Taxonomic revision of the genus Dicephalospora, Helotiales in China. Mycosystem 2016, 35, 791–801. [Google Scholar] [CrossRef]
- Hosoya, T.; Hamano, K.; Sugano, M.; Ogura, Y.; Hatano, E.; Hamada, T. Discovery of Dicephalosterol, a new testosterone 5α-reductase inhibitor, and some new mycological aspects of its producer, Dicephalospora rufocornea (Sclerotiniaceae, Discomycetes). Mycoscience 1999, 40, 525–529. [Google Scholar] [CrossRef]
- Index Fungorum—Search Page. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 3 March 2022).
- Zheng, H.-D.; Zhuang, W.-Y. Three new species of Dicephalospora from China as revealed by morphological and molecular evidences. MycoKeys 2019, 55, 87–99. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Hall, T.; BioEdit. Ibis Therapeutics, Carlsbad. 2004. Available online: http://www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 24 January 2021).
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented conversion of dna and protein alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree: Tree Figure Drawing Tool Version 1.4.0; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2016; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 24 January 2021).
- Villesen, P. FaBox: An online toolbox for fasta sequences. Mol. Ecol. Notes 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2. program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal dna barcodes boosts coverage for Kingdom Fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Rämä, T.; Mathiassen, G.H.; Kauserud, H. Marine fungi new to Norway, with an outlook to the overall diversity. Agarica 2014, 35, 35–47. [Google Scholar]
- Stenroos, S.; Laukka, T.; Huhtinen, S.; Döbbeler, P.; Myllys, L.; Syrjänen, K.; Hyvönen, J. Multiple origins of symbioses between ascomycetes and bryophytes suggested by a five-gene phylogeny. Cladistics 2010, 26, 281–300. [Google Scholar] [CrossRef]
- Suh, S.-O.; Blackwell, M. Molecular phylogeny of the cleistothecial fungi placed in Cephalothecaceae and Pseudeurotiaceae. Mycologia 1999, 91, 836. [Google Scholar] [CrossRef]
- Peláez, F.; Collado, J.; Platas, G.; Overy, D.P.; Martín, J.; Vicente, F.; González del Val, A.; Basilio, A.; De la Cruz, M.; Tormo, J.R. Phylogeny and intercontinental distribution of the pneumocandin-producing anamorphic fungus Glarea lozoyensis. Mycology 2011, 2, 1–17. [Google Scholar] [CrossRef][Green Version]
- Spatafora, J.W.; Sung, G.-H.; Johnson, D.; Hesse, C.; O’Rourke, B.; Serdani, M.; Spotts, R.; Lutzoni, F.; Hofstetter, V.; Miadlikowska, J. A five-gene phylogeny of Pezizomycotina. Mycologia 2006, 98, 1018–1028. [Google Scholar] [CrossRef]
- Baral, H.-O.; De Sloover, J.; Huhtinen, S.; Laukka, T.; Stenroos, S. An emendation of the genus Hyaloscypha to include Fuscoscypha (Hyaloscyphaceae, Helotiales, Ascomycotina). Karstenia 2009, 49, 1–17. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, C.; Lee, H.B. Eight previously unreported species of fungi identified in Mt. Manggyeong, Korea. Korean J. Mycol. 2014, 42, 344–348. [Google Scholar] [CrossRef]
- Bills, G.F.; Platas, G.; Peláez, F.; Masurekar, P. Reclassification of a pneumocandin-producing anamorph, Glarea lozoyensis gen. et. sp. nov., previously identified as Zalerion arboricola. Mycol. Res. 1999, 103, 179–192. [Google Scholar] [CrossRef]
- Yokoya, K.; Postel, S.; Fang, R.; Sarasan, V. Endophytic fungal diversity of Fragaria vesca, a crop wild relative of strawberry, along environmental gradients within a small geographical area. PeerJ 2017, 5, e2860. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Han, J.G. Hymenoscyphus fraxineus and two new Hymenoscyphus species identified in Korea. Mycol. Prog. 2015, 14, 19. [Google Scholar] [CrossRef]
- Gross, A.; Hosoya, T.; Queloz, V. Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol. Ecol. 2014, 23, 2943–2960. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.R.; Park, D. The phylogenetic position of Lanzia berggrenii and its sister species. Mycosystema 2013, 32, 366–385. [Google Scholar]
- Wang, Z.; Binder, M.; Hibbett, D. Life history and systematics of the aquatic discomycete Mitrula (Helotiales, Ascomycota) based on cultural, morphological, and molecular studies. Am. J. Bot. 2005, 92, 1565–1574. [Google Scholar] [CrossRef]
- Zhao, Y.-J. Taxonomic Study of Lambertella (Rutstroemiaceae, Helotiales) and Allied Substratal Stroma Forming Fungi from Japan; University of Tsukuba: Tsukuba, Japan, 2014. [Google Scholar]
- Malloch, D.; Sigler, L.; Hambleton, S.; Vanderwolf, K.J.; Gibas, C.F.C.; McAlpine, D.F. Fungi associated with hibernating bats in New Brunswick caves: The genus Leuconeurospora. Botany 2016, 94, 1171–1181. [Google Scholar] [CrossRef]
- Kirchmair, M.; Neuhauser, S.; Buzina, W.; Huber, L. The taxonomic position of Roesleria subterranea. Mycol. Res. 2008, 112, 1210–1219. [Google Scholar] [CrossRef]
- Jeewon, R. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Chaiwan, N.; Gomdola, D.; Wang, S.; Monkai, J.; Tibpromma, S.; Doilom, M.; Wanasingh, D.N.; Mortime, P.E.; Lumyong, S.; Hyde, K.D. https://Gmsmicrofungi.Org: An Online Database Providing Updated Information of Microfungi in the Greater Mekong Subregion. Mycosphere 2021, 12, 1513–1526. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).