Abstract
Portunus trituberculatus is an important economic species of crab that is artificially bred and released in the Yangtze River Estuary and its adjacent sea areas. Based on six microsatellite markers, we investigate the genetic diversity and structure of 101 P. trituberculatus specimens collected from two hatcheries in Nantong and Zhoushan that participated in stock enhancement in the year 2019. We compared these with 124 wild specimens caught from 13 localities in the estuary. Analysis of several genetic diversity parameters (NA, RS, I, HO, HE, FIS, and FST) for the 15 populations demonstrates that both released and wild populations possess relatively rich genetic diversity. Furthermore, the released groups demonstrate no less genetic variation between themselves than do the wild crabs. Most FIS values are greater than zero, which shows inbreeding is common among specimens with geographically open sites. However, insufficient sampling may have led to a wide distribution of null alleles, a Hardy–Weinberg test disequilibrium in microsatellite markers PN22 and P04, and a lack of crab genetic diversity in site 14. All populations (except locality 14) have not suffered the bottleneck effect. Four subgroups can be seen to roughly spread longitudinally along the sample area by performing pairwise comparisons of genetic distance and FST values among the populations. No obvious topological heterogeneity is discovered among the four subgroups in a phylogenetic tree. The existence of genetic exchange and differentiation among the subgroups is also verified using structure analysis. Therefore, based on this evidence, we propose that the hatchery stock enhancements performed in Nantong and Zhoushan result in no reduction in genetic diversity for wild populations in the Yangtze Estuary in 2019.
1. Introduction
Portunus trituberculatus (Crustacea:Decapoda: Brachyura) (Miers, 1876) is a swimming crab widely distributed on the East Asian coastal seabed [1]. It is a well-known and very popular edible species that spreads from Bohai to the South China Sea in China [2]. Unfortunately, wild P. trituberculatus resources have sharply declined over the last several decades because of environmental pollution, habitat destruction, and unsustainable fishing [3,4]. Both artificial propagation and release, and restocking and stock enhancement have proven to be effective measures for restoring fishery resources since 1986 in Yingkou and have been widely adopted and used throughout the world [5,6,7]. The Yangtze Estuary and its adjacent sea area, which ranges across the coastlines of Jiangsu, Shanghai, and Zhejiang, from north to south, is the largest estuary area in China. The runoff of the Yangtze River contains a very large number of nutrients. This makes it the most productive offshore water area in China and the largest estuarine fishery closely connected with the Zhoushan and Lvsi Fisheries [8].
Swimming crabs are the dominant invertebrate fauna in the Yangtze Estuary [9]. Half of the national annual fishery harvest output can be accounted for by euryhaline crabs from this region, whether through wild fishing or artificial breeding [10,11]. In 2014, more than 18 billion juvenile swimming crabs were released in the Dongtou sea area of Zhejiang Province to help the fishery resources there recover [12]. It is almost impossible to accurately collect annual release stock enhancement data in this vast marine area. Nevertheless, the pouring of such a large number of artificially bred crabs into the natural environment can certainly have a great impact on the genetic characteristics and structure of wild populations [13,14]. As a powerful molecular marker, microsatellite DNA markers (simple sequence repeats) are widely used in population genetics and, in particular, for detecting genetic differences between wild and hatchery fish [15,16].
In this study, six polymorphic microsatellite loci were used to analyze genetic variation, and population genetic structure within wild P. trituberculatus collected from different locations in the Yangtze Estuary and crabs caught from hatcheries in Nantong and Zhoushan, which are situated in the north and the south boundary of the estuary, respectively.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
In total, 101 artificially bred crabs were collected in March 2019 from two hatcheries that participated in stock enhancement in the year 2019, as follows: Rudong Xinlei in Nantong (n = 52) and Daishan Risheng in Zhoushan (n = 49). In total, 124 specimens (Table 1) were captured at identified sites in the Yangtze Estuary (Figure 1) in October 2019. All samples were adult individuals with equal male and female depending on the catch, weighing between 300 and 400 g, and their lengths of carapace ranged from 7 to 9 cm. Total DNA was extracted from excised muscle using a DNeasy Blood and Tissue Kit (Qiagen, Dusseldorf, Germany) as per manufacturer’s instructions. The extracted DNA was quantified using a spectrophotometer (Thermo Scientific NanoDrop 2000, Waltham, MA, USA).

Table 1.
Summary information on the 13 sampled localities.
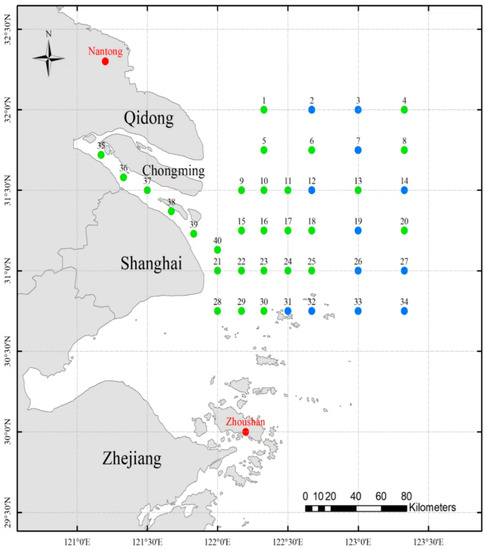
Figure 1.
Sample localities of P. trituberculatus. crabs were collected from 13 sites of the Yangtze Estuary in wild. Blue and red circles indicate sampling sites for wild and cultivated crabs, respectively.
2.2. Microsatellite Polymerase Chain Reaction Amplification
Six primer pairs based on our targeted microsatellite sequences were synthesized with fluorescence dyes (FAM and HEX) labeled at the 5′ end of each pair (Table 2). Polymerase chain reaction (PCR) amplifications were performed in 50 µL reactions containing 10 × PCR Buffer 2.5 μL, dNTP 2 μL, each primer 1 μL, Taq DNA polymerase 0.15 μL, ddH2O 17.5 μL, and 100 ng DNA templates using the following conditions: 95 °C for 5 min, 35 cycles of 95 °C for 1 min, using this Tm temperature for 1 min, then 72 °C for 1 min, followed by elongation at 72 °C for 9 min. All PCR products were electrophoresed on a 1.5% agarose gel and visualized under UV light with DuRed (Fanbo, Beijing, China) staining, and then sent to the Personalbio Company (Shanghai, China) for capillary electrophoresis detection.

Table 2.
Summary information of 6 microsatellite loci of P. trituberculatus.
2.3. Data Analysis
GeneMarker v. 2.2.0 was used to read the data, supplemented by manual correction [18]. The number of alleles (NA), genetic diversity index (Shannon Index, I), observed heterozygosity (HO), and expected heterozygosity (HE) of each microsatellite locus were counted by PopGene v.1.32 [19]. FSTAT v. 2.9.3 was used to calculate the inbreeding coefficient (FIS), allelic richness (RS), and genetic differentiation index (FST) between every two populations, and 1000 substitution tests were carried out to analyze significance [20]. Genetic distances between populations were calculated with GENEPOPv. 4.0 [21] based on Nei’s standard (1983), and a phylogenetic tree was implemented in MEGA X v.10.1.8 from this data using the neighbor-joining method [22]. Bootstrapping with 1000 iterations of data matrix was tested to give support for tree topology [23]. Micro-Checker v 2. 2. 3 was applied to detect whether null alleles occurred at each point [24]. A Hardy–Weinberg equilibrium test was performed for each locus using GENEPOP v. 4.0 [21]. Bottleneck effect detection for each population was accomplished by BOTTLENECK v. 1.2.02 [25] using the infinite allele model (IAM), two-phased model (TPM), and stepwise mutation model (SMM) [26,27]. We determined mode shift allele frequency distributions [28] for all 14 populations, with 5% variance for IAM and 95% proportions of SMM in the TPM test. One-tailed Wilcoxon signed-rank test and Bonferroni corrections were implemented for the results of the test [25,29]. The LEA R-language-based package (v. 3. 5. 1) was used for structure analysis to estimate the most likely value of the genetic clusters (K) in the estuary [30]. The Sea-born Python-based package was used to draw heatmap of genetic differentiation indices and genetic distances.
3. Results
3.1. Genetic Diversity Analysis for Wild and Aritificially Bred Crabs Caught in Yangtze Estuary
Six microsatellite loci were checked in 225 P. trituberculatus specimens caught from either wild environments or aquaculture. Our findings (Table 3) show that the total NA number ranges from 20 to 90, with an average NA number between 3.33 and 15, and the average RS values range from 3.33 to 5.34. There is little numerical difference in the Shannon Indices (I) between populations. The same situation appears in the HO and HE statistics. Inbreeding is widely spread among all populations, as calculated by FIS. The expected heterozygosity is a measure of genetic diversity and provides the probability that two alleles randomly drawn from a population differ in type [31]. It is worth noting that sufficient broodstocks for artificial breeding were selected from wild sea areas and that the artificially bred and wild populations both possess the same level of genetic diversity. Furthermore, the swimming crab population genetics parameters from Nantong and Zhoushan that we observed are generally consistent with Cui et al. [17]. The exceptional situation we observed at site 14 is likely related to a limited sample size. The widespread null allele distribution we saw in six of the loci results in a loss of heterozygosity and provides valuable insight for further crab population genetic diversity analyses. It may also help explain Hardy–Weinberg equilibrium deviations seen in our survey and why the HO values are much lower than HE, especially for the gene fragments PN22 and P04. As demonstrated in Figure 2, the FST values between the two released populations and the other thirteen populations captured wild in the estuary are less than 0.15, suggesting that genetic differentiation among these populations is not very obvious. Low levels of genetic differentiation are observed among the fourteen populations. Statistical data on the genetic distances (Figure 2) and our phylogenetic tree (Figure 3) also confirmed these conclusions. There are four subgroups in this estuary from the neighbor-joining analysis displayed in Figure 3. The specimens from aquaculture located in the north (Nantong) and south (Zhoushan) of the target sea area have similar genetic characteristics. Furthermore, samples from sites 12, 14, 32, and 34 have a closer genetic relationship to each other than to the others. Similarly, the catches from sites 3, 7, 19, 26, 27, 31, and 33 can be regarded as another sub-branch.

Table 3.
Main polymorphic parameters for six microsatellite loci utilized in this study.
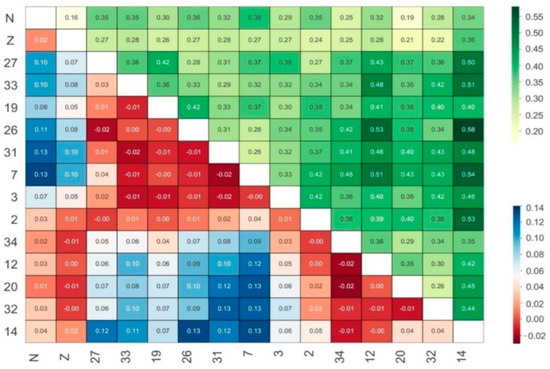
Figure 2.
Pairwise Nei’s genetic distance (above diagonal) and genetic differentiation index (below diagonal) among P. trituberculatus populations. “N” and “Z” stand for crabs caught from Nantong and Zhoushan.
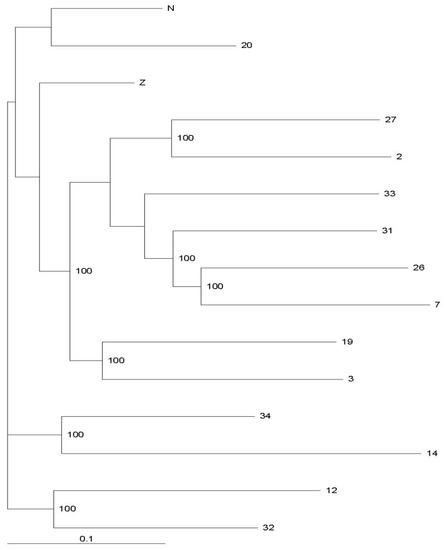
Figure 3.
Phylogenetic analysis of the P. trituberculatus collected from 13 sites of Yangtze Estuary and 2 hatcheries located in Nantong (N) and Zhoushan (Z).
3.2. Genctic Population Structure Analysis for Wild and Bred Populations of P. trituberculatus
Bottleneck effects show all populations (except site 14) exhibit a conventional L-type distribution (Figure 4). Thus, we infer that swimming crab populations have not experienced any recent bottleneck events and that the abnormality of site 14 may relate to its small sample size (n = 4). Assuming that the values of K can be 1~15, each hypothetical K value was subjected to 10 independently repeated operations in Bayesian clustering analysis, and then the best K with the highest averaged maximum log-likelihood [32] was estimated to be 4 by the least square method (Figure 5). The bar graph obtained from the structure analysis (Figure 6) reveals that genetic differentiation does not completely isolate the four subgroups of crabs in this estuary.
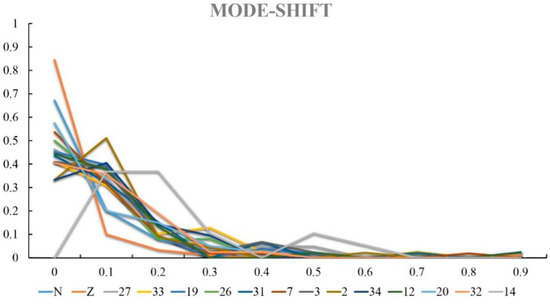
Figure 4.
Distribution map of allele frequency. “N” and “Z” stand for crabs caught from Nantong and Zhoushan.
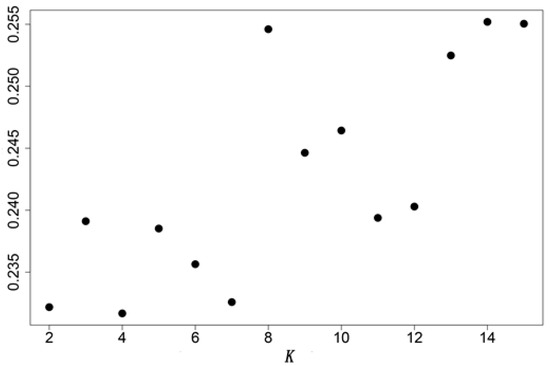
Figure 5.
Cross-entropy at different values of number of genetic cluster (K).
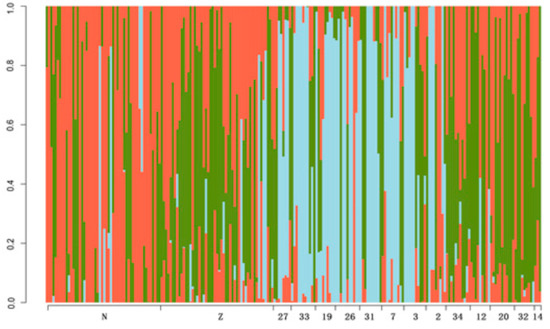
Figure 6.
Structure bar plots for 15 populations of P. trituberculatus (K = 4). “N” and “Z” stand for crabs caught from Nantong and Zhoushan.
4. Discussion
Due to its commercial importance, P. trituberculatus has suffered unprecedented over-exploitation in the East China Sea since the 1960s, causing a sharp decrease in its biomass [33]. Hence, artificial breeding of the crab was approved to meet the demand for the crab as human food and to resupply the marine swimming crab fishery resource in China [34]. The genetic characteristics of wild populations were inevitably affected by the numerous stocked specimens. Many studies began to focus on effective and scientific analyses of these impacts [35,36,37].
Conventional physical markers are not suitable for species that undergo molting processes. Thus, microsatellite data has become a practical tool, offering the following several advantages in genetic analyses: rich polymorphism, good stability, and high recognition [38]. Imai et al. used restriction fragment length polymorphism analysis to investigate genetic variation in the mtDNA of P. trituberculatus caught in Japan, but their results were inconclusive [39]. Wang et al. using isozyme polymorphism analysis, found that wild populations possess higher levels of polymorphism than do aquaculture Portunus in Zhoushan [40]. Liu et al. and Xu et al. examined genetic diversity among geographically defined swimming crab populations based on mtDNA gene sequence fragments [41,42], including COI and 16S rRNA. Guo et al. verified the correlations between genetic and geographic distances by comparing the mtDNA control region of crab samples from different locations in China [43]. Liu et al. sequenced mtDNA control regions to prove that four wild populations and two stocked populations located in Liaodong Bay all have rich genetic diversity [44]. As reported by Supmee et al. [45], in addition to the sequencing of the control region, more sensitive markers should be suggested to be put into use to explore the genetic structure of the swimming crab. Lee et al. confirmed that there were no significant genetic differences between wild populations collected from the Yellow Sea islands based on 20 microsatellite loci [46]. By the means of microsatellite analysis for P. trituberculatus collected in localities in Bohai Bay, the study by Duan et al. [47] showed there was high genetic diversity and similar genetic structure between wild and cultivated populations, which is in good agreement with our conclusion. However, based on microsatellite research, Liu et al. predicted that large-scale stock enhancement of crabs in Panjin could present a strong genetic risk for the local population [48].
Stocked populations in our study, such as those from Nantong and Zhoushan, have rich genetic diversity levels, about the same as the wild community. Our stock population values are consistent with wild P. trituberculatus collected in Yingkou by Cui et al. [17], which suggests to us that aquaculture specimens have no reduction in genetic variation compared to wild populations and that the stock enhancement executed by the two hatcheries did not lead to a genetic diversity decline in 2019. Although our surveyed populations were divided into four gene pools, and more similar genetic characteristics were discovered between populations with smaller distances, consistent with Guo’s research [43], we deduce that genetic differentiation between the four subgroups is quite limited and observe that there is no obvious topological heterogeneity in our phylogenetic tree. Genotyping-by-sequencing was carried out for nine populations obtained from the coast of China [49]. The result also displayed little genetic differentiation existed among samples; meanwhile, genetic connectivity combined with biological mixing consisted of the three subgroups divided from the nine localities.
Samples from sites far away from a coastline, such as 3, 19, 26, 17, and 33, may have more gene exchange and gene flow with specimens from the open sea. Subpopulation distributions are roughly related to the longitude of sites. We propose that the life cycle of the crab, the nearshore ocean current, and other environmental conditions may potentially influence this trend. P. trituberculatus is characterized by long-distance migrations for spawning and over-wintering [50]. The Yangtze Estuary and Zhoushan Islands are primary spawning grounds for swimming crabs and also serve as destinations and relay stations for over-wintering crabs [51]. Liu et al. [52] monitored the temporal variation in genetic architecture in Panjing and Yingkou with eighteen microsatellite loci across six years. They also found the crab life cycle and mass anthropogenic release of P. trituberculatus would lead to fluctuations in and differences in the genetic status of the local population. A high level of gene flow among most populations from offshore China was discovered by Hui et al. [53]. They indicated that sea-level changes and ocean currents would be the underlying factors in shaping the genetic structure of P. trituberculatus.
However, more sampling and sequencing data are needed to verify the scope and extent of the influences on the genetic exchange created by the life cycle or the migration route of the crab. Fortunately, bottleneck effects have not extensively occurred in open sea areas; hence, broodstock selection and repeated sampling for captive stock should be encouraged to screen out wild specimens possessing high variations to avoid inbreeding degradation of stocked specimens.
The primary purpose of P. trituberculatus artificial propagation and release methods cannot be to restore original biomass in its habitat, nor to meet specific human food needs for crab recapture, but rather to ensure the sustainable quantitative and genetic stability of the wild species. Insufficient crab samples captured in wild sea areas may artificially cause the loss of heterozygotes and deviation from Hardy–Weinberg equilibrium. As a result, P. trituberculatus sequencing data caught from the wild and stock enhancements in the Yangtze Estuary should be systematically tracked and accumulated in the following years to evaluate the genetic diversity and structure of each population scientifically and, thereby, provide reasonable suggestions and opinions for the management of stock enhancement.
Author Contributions
Data collection and analysis, writing—original draft preparation, L.Y.; data analysis and software, Y.W.; supervision, project administration, writing—review and editing W.X. and H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was funded by Youth Innovation Promotion Association CAS (Grant Number: 2020211).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data or models generated or used during the study are available from the corresponding author by request.
Acknowledgments
We thank Steven M. Thompson for improving the English language of a draft of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sakai, T. Crabs of Japan and the Adjacent Seas; Kodansha Ltd.: Tokyo, Japan, 1976; pp. 1–773. [Google Scholar]
- Yang, F.; Xu, H.T.; Dai, Z.M.; Yang, W.J. Molecular characterization and expression analysis of vitellogenin in the marine crab Portunus trituberculatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2005, 142, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Dai, A. Primary investigation on the fishery biology of Portunus trituberculatus. Mar. Fish. 1997, 25, 136–141. (In Chinese) [Google Scholar]
- Huang, Q.; Ling, J.; Sheng, F. Composition and distribution of shrimps in the coastal area of the Northern East China Sea in summer. Mar. Fish. 2009, 3, 237–242. (In Chinese) [Google Scholar]
- Bartley, D.M.; Bell, J. Restocking. Stock enhancement, and sea ranching: Arenas of progress. Rev. Fish. Sci. 2008, 16, 357–365. [Google Scholar] [CrossRef]
- Bell, J.D.; Bartley, D.M.; Lorenzen, K.; Loneragan, N.R. Restocking and stock enhancement of coastal fisheries: Potential, problems and progress. Fish. Res. 2006, 80, 1–8. [Google Scholar] [CrossRef]
- Chen, Y. Discussion on the mark method of juvenile crab with Portunus trituberculatus. Fish. Sci. 1991, 10, 26–28. (In Chinese) [Google Scholar]
- Liang, C. A Preliminary Study of the Yangtze River Terrestrial Input Change and Response of the Estuarine Ecological Environment. Master’s Thesis, Chinese Academy of Sciences (Institute of Oceanology), Qingdao, China, 2013. (In Chinese). [Google Scholar]
- Ren, Q.; Xian, W.; Zhang, Y.; Liu, C.; Li, W. Invertebrate assemblage structure associated with key environmental factors in the waters across the Yangtze River Estuary. China. Chin. J. Appl. Ecol. 2018, 29, 3067–3077. (In Chinese) [Google Scholar] [CrossRef]
- Yu, C.; Song, H.; Yao, G. Geographical distribution and faunal analysis of crab resources in the East China Sea. J. Zhejiang Ocean. Univ. 2003, 22, 108–113. (In Chinese) [Google Scholar]
- China Fishery Statistical Yearbook; Ministry of Agriculture and Rural Affairs of the Peoples’ Republic of China: Beijing, China, 2018; pp. 22–27.
- Xu, K.; Zhou, Y.; Zhu, W.; Jiang, R.; Wang, Y.; Li, P.; Zhou, S.; Zhang, Y.; Zhang, H.; Hu, C. Stocking effectiveness of hatchery-released Portunus trituberculatus in the Dongtou sea area of Zhejiang Province. J. Zhejiang Univ. (Agric. Life Sci.) 2018, 44, 373–380. (In Chinese) [Google Scholar] [CrossRef]
- Ortega-Villaizn Romo, M.D.M.; Suzuki, S.; Nakajima, M.; Taniguchi, N. Genetic evaluation of inter individual relatedness for brood stock management of the rare species barfin flounder Verasper moseri using microsatellite DNA markers. Fish. Sci. 2006, 72, 33–39. [Google Scholar] [CrossRef]
- Escalante, M.A.; García-De-León, F.J.; Dillman, C.B.; Anabelia, D.L.S.C.; George, A.; Irene, B.S.; Ruiz-Luna, A.; Mayden, R.L.; Manel, S. Genetic introgression of cultured rainbow trout in the Mexican native trout complex. Conserv. Genet. 2014, 15, 1063–1071. [Google Scholar] [CrossRef]
- Borrell, Y.J.; Alvarez, J.; Blanco, G.; Murguía, A.; Lee, D.; Fernández, C.; Martinez, C.; Cotano, C.; Alvarez, P.; Prado, J. A parentage study using microsatellite loci in a pilot project for aquaculture of the European anchovy Engraulis encrasicolus L. Aquaculture 2011, 310, 305–311. [Google Scholar] [CrossRef]
- Sekino, M.; Hara, M. Microsatellite DNA loci in Pacific abalone Haliotis discus discus (Mollusca, Gastropoda, Haliotidae). Mol. Ecol. Resour. 2010, 1, 8–10. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Wang, H.; Wu, D.; Luan, W.; Tan, F.; Huang, M. Isolation and characterization of microsatellites in Portunus trituberculatus. Conserv. Genet. Resour. 2012, 4, 251–255. [Google Scholar] [CrossRef]
- Kellander, M.; Riley, M.; Liu, C. GeneMarker®® Software for Multiplex Ligation-Dependent Probe Amplification (MLPA™); SoftGenetics LLC: State College, PA, USA, 2002. [Google Scholar]
- Francis, C.Y.; Rong, C.Y.; Boyle, T. POPGENE, Microsoft Window-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Goudet, J. FSTAT Version 2.9.3.: A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Rousset, F. Genepop’007: A complete reimplementation of the Genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Peterson, A.C.; Garza, J.C.; Valdes, A.M.; Slatkin, M.; Freimer, N.B. Mutational processes of simple-sequence repeat loci in human populations. Proc. Natl. Acad. Sci. USA 1994, 91, 3166–3170. [Google Scholar] [CrossRef] [Green Version]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Luikart, G.; Cornuet, J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1998, 12, 228–237. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Frichot, E.; François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Boca, S.M.; Huang, L.; Rosenberg, N.A. On the heterozygosity of an admixed population. J. Math. Biol. 2020, 81, 1217–1250. [Google Scholar] [CrossRef]
- Yang, X.; Qian, L.; Wu, H.; Fan, Z.; Wang, C. Population differentiation, bottleneck and selection of Eurasian perch (Perca fluviatilis L.) at the Asian edge of its natural range. Biochem. Syst. Ecol. 2012, 40, 6–12. [Google Scholar] [CrossRef]
- Yu, C.; Song, H.; Yao, G. Assessment of the crab stock biomass in the continental shelf waters of the East China Sea. J. Fish. China 2004, 28, 41–46. (In Chinese) [Google Scholar]
- Sun, Y.; Yan, Y.; Sun, J. Larvae development of the swimming crab Portunus trituberculatus. J. Fish. China 1984, 8, 219–226. (In Chinese) [Google Scholar]
- Kitada, S.; Kishino, H. Lessons learned from Japanese marine finfish stock enhancement programmes. Fish. Res. 2006, 80, 101–112. [Google Scholar] [CrossRef]
- Segovia-Viadero, M.; Serrão, E.; Canteras-Jordana, J.; González-Wanguemert, M. Do hatchery-reared sea urchins pose a threat to genetic diversity in wild populations? Heredity 2016, 116, 378–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitada, S. Economic, ecological and genetic impacts of marine stock enhancement and sea ranching: A systematic review. Fish Fish. 2018, 19, 511–532. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Kong, J.; Zhang, Q.; Liu, P.; Wang, R. Pedigree tracing of Fenneropenaeus chinensis by microsatellite DNA marker genotyping. Acta Oceanol. Sin. 2006, 5, 151–157. [Google Scholar] [CrossRef]
- Imai, H.; Fujii, Y.; Karakawa, J.; Yamamoto, S.; Numachi, K.I. Analysis of the population structure of the swimming crab, Portunus trituberculatus in the coastal waters of Okayama Prefecture, by RFLPs in the whole region of mitochondrial DNA. Fish. Sci. 1999, 65, 655–656. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Jin, S.; Zheng, L.; Chen, Y. Tissue specificity and biochemical genetic analysis of isozyme on cultured Portunus trituberculatus stock. J. Oceanogr. Taiwan Strait 2005, 24, 474–480. [Google Scholar]
- Liu, Y.; Liu, R.; Ye, L.; Liang, J.; Xuan, F.; Xu, Q. Genetic differentiation between populations of swimming crab Portunus trituberculatus along the coastal waters of the East China Sea. Hydrobiologia 2009, 618, 125–137. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, R.; Liu, Y. Genetic population structure of the swimming crab, Portunus trituberculatus in the East China Sea based on mtDNA 16S rRNA sequences. J. Exp. Mar. Biol. Ecol. 2009, 371, 121–129. [Google Scholar] [CrossRef]
- Guo, E.; Liu, Y.; Cui, Z.; Li, X.; Cheng, Y.; Wu, X. Genetic variation and population structure of swimming crab (Portunus trituberculatus) inferred from mitochondrial control region. Mol. Biol. Rep. 2012, 39, 1453–1463. [Google Scholar] [CrossRef]
- Liu, H.; Lv, H.; Cui, F. Parental contribution and genetic diversity between broodstock and offsprings in swimming crab (Portunus trituberculatus) releasing into natural waters. Fish. Sci. 2016, 35, 614–619. (In Chinese) [Google Scholar] [CrossRef]
- Supmee, V.; Sawusdee, A.; Sangthong, P.; Suppapan, J. Population genetic structure of blue swimming crab (Portunus pelagicus) in the Gulf of Thailand. Biodiversitas J. Biol. Divers. 2020, 21, 4260–4268. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, D.H.; Yoon, S.J.; Kim, D.H.; Kim, S.G.; Hyun, Y.S.; Min, G.S.; Chung, K.W. Characterization of 20 microsatellite loci by multiplex PCR in swimming crab, Portunus trituberculatus. Genes Genom. 2013, 35, 77–85. [Google Scholar] [CrossRef]
- Duan, B.; Liu, W.; Li, S.; Yu, Y.; Guan, Y.; Mu, S.; Li, Z.; Ji, X.; Kang, X. Microsatellite analysis of genetic diversity in wild and cultivated Portunus trituberculatus in Bohai Bay. Mol. Biol. Rep. 2022, 49, 2543–2551. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cui, F.; Hu, P.; Yi, G.; Ge, Y.; Liu, W.; Yan, H.; Wang, L.; Liu, H.; Song, J. Using of microsatellite DNA profiling to identify hatchery-reared seed and assess potential genetic risks associated with large-scale release of swimming crab, Portunus trituberculatus, in Panjin, China. Fish. Res. 2018, 207, 187–196. [Google Scholar] [CrossRef]
- Duan, B.; Mu, S.; Guan, Y.; Li, S.; Yu, Y.; Liu, W.; Li, Z.; Ji, X.; Kang, X. Genetic diversity and population structure of the swimming crab (Portunus trituberculatus) in China seas determined by genotyping-by-sequencing (GBS). Aquaculture 2022, 555, 738233. [Google Scholar] [CrossRef]
- Dai, A.; Yang, S.; Song, Y. Marine Crabs in China Sea; Marine Publishing Company: Beijing, China, 1986; pp. 194–196. [Google Scholar]
- Xue, J.; Du, N.; Nai, W. The researches on the Portunus trituberculatus in China. East China Sea 1997, 15, 60–64. (In Chinese) [Google Scholar]
- Liu, B.; Zhang, X.; Wang, Z.; Li, W.; Zhang, Q.; Liu, Q.; Liu, W.; Zhang, L.; Liu, Y.; Wang, C. Genetic pattern fluctuations in wild swimming crab populations, under the influence of continuous mass stock enhancement. Fish. Res. 2021, 243, 106075. [Google Scholar] [CrossRef]
- Hui, M.; Shi, G.; Sha, Z.; Liu, Y.; Cui, Z. Genetic population structure in the swimming crab, Portunus trituberculatus and its implications for fishery management. J. Mar. Biol. Assoc. UK 2019, 99, 891–899. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).