Abstract
Two new species of Globisporangium, G. pengfuense and G. tenuihyphum, that were isolated from soybean in China are described and illustrated based on their morphological characters and molecular evidence. The characteristics of G. pengfuense include globose to sub-globose sporangia that are: catenulate, terminal, occasionally with apical papillae or intercalary, smooth oogonia, mostly monoclinous, occasionally diclinous antheridia, fist-shaped to crook-necked, with straight or curved antheridial cells, and plerotic or nearly plerotic with thin-walled oospores (0.5–1.0 µm). Globisporangium tenuihyphum differs from other species in this genus by its relatively narrow hyphae that are: mostly sub-globose to globose, occasionally ovoid-obpyriform sporangia, smooth oogonia, monoclinous antheridia, subclavate, falcate or semicircular to subcircular antheridial cells, and with aplerotic oospores. Phylogenetically, data from the combined internal transcribed spacer (ITS) and cytochrome c oxidase subunit I (Cox1) sequences support the designation of G. pengfuense and G. tenuihyphum as two distinct species of Globisporangium (Pythium sensu lato clades G and J). In addition, the two new species were compared to closely related Globisporangium species, to delineate their phylogenetic positions and morphological features.
1. Introduction
Pythium Pringsheim was established by Pringsheim [], based on P. monospermum Pringsh., and accommodates oomycetes that produce hyaline and coenocytic hyphae, without septa and zoospores, which are developed in a vesicle that forms at the tip of a discharge tube derived from a sporangium []. To date, there are more than 160 species in the genus Pythium sensu lato (s.l.), which include plant pathogens found worldwide [,,,]. Some Pythium species are important pathogens of animals, and others can be beneficial as biological control agents that protect against pathogenic fungi [,]. According to Lévesque and de Cock [], Pythium s.l. could be split into eleven clades (A–K) based on the phylogenetic analysis. After several rearrangement, Pythium s.l. were divided into five genera including Pythium sensu stricto (s.s.) (clades A–D); Elongisporangium by Uzuhashi, Tojo and Kakish (clade H); Globisporangium by Uzuhashi, Tojo and Kakish (clades E–G, I, and J); Pilasporangium Uzuhashi by Tojo and Kakish (distinct from the 11 lettered clades); and Phytopythium by Abad, De Cock, Bala, Robideau, A.M. Lodhi, and Lévesque (clade K) [].
During studies on the diversity of Pythium s.l. species in southern China, two new species of Globisporangium (Pythium s.l. clades G and J) were identified, based on morphological characters and molecular phylogenetic analyses of the internal transcribed spacer (ITS) and cytochrome c oxidase subunit I (Cox1) sequence data. These two species are described and illustrated in this study. Moreover, comparisons of these two species and their morphological and phylogenetically related species are also provided.
2. Materials and Methods
2.1. Isolates
Thirty specimens of the soybean cultivar Hefeng 47, a common cultivar in the Huang-Huai region of China, which exhibited seedling blight, damping off, and root rot, were collected from Nanjing, Jiangsu province, in April 2016. The soybean plants were sampled from fields at approximately 10 m intervals along a 150 m transect laid out in a “W” pattern.
The soybean plants were washed three times with sterile water, and six sections that were 0.5–1 cm long were cut from each root with a sterile scalpel, including one at the root tip, one at the interface between hypocotyl and soil, and the others at either the middle of root or at a symptomatic area along the length of root. The sections were blotted dry and embedded in selective V8 juice agar (V8A) that contained rifampicin (50 mg/L), phenamacril (5 mg/L), ampicillin (50 mg/L), and pentachloronitrobenzene (50 mg/L), and were incubated in the dark at 25 °C for 2–3 d. When mycelial growth was observed, the isolates were purified by cutting a small piece of medium with mycelia at the edge of a colony or by transferring a single hyphal tip of colonies onto water agar in triplicate and transferring the parts that were cut into new agar plates.
2.2. Morphology and Growth Rate
The cultures were deposited in the herbaria of the Institute of Microbiology, Beijing Forestry University (BJFC), Beijing, China; the College of Plant Protection, Nanjing Agricultural University (NJAU), Najing China; and the College of Landscape Architecture, Jiangsu Polytechnic College of Agriculture and Forestry, Zhenjiang (JAFLA), Zhenjiang, China. The purified isolates were examined after incubation on V8A in the dark for 2–3 d at 25 °C. The colony patterns of two representative isolates of the new species were examined after incubation for 3 d at 25 °C on V8A plates prepared as previously described []. The isolates were transferred to sterilized distilled water for sporulation. Fifty measurements were taken for each morphological feature, such as sporangia, oogonia, and oospores. The cardinal temperatures were examined on potato carrot agar (PCA), as described by van der Plaäts-Niterink [], and the growth rates were measured after 24 h of incubation. Each isolate was incubated at 5–40 °C at intervals of 5 °C on PCA media. When no growth was observed, the intervals were reduced from 5 to 2 or 1 °C, and the culture was returned to room temperature to check the revival of the growth. The experiment was repeated two times using a single plate per repetition.
2.3. DNA Extraction, PCR and Sequencing
Acetyl trimethylammonium bromide rapid plant genome extraction kit (Demeter Biotechnologies Co., Ltd., Beijing, China) was used to extract the total genomic DNA from purified isolates, and PCR was performed in accordance with the instructions of the manufacturer [].
The ITS region was amplified with the primers ITS4 and ITS5 []. The Cox1 gene was amplified with the primers OomCoxI-Levlo (CYTCHGGRTGWCCRAAAAACCAAA) and OomCoxI-Levup (TCAWCWMGATGGCTTTTTTCAAC) []. The PCR procedure for the ITS was as follows: initial denaturation at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 54 °C for 45 s and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR procedure for Cox1 was as follows: initial denaturation at 94 °C for 2–5 min, followed by 35 cycles at 94 °C for 30 s, 52 °C for 30 s and 72 °C for 1–2 min, and a final extension of 72 °C for 5–10 min []. The PCR products were purified and sequenced by the GenScript Biotech Company (Nanjing, China) with the same primers.
The sequences generated in this study were aligned with additional sequences downloaded from GenBank (Table 1) using ClustalX [] and manually adjusted in BioEdit [].

Table 1.
A list of species, cultures, and GenBank accession numbers of sequences used in this study.
2.4. Phylogenetic Analyses
ITS sequences and the Cox1 gene have become powerful tools to identify Pythium species [,]. Therefore, a phylogenetic analysis was inferred from the combined ITS and Cox1 dataset to explore the relationships of uncertain species within taxa in Pythium. The samples were analyzed phylogenetically as described by Cui et al. []. Maximum parsimony (MP) analysis was applied to the combined datasets of ITS and Cox1 sequences. Two isolates of Saprolegnia parasitica Coker were used as outgroups []. The tree-construction procedure was performed in PAUP* version 4.0b10 []. All the characters were equally weighted, and gaps were treated as missing data. Phylogenetic trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. The max-trees were set to 5000; branches of zero length were collapsed, and all the parsimonious trees were saved. The clade robustness was assessed using a bootstrap (BT) analysis with 1000 replicates []. The descriptive tree statistics tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC), and homoplasy index (HI) were calculated for each maximum parsimonious tree (MPT) generated.
MrModeltest 2.3 [] was used to determine the best-fit evolution model for the dataset for Bayesian inference (BI). The BI was calculated using MrBayes3.1.2 [] with a general time reversible (GTR) model of DNA substitution and an inverse gamma distribution rate variation across the sites. Four Markov chains were run for 2 million generations until the split-deviation-frequency value < 0.01 and sampled every 100 generations. The burn-in was set to discard the first 25% of the trees. A majority rule-consensus tree of all the remaining trees was calculated. The branches that received bootstrap support for maximum likelihood (BS), maximum parsimony (MP), and Bayesian posterior probabilities (BPP) ≥ 75% (MP) and 0.95 (BPP) were considered as significantly supported, respectively. The phylogenetic trees were visualized using FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 9 July 2014).
2.5. Pathogenicity
Pathogenicity was confirmed using 3-day-old V8A plugs (1.5 cm diam) of the isolates Chen 93 and Chen 268 using the hypocotyl slit inoculation method [] on the soybean cultivar Hefeng 47, which is commonly grown in Huang-Huai Valley. Five soybean seedlings inoculated with uncolonized agar plugs served as the controls. Five soybean seedlings were inoculated for each isolate and incubated at 25 °C with a 12 h photoperiod in a greenhouse for 4–5 d. The experiments were repeated three times.
3. Results
3.1. Isolates
In April 2016, isolation from the samples of soybean plants collected from three fields in three cities yielded five cultures of Globisporangium cultures (Chen 93 and 98, Chen 268, 271, and 272), which represented two unknown Globisporangium species.
3.2. Phylogenetic Analyses
BLAST analyses of ITS or Cox1 sequences of the five isolates recovered from the roots of soybean, described here as Globisporangium pengfuense and G. tenuihyphum, showed that the best matches of the five isolates were analogous with the species of Globisporangium (Pythium s.l. clades G and J, e-value = 1 × 10−6) [].
The combined ITS + Cox1 dataset of the Pythium s.l. species included sequences from 51 isolates that represented 47 taxa. The dataset had an aligned length of 1781 characters. A total of 689 of these characters were constant: 126 were variable and parsimony-uninformative, and 966 were parsimony-informative. Maximum parsimony analysis yielded one equally parsimonious tree (TL = 4735, CI = 0.441, RI = 0.722, RC = 0.319, and HI = 0.559). The best model for the combined ITS + Cox1 sequences dataset estimated and applied in the Bayesian analysis: GTR+I+G, lset nst = 6, rates = inverse gamma; and prset statefreqpr = dirichlet (1,1,1,1). A Bayesian analysis resulted in the same topology with an average standard deviation of split frequencies = 0.008893. Two sampled isolates of the new species, Globisporangium pengfuense, formed a well-supported lineage (100% MP and 1 BPPs), indicating that they are phylogenetically distinct from the other species in Pythium s.l. clade G (Figure 1). The sequences from three sampled cultures of G. tenuihyphum formed a well-supported lineage (100% ML and 1 BPPs), indicating that they represent species that are phylogenetically distinct from other Pythium s.l. clade J.
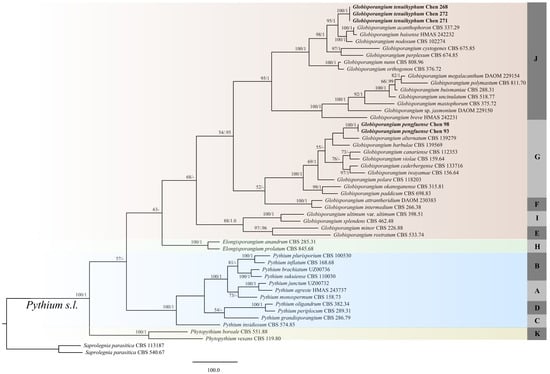
Figure 1.
Phylogeny of Globisporangium pengfuense, G. tenuihyphum, and related species generated by maximum parsimony based on ITS + Cox1 sequences. The branches are labeled with parsimony bootstrap proportions (before the slanting line) higher than 50% and Bayesian posterior probabilities (after the slanting line) ≥ 0.95. The new species are indicated in bold. Ph. refers to Phytopythium.
3.3. Taxonomy
Globisporangium pengfuense Jia J. Chen, sp. nov. Figures 2 and 3.
MycoBank number: MB 844572.
Diagnosis—Globisporangium pengfuense can be distinguished morphologically from its closest relatives, including G. alternatum (M.Z. Rahman, Abdelz, and Kageyama) by H.D.T. Nguyen and C.F.J. Spies; G. barbulae (Ueta and Tojo) by H.D.T. Nguyen and C.F.J. Spies; G. canariense (B. Paul) by Uzuhashi, Tojo, and Kakish; G. cederbergense (Bahram., Botha and Lampr.) by H.D.T. Nguyen and C.F.J. Spies; G. iwayamae (S. Ito) Uzuhashi, Tojo, andKakish; and G. violae (Chesters and Hickman) by Uzuhashi, Tojo, and Kakish., by its narrower hyphae and the presence of apical papillae on its sporangia. It is ecologically characterized by a weak ability to infect the seeds, seedings, and roots of soybean.
Etymology—Pengfuense (Lat.): referring to the location in China where the holotype was isolated.
Type—China, Jiangsu province, Nanjing, Jiangning district, Pengfu village, from the roots of Glycine max, 1 April 2016, J.J. Chen 93 (holotype, BJFC-C 1994; isotype, NJAU and JAFLA; ex-type cultures, BJFC, NJAU, and JAFLA).
Colonies were submerged, with a stellate pattern on the 10% V8A. The average growth rates were 14 mm/d at 25 °C on PCA. The cardinal temperatures were minimum 5 °C, optimum 25 °C, and maximum 35 °C (Figure 2). The main hyphae hyaline, aseptate, was up to 5.0 µm wide. There were no hyphal swellings. They were sporangia globose to sub-globose, catenulate, terminal, and occasionally with apical papillae or intercalary that were 10–20.5 (mean 17.5) μm. Zoospores were not observed. They were homothallic, with oogonia globose that were smooth, terminal, or intercalary and 12.5–22.5 µm (mean 17.5 µm) in diameter. Antheridia were mostly monoclinous, occasionally diclinous, totaling one to three per oogonium; the antheridial stalks were unbranched; the antheridial cells were sub-globose, fist-shaped to crook-necked, and straight or curved, making apical contact with the oogonia (Figure 3). The oospores were plerotic or nearly plerotic and globose, and they were 10.5–21.5 μm (mean 16.5 µm) in diameter as well as hyaline. The oospore wall was 0.5–1.0 µm (mean 0.7 µm) thick.
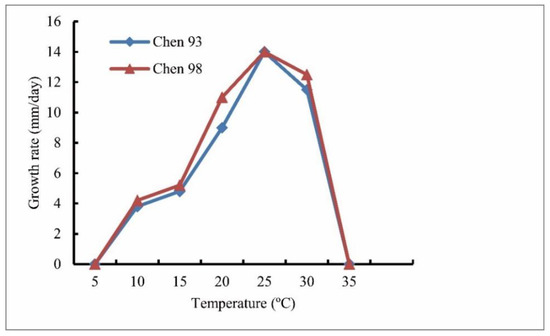
Figure 2.
Mycelial growth rate of Globisporangium pengfuense isolates Chen 93 and 98 on potato carrot agar at different temperatures.
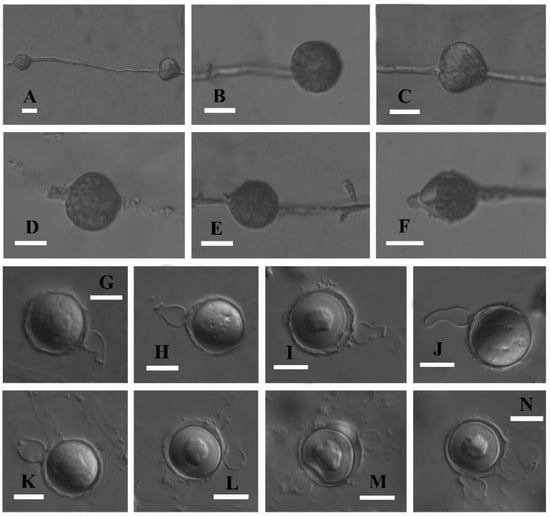
Figure 3.
Asexual and sexual reproductive bodies of Globisporangium pengfuense (holotype, Chen 93). (A) Catenulate sporangia. (B) Globose and terminal sporangium. (C–E) Intercalary sporangia. (F) Sporangium with apical papillae. (G) Diclinous antheridium and plerotic oospore. (H,I) Fist-shaped antheridial cells. (J) Crook-necked antheridial cell. (K) Monoclinous antheridium. (L) Intercalary oogonium. (M) Nearly plerotic oospore and two antheridia. (N) Plerotic oospore and three antheridia. Scale Bars (A–N) = 10 μm.
Additional cultures examined (paratype) were of type China, Jiangsu province, Nanjing, Jiangning district, Pengfu village, from the roots of Glycine max, 1 April 2016, J.J. Chen 98 (NJAU and JAFLA).
Globisporangium tenuihyphum Jia J. Chen, sp. nov. Figures 4 and 5.
MycoBank number: MB 844573.
Diagnosis—Globisporangium tenuihyphum is characterized by relatively narrow hyphae that are up to 4.0 µm wide, mostly sub-globose to globose, occasionally ovoid-obpyriform sporangia, with smooth oogonia, monoclinous antheridia, subclavate, with falcate or semicircular to subcircular antheridial cells, and aplerotic oospores. It is ecologically characterized by a weak ability to infect the seeds, seedings, and roots of soybean.
Etymology—Tenuihyphum (Lat.): referring to relatively narrow hyphae.
Type—China, Jiangsu province, Nanjing, Jiangpu farm, from the roots of Glycine max, 1 April 2016, J.J. Chen 268 (holotype JAFLA-C 268; isotype, NJAU; ex-type cultures, JAFLA and NJAU).
Colonies were submerged, with a radiate pattern on 10% V8A. The average growth rates were 25.5 mm day−1 at 25 °C on PCA. The cardinal temperatures were minimum 5 °C, optimum 30 °C, and maximum 39 °C (Figure 4). The main hyphae hyaline, aseptate, was up to 4.0 µm wide. There were no hyphal swellings. The sporangia were mostly sub-globose to globose, occasionally ovoid-obpyriform, terminal, or intercalary, that were 15–27.5 × 12.5–25 (mean 22.5 × 21.5) μm. They were omothallic, with oogonia globose that were smooth, terminal, and 20–25 µm (mean 23.5 µm) in diameter. Antheridia monoclinous were one to two per oogonium (Figure 5); the antheridial stalks were unbranched; the antheridial cells were slightly elongated along the oogonial stalk, subclavate, and falcate or semicircular to subcircular. The oospores were aplerotic and globose, and they were 15–21.5 μm (mean 18.5 µm) in diameter as well as hyaline. The oospore wall was 1.5–2.5 µm (mean 2 µm) thick.
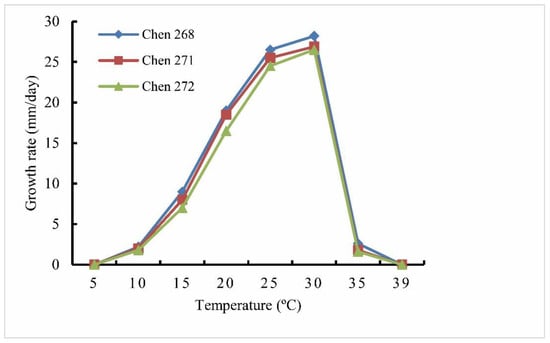
Figure 4.
Mycelial growth rate of Globisporangium tenuihyphum isolates Chen 268, 271, and 272 on potato carrot agar at different temperatures.
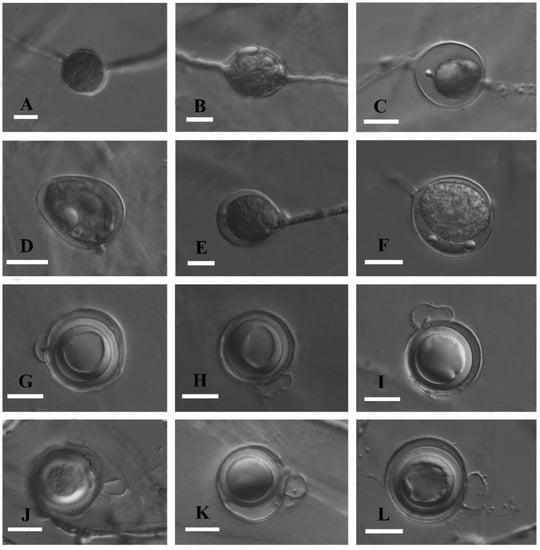
Figure 5.
Asexual and sexual reproductive bodies of Globisporangium tenuihyphum (holotype, Chen 268). (A–C) sub-globose, globose, and terminal sporangia. (D). Ovoid-obpyriform sporangium. (E,F). Intercalary sporangia. (G). Semicircle antheridial cell. (H,I). Elongated antheridial cells wavy in contour. (J,K). Subcircular antheridial cell. (L). Aplerotic oospore and two antheridia. Scale bars (A–L) = 10 μm.
Additional cultures examined (paratypes) were of type: China, Jiangsu province, Nanjing, Jiangpu farm, from roots of Glycine max, 1 April 2016, J.J. Chen 271 and 272 (NJAU and JAFLA).
3.4. Pathogenicity
Five soybean seedlings were inoculated with Globisporangium pengfuense strains (Chen 93) and an additional five were inoculated with G. tenuihyphum strains (Chen 268), which showed light-brown-color symptoms on the inoculated positions compared with the control (Figure 6). Globisporangium pengfuense and G. tenuihyphum, respectively, could be recovered from the slightly diseased soybean seedlings and were identified based on their morphological characteristics and the comparisons with ITS and Cox1 sequences.

Figure 6.
Pathogenicity of Globisporangium pengfuense and G. tenuihyphum isolates on the soybean cultivar Hefeng 47. (A) Control. (B) Disease symptoms caused by G. pengfuense (Chen 93). (C) Disease symptoms caused by G. tenuihyphum (Chen 268).
4. Discussion
Pythium s.l. species are cosmopolitan with a variety of diverse habitats []. To date, 75 species of Pythium s.l. have been reported in China [,,]. As part of an ongoing study on the diversity of the Pythium s.l. species associated with soybean in China, two new species of Globisporangium (Pythium s.l. clades G and J), G. pengfuense and G. tenuihyphum, were isolated and identified based on the phenotypic and phylogenetic characteristics in this study.
Globisporangium pengfuense differs from other Globisporangium species by its globose to sub-globose sporangia that are: catenulate, terminal, occasionally with apical papillae or intercalary, smooth oogonia, mostly monoclinous, occasionally diclinous antheridia, fist-shaped to crook-necked, straight or curved antheridial cells, and plerotic or nearly plerotic with thin-walled oospores (0.5–1.0 µm). In addition, G. tenuihyphum is characterized by relatively narrow hyphae that are: up to 4.0 µm wide, mostly sub-globose to globose, occasionally ovoid-obpyriform sporangia, smooth oogonia, monoclinous antheridia, subclavate, falcate or semicircle to subcircular antheridial cells, and with aplerotic oospores.
Comparisons of these two new species and their morphological and/or phylogenetically related species are also provided in Table 2 and Table 3.

Table 2.
Morphological description of Globisporangium pengfuense and the most closely related species.

Table 3.
Morphological description of Globisporangium tenuihyphum and the most closely related species.
Phylogenetically, Globisporangium pengfuense is a member of Globisporangium in the phylogenetic tree, and forms a lineage that is independent from other lineages of Pythium s.l. clade G. The principal characteristics of the species in Pythium s.l. clade G include a combination of ovoid, internally proliferating sporangia, and smooth oogonia (only P. paddicum has oogonia with ornamentation). Globisporangium pengfuense is closely related to G. alternatum in our phylogeny with low support (25% MP and 0.51 BPPs), but G. pengfuense differs from G. alternatum owing to its plerotic or nearly plerotic oospores and fast growth rate (14 mm/d vs. 8 mm/d) []. Globisporangium pengfuense is very similar to G. barbulae in morphology by the globose to sub-globose sporangia, mostly monoclinous antheridia, and similar growth (13.4 mm/d in G. barbulae) []. However, G. barbulae differs from G. pengfuense in lacking catenulate sporangia. In addition, G. barbulae can grow at 0 °C, while G. pengfuense has a minimum growth temperature of 5 °C. Globisporangium pengfuense differs from G. cederbergense and G. canariense by having thicker oospore walls [,]. Furthermore, G. pengfuense is distinguished from G. iwayamae and G. violae by having smaller oogonia []. Globisporangium pengfuense is ecologically characterized by a weak ability to infect the seeds, seedings, and roots of soybean.
Phylogenetically, Globisporangium tenuihyphum belongs to clade J of Pythium s.l. in the phylogenetic tree. Globisporangium acanthophoron and G. baisense resemble G. tenuihyphum, and the combined ITS and cox1 sequences also suggest a close relationship between these two species and G. tenuihyphum (Figure 1). However, G. acanthophoron has ornamented oogonia and a thinner-walled oospore wall (up to 1.5 µm) [], while G. baisense is distinguished from G. tenuihyphum by producing variously sac-like oogonia and the presence of diclinous sessile antheridia and double oospores (Table 3) [].
Author Contributions
Conceptualization, J.-J.C.; methodology, S.-B.Z.; performing the experiment, X.-X.H.; formal analysis, J.Y.; validation, J.-J.C., X.-X.H. and H.-J.Y.; resources, J.-J.C.; writing—original draft preparation, J.-J.C.; writing—review and editing, J.-J.C. and S.-B.Z.; visualization, S.-B.Z.; supervision, J.-J.C. and W.-W.Y.; project administration, J.-J.C.; funding acquisition, J.-J.C. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (32070022), and the Science Fund of the Jiangsu Vocational College of Agriculture and Forestry (2020kj003 and 2021kj91).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://www.ncbi.nlm.nih.gov/; https://www.mycobank.org/page/Simple%20names%20search.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Pringsheim, N. Beiträge zur morphology and systematik der algen. 2. Die Saprolegníeen [Contributions to the morphology and systematics of algae]. Jb. Wiss. Bot. 1858, 1, 284–306. [Google Scholar]
- van der Plaäts-Niterink, A.J. Monograph of the genus Pythium. Stud. Mycol. 1981, 21, 242. [Google Scholar]
- Uzuhashi, S.; Okada, G.; Ohkuma, M. Four new Pythium species from aquatic environments in Japan. Anton. Leeuw. Int. J. G 2015, 107, 375–391. [Google Scholar] [CrossRef] [PubMed]
- Ueta, S.; Tojo, M. Pythium barbulae sp. nov. isolated from the moss, Barbula unguiculata; morphology, molecular phylogeny and pathogenicity. Mycoscience 2016, 57, 11–19. [Google Scholar] [CrossRef]
- Salmaninezhad, F.; Mostowfizadeh-Ghalamfarsa, R. Three new Pythium species from rice paddy fields. Mycologia 2019, 111, 274–290. [Google Scholar] [CrossRef]
- Chen, J.J.; Feng, H.; Yu, J.; Ye, W.W.; Zheng, X.B. Pythium huanghuaiense sp. nov. isolated from soybean: Morphology, molecular phylogeny and pathogenicity. Biodivers. Data J. 2021, 9, e65227. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.S.; Saleh, A.S.F. Isolation of Pythium acanthicum, P. oligandrum, and P. periplocum from soil and evaluation of their mycoparasitic activity and biocontrol efficacy against selected phytopathogenic Pythium species. Mycopathologia 1999, 145, 143–153. [Google Scholar] [CrossRef]
- Lévesque, C.A.; de Cock, A.W.A.M. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 2004, 108, 1363–1383. [Google Scholar] [CrossRef]
- Nguyen, H.D.T.; Dodge, A.; Dadej, K.; Rintoul, T.L.; Ponomareva, E.; Martin, F.N.; de Cock, A.W.A.M.; Lévesque, C.A.; Redhead, S.A.; Spies, C.F.J. Whole genome sequencing and phylogenomic analysis show support for the splitting of genus Pythium. Mycologia 2022, 114, 501–515. [Google Scholar] [CrossRef]
- Miller, P.M. V-8 juice agar as a general purpose medium for fungi and bacteria. Phytopathology 1955, 145, 461–462. [Google Scholar]
- Cui, B.K.; Li, H.J.; Ji, X.; Zhou, J.L.; Song, J.; Si, J.; Yang, Z.L.; Dai, Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019, 97, 137–392. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gefand, D.H., Sninsky, J.J., White, J.T., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Robideau, G.P.; de Cock, A.W.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; Désaulniers, N.; Eggertson, Q.A.; Gachon, C.M.; et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 2011, 11, 1002–1011. [Google Scholar] [CrossRef]
- Rahman, M.Z.; Uematsu, S.; Coffey, M.D.; Uzuhashi, S.; Suga, H.; Kageyama, K. Re-evaluation of Japanese Phytophthora isolates based on molecular phylogenetic analyses. Mycoscience 2014, 55, 314–327. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Villa, N.O.; Kageyama, K.; Asano, T.; Suga, H. Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and tubulin gene sequences. Mycologia 2006, 98, 410–422. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic Analysis Using Parsimony Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Felsenstein, J. Confidence intervals on phylogenetics: An approach using bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Mark, P.; Avres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes3.2: Efficient Bayesian phylogenetic inference and model choice, across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Dorrance, A.E.; Berry, S.A.; Anderson, T.R.; Meharg, C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant health Prog. 2008, 9, 35. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W. Basic local alignment search tool. J. Mol. Biol. 1990, 25, 403–410. [Google Scholar] [CrossRef]
- Long, Y.Y.; Wei, J.G.; Sun, X.; He, Y.Q.; Luo, J.T.; Guo, L.D. Two new Pythium species from China based on the morphology and DNA sequence data. Mycol. Prog. 2012, 11, 689–698. [Google Scholar] [CrossRef]
- Ho, H.H. The genus Pythium in mainland China. Mycosystema 2013, 32, 20–44. [Google Scholar]
- Rahman, M.H.; Abdelzaher, H.M.A.; Mingzhu, L.; Motohashi, K.; Suga, H.; Kageyama, K. Pythium rishiriense sp. nov. from water and P. alternatum sp. nov. from soil, two new species from Japan. FEMS Microbiol. Lett. 2015, 362, fnv086. [Google Scholar]
- Paul, B. ITS region of Pythium canariense sp. nov., its morphology and its interaction with Botrytis cinerea. FEMS Microbiol. Lett. 2002, 208, 135–141. [Google Scholar] [CrossRef][Green Version]
- Bahramisharif, A.; Lamprecht, S.C.; Spies, C.F.J.; Botha, W.J. Pythium cederbergense sp. nov. and related taxa from Pythium clade G associated with the South African indigenous plant Aspalathus linearis (rooibos). FEMS Microbiol. Lett. 2013, 105, 1174–1189. [Google Scholar]
- Paul, B.; Galland, D.; Bhatnagar, T.; Dulieu, H. A new species of Pythium isolated from the Burgundy region in France. FEMS Microbiol. Lett. 1998, 158, 207–213. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).