Abstract
The Western Palearctic is one of the most investigated regions for avian haemosporidian parasites (Haemoproteus, Plasmodium and Leucocytozoon), yet geographic gaps in our regional knowledge remain. Here, we report the first haemosporidian screening of the breeding birds from Sardinia (the second-largest Mediterranean Island and a biodiversity hotspot), and the first for the insular Mediterranean in general. We examined the occurrence of haemosporidians by amplifying their mtDNA cytb gene in 217 breeding birds, belonging to 32 species. The total prevalence of infected birds was 55.3%, and of the 116 haplotypes recovered, 84 were novel. Despite the high number of novel lineages, phylogenetic analysis did not highlight Sardinia-specific clades; instead, some Sardinian lineages were more closely related to lineages previously recovered from continental Europe. Host-parasite network analysis indicated a specialized host-parasite community. Binomial generalized linear models (GLMs), performed at the community level, suggested an elevational effect on haemosporidian occurrence probability (negative for Haemoproteus; positive for Leucocytozoon) likely due to differences in the abundance of insect vectors at different elevations. Furthermore, a GLM revealed that sedentary birds showed a higher probability of being infected by novel haplotypes and long-distance migrants showed a lower probability of novel haplotype infection. We hypothesize that the high diversity of haemosporidians is linked to the isolation of breeding bird populations on Sardinia. This study adds to the growing knowledge on haemosporidians lineage diversity and distribution in insular environments and presents new insights on potential host-parasite associations.
1. Introduction
Blood parasite pathogenic potential can negatively affect wild bird’s survival probability and consequently can also affect population dynamics [1,2,3,4]. By extension, parasite infections can have conservation impacts, particularly for small populations or declining bird species [3]. Avian blood parasites (Apicomplexa: order Haemosporida) represent one of most studied vector-borne parasite systems in birds and studies of avian haemosporidians have noticeably increased since the 2000′s. This increase is in no small part due to the development of molecular techniques for haemosporidian detection, such as specific PCR protocols, which usually concern the three genera Plasmodium, Haemoproteus and Leucocytozoon [5]. The insect vectors of these haemosporidian genera belong to different dipteran families: mosquitos (Culicidae) are known to be Plasmodium vectors, both louse flies (Hippoboscidae) and biting midges (Ceratopogonidae) are Haemoproteus vectors, and black flies (Simuliidae) are vectors for Leucocytozoon [6,7]. These insect vectors have a near global distribution and have different ecological requirements (e.g., related to variations in water availability or temperature). In addition, it appears that some haemosporidian lineages are either generalists (found in many avian host species) or specialists (found in very few, or single host species) [8]. These factors collectively mean that different geographic regions can have very different parasite communities infecting avian hosts. Indeed, the remarkable genetic richness in haemosporidian lineages (i.e., n = 4169 MalAvi haplotypes, as of November 2020 [5]) has provided an ideal framework to assess host-parasite dynamics. For example, genetic diversity reaches higher values in areas with high environmental heterogeneity and rich avian host communities [9], suggesting that there is still much diversity to discover and describe in such environments. An extensive literature which comprises both single-species [9,10] and multi-species/communities’ studies [11,12] also points to a very complicated framework affected by intertwined ecological factors, to include host-parasite relationships [13,14], host ecological or ethological traits [15], insect vector abundance [16] and climate influence [17]. Moreover, the framework is further complicated by unique dynamics, inherent to the biogeographic region in which studies are conducted, such as isolation-colonization events [18,19]. Thus, studies from new regions may be useful in disentangling the determining factors influencing haemosporidian infections and allowing assessments of whether those factors are generalized (e.g., similar across regions) or unique [18].
The Western Palearctic, from Iberia and Morocco in the west to the Caucasus and Ural Mountain regions in the east, is among the most investigated biogeographic regions for genetic diversity of avian haemosporidians [18,20,21,22,23,24]. However, there are still numerous gaps within this extensive geographic range. This includes very poorly sampled areas such as the Italian Peninsula, where molecular diversity data for haemosporidians are limited to a few host species [11,25,26], and areas completely overlooked as is the case for insular land masses in the Mediterranean Sea. Indeed, to our knowledge, no bird community from the insular Mediterranean has ever been investigated for avian haemosporidians via molecular methods, although a few studies focused on single host species [27] or migrating birds [28] have been conducted on very small islands or islets.
One such overlooked island is Sardinia, the second-largest (after Sicily) in the Mediterranean Sea. This island is ecologically diverse, ranging from coastal habitats to a mountainous interior, which results in varying microclimates. During the Pleistocene, the Western Mediterranean islands (i.e., Balearic Islands, Corsica, Tuscan Archipelago and Sardinia) acted as a glacial refugium for several organisms [29], and through subsequent isolation mechanisms became an important biodiversity hotspot in Europe [30]. Sardinia hosts a very rich avifauna including 163 breeding bird species [31], of which several are endemic taxa or have unique genotypes relative to mainland populations [32,33,34,35,36]. In addition, the breeding avifauna is comprised of both sedentary species (or populations) and migratory species.
In this study we investigated, for the first time, the avian malaria parasites of Sardinian breeding bird communities through the use of molecular tools, in order to: (i) describe the prevalence and the local genetic diversity of parasites; (ii) assess the uniqueness of Sardinian lineages by comparison to described lineages and their distribution using MalAvi [5]; and (iii) analyze the host-parasite relationships that occur in this area and test the influence of possible environmental or host-species traits that may affect infection occurrence (i.e., individual infection status). Given its insular nature and largely endemic biodiversity, we hypothesize that the Sardinian breeding bird community will harbor a high diversity of avian haemosporidian lineages. We further expect this haemosporidian community will include novel haplotypes, but also expect the occurrence of widespread generalist parasite lineages from the mainland.
2. Methods
2.1. Study Area and Fieldwork
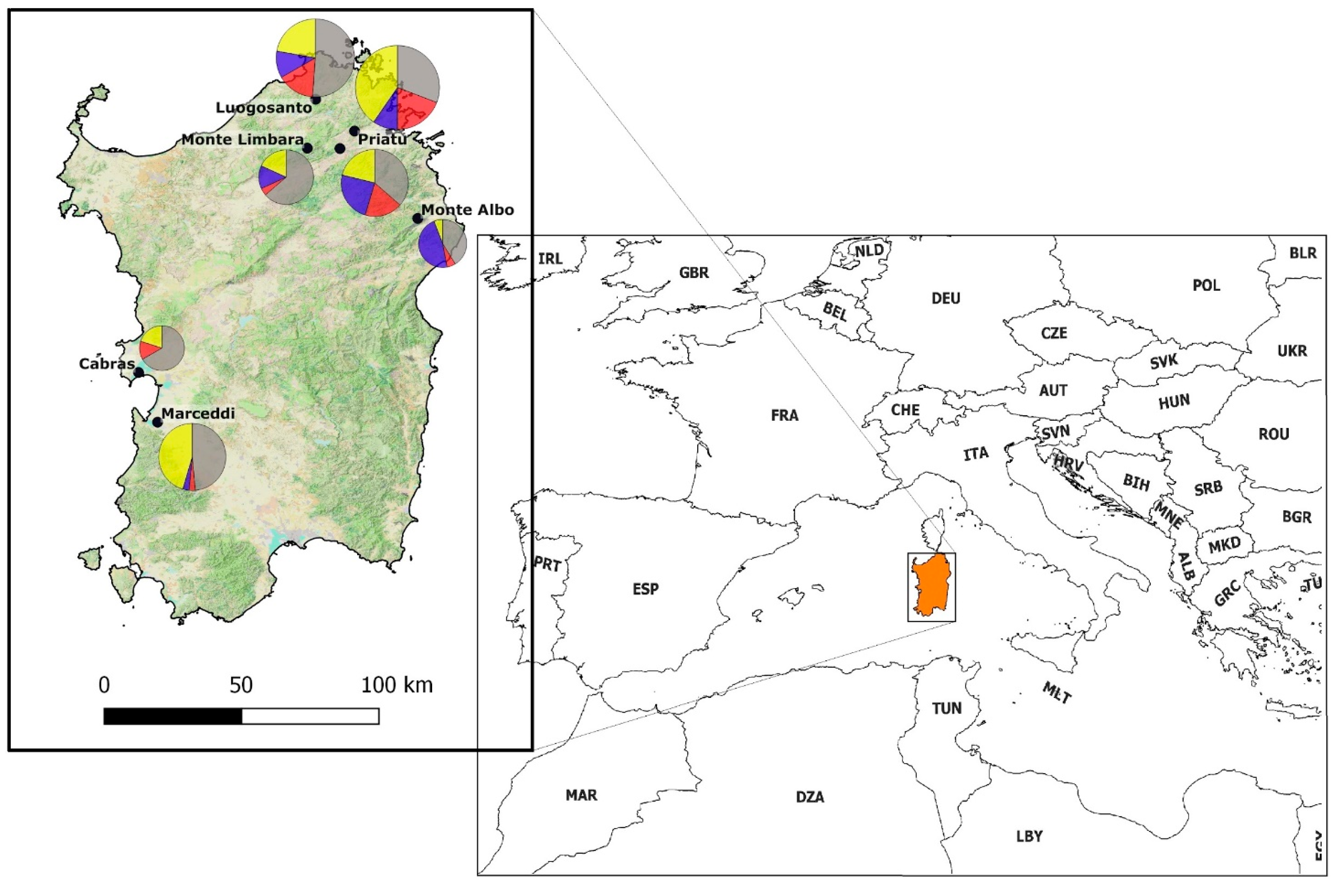
Sardinia is the second-largest Mediterranean island (23,821 km2), positioned in the western part of the basin. This insular land mass is about 180 km from continental land masses (both Europe and Africa). Blood samples were collected from wild birds (217 individuals) in seven localities (Figure 1). Sample sites were selected to be representative of the island’s elevational gradient (ranging from 3 m asl to 1309 m asl) and different environments to include reed-dominated wetland, woodland (e.g., Mediterranean sclerophyllous and Tyrrhenian cork-oak forests), agricultural mosaics and montane shrublands. Fieldwork took place during the breeding season in 2018, in the last two weeks of May. Birds were captured using mist nets and were ringed in order to avoid double sampling, under permits of the Sardinian Assessorato della Difesa dell’Ambiente (Prot. 822/18 of 15 Jan 2018, Det. 10616/288 of 11 May 2018) and of the Italian Istituto Superiore per la Protezione e la Ricerca Ambientale (Prot. 9447 of 26 Apr 2018). Approximately 50–100 μL of whole blood was drawn from the brachial vein by puncturing it with a sterile 25-gauge needle and preserved in Queens’s Lysis buffer [37]. Sampled birds represented 20% of Sardinian breeding species [31]. As the Sardinian breeding bird community is composed of species with very different migratory patterns (sedentary, short-distance (intra-Palearctic), or long-distance (trans-Saharan), we collected samples only from those individuals showing morphological evidence of breeding status (i.e., cloacal protuberance or brood patch), thus avoiding the inclusion of birds that were actively migrating in the analysis. Nomenclature used is in accordance with IOC World Bird List [38]. All voucher specimens (i.e., collected blood, See Table S1) collected are deposited in the Lab of Zoology at the University of Piemonte Orientale, Vercelli, Italy.
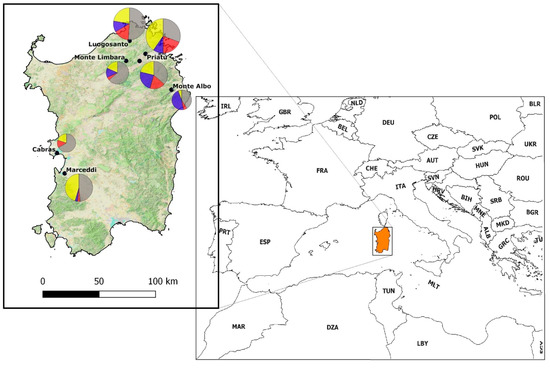
Figure 1.
Study area location in the Mediterranean basin. Sampling localities (black dots) are Cabras (3 m asl), Luogosanto (216 m asl), Marceddì (3 m asl), Monte Albo (780 m asl), Monte Limbara (1309 m asl), Priatu (308 m slm), Telti (368 m asl). Pie diagrams show the proportion for each site of uninfected birds (grey) and birds infected by Haemoproteus (yellow), Leucocytozoon (blue) and Plasmodium (red).
2.2. Molecular Analyses
DNA was extracted using the E.Z.N.A. tissue extraction kit (Omega Bio-Tek, Norcross, GA, USA) following the standard protocols, with a final elution of 75 μL to increase overall DNA yield. Polymerase chain reaction (PCR) was used to identify haemosporidian infection by amplifying a fragment of the mtDNA cytochrome b (cytb) gene, and multiple primer pairs [8] were used to amplify across known avian haemosporidian genetic diversity following molecular protocols previously described in Harvey and Voelker [17]. A single forward primer, UNIVF, was combined with each of three reverse primers UNIVR1, UNIVR2, UNIVR3, to amplify all the three genera of avian haemosporidians: Haemoproteus, Leucocytozoon, and Plasmodium. To verify an infection, 4 μL of the final PCR product was run on 1.8% agarose gel. All reactions were performed with negative (double-distilled H2O) and positive controls (infected individuals determined after three repeated PCR) to evaluate the validity of the PCR and detect possible contamination. PCR fragments were sequenced in both directions by Macrogen Europe labs (Amsterdam, The Netherlands).
2.3. Phylogenetic and Statistical Analysis
The sequences of 479 base pairs of the cytb gene obtained were controlled through the visualization of electropherograms using FinchTV (Geospiza Inc., Seattle, WA, USA http://www.geospiza.com (accessed on January 2020)). Any sequences containing ambiguities or evident sequencing errors (e.g., insertions, deletions, and faulty end-parts) were discarded and the samples were reprocessed with a new PCR and sequencing. We utilized MEGA X [39,40] to align and trim the sequences, and DnaSP v5.1 [41] to identify unique haplotypes and to reconstruct single infection haplotypes [42]. Double infections were identified by the presence of multiple peaks in chromatograms, these sequences were phased using the Phase 2.1 [43] implementation in DnaSP 5.10.1 following the procedure described by Harvey and Voelker [17]. The reconstructed single infection haplotypes were subjected to BLAST analysis on the MalAvi database (http://mbio-serv2.mbioekol.lu.se/Malavi/blast.html (accessed on May 2020)) and any sequences that had a match <97% were excluded from the data set.
The obtained haplotypes were blasted in GenBank and MalAvi [5] databases using the NCBI BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on May 2020)), and the sequences with one or more nucleotide substitutions were identified as new lineages according to the MalAvi Public Database. All comparisons to MalAvi are based on records accessed in November 2020 (Version 2.4.7, 6 October 2020). All sequences were deposited in both the MalAvi database (http://mbio-serv4.mbioekol.lu.se/avianmalaria (accessed on May 2020)) and GenBank (Table S1).
Genetic distances (p-distance) within and between haemosporidian genera were calculated, and phylogenetic relationships were reconstructed in MEGA X. Phylogenetic trees were calculated by Neighbor-Joining with 1000 bootstrap replicates (pairwise deletion), and by Bayesian Inference utilizing the Markov Chain Monte Carlo (MCMC) method implemented in MrBayes 3.1 [44], and implementing the best model estimate of sequence evolution (GTR+I+G) acquired from jModeltest v2.1 [45]. Our MrBayes analysis was run for 4,000,000 generations with a burn-in of 25%. A cytb sequence from Hepatocystis sp. (GenBank KC262858) was used as an outgroup. Phylogenetic trees were edited in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on May 2020)).
To highlight the host-parasite degree of specialization we calculated the network level specificity index (H2′) for bird-haemosporidian communities and the species-level specificity index (d′) for each parasite lineage using the bipartite package [46]. These indices range from 0 to 1, indicating a generalized network for H2′ values close to 0, and a specialized network for H2′ close to 1, and no specialization (d’ = 0) and high specialization (d’ = 1) at the parasite lineage level. We compared observed (calculated on our data) with expected H2′ and d’ (estimated for random interactions) after generating 10,000 null models of the network, using the t-test in bipartite.
In order to evaluate the relationships between haemosporidian occurrence and environmental or species features within the Sardinian bird community, we ran binomial Generalized Linear Models (GLM) using the logit link function as implemented in R 3.6.1 [47]. Firstly, we assessed sampling site elevation as an environmental explanatory variable, in affecting the occurrence probability for each haemosporidian genus (Plasmodium, Haemoproteus and Leucocytozoon) as response variables (i.e., individual infection status; n = 217). Secondly, we tested species migratory patterns (sedentary, short-distance migrant or long-distance migrant), as an explanatory variable in predicting the occurrence probability of Sardinian-only haplotypes (i.e., those lineages that were found for the first time in this study). We considered novel Sardinian haplotype occurrence as a trait that points to haemosporidian geographic uniqueness. In this analysis, we included only infected individuals (n = 119), as haemosporidian-free individuals are not informative in terms of environmental or geographic features that may drive haplotype distribution. Among all performed models, no data overdispersion was detected. Models plots were made using the visreg R package [48].
3. Results
3.1. Prevalences of Haemosporidian Parasites
In total, we analyzed 217 individuals representing 32 bird species (three non-Passeriformes and 29 Passeriformes) belonging to 19 avian families. All of the individuals of the non-Passeriformes species were negative for infection (Caprimulgus europaeus, n = 1; Dendrocopos major, n = 4; Falco tinnunculus, n = 1), as were all individuals of six species of Passeriformes (Carduelis carduelis, n = 1; Hirundo rustica, n = 1; Lullula arborea, n = 1; Phylloscopus collybita, n = 1; Regulus ignicapilla, n = 2; Curruca sarda, n = 3).
Across all other species we found 119 positive individuals (total prevalence of 55.3%). Rates of infection varied from 14.3% (Emberiza calandra, n = 7 total samples) to 100% (Lanius senator, n = 2; Muscicapa tyrrhenica, n = 1; Garrulus glandarius, n = 7; Turdus viscivorus, n = 1) (Table 1). The prevalence in Passeriformes families for which we analyzed more than five individuals significantly varies from 38.5% in Emberizidae to 100% in Corvidae (Pearson’s Chi2-test: χ2 = 50.953, df = 9, p ≤ 0.001).

Table 1.
List of avian families and species tested for infection by haemosporidians, along with migratory patterns (S = Sedentary; SD = Short-distance migrant; LD = Long-distance migrant); number of sampled individuals (n); haemosporidian prevalence (prev.); number of positive (+) and negative (−) individuals for haemosporidian infection; number of infection relative to Haemoproteus (H), Leucocytozoon (L) Plasmodium (P) haplotypes; and list of haemosporidian haplotypes detected. SA precedes novel Sardinian haplotypes, followed by a unique number, and a designation of which parasite lineage is represented (e.g., SA14_H is the 14th novel lineage we identified, representing Haemoproteus). Haplotypes in bold highlight new host-parasite associations for known MalAvi lineages.
The number of haemosporidian lineages detected in each host species ranged from one (several species), to 19 in Sylvia atricapilla; other species with a high number of lineages are Parus major (15) and Passer hispaniolensis (14).
In total, we detected 116 unique haplotypes, 84 (72.4%) of which were novel, as they had at least a one base-pair difference from the lineages represented in the MalAvi database, while 32 (27.6%) haplotypes have previously been deposited in MalAvi (Table S1). The identified lineages were attributed to the genera of Haemoproteus (n = 55: 36 novel; 19 on MalAvi), Plasmodium (n = 20: 17 novel; 3 on MalAvi), and Leucocytozoon (n = 41: 31 novel; 10 on MalAvi).
Co-infections were recovered for 13 individuals of four species. Haemoproteus and Plasmodium co-infections were recovered from two individuals (Curruca melanocephala and Erithacus rubecula). Leucocytozoon and Plasmodium were recovered simultaneously in eight individuals (one Emberiza cirlus, three in both Parus major and Passer hispaniolensis and one Periparus ater). Haemoproteus and Leucocytozoon were recovered in three individuals (two Cyanistes caeruleus and one Muscicapa tyrrhenica) (Table S1).
Prevalence of Haemosporidian infections in the sampled localities (Table 2; Figure 1) varied from 33.3% (Cabras) to 69.2% (Priatu), and in two localities (Cabras and Marceddì, both about 20 km from the coast and effectively at sea level), we did not find Leucocytozoon infections. Haemoproteus infections were the most common in all sampling localities except in Monte Albo (780 m asl), where we identified an evident dominance of Leucocytozoon infections (Pearson’s Chi2-test: χ2 = 9.8, df = 2, p = 0.0074) of 80% versus 10% each for Plasmodium and Haemoproteus.

Table 2.
Sampling localities with information about elevation, environmental features, number of sampled host individuals (n); overall haemosporidian prevalence (prev.); haplotypes number found for Haemoproteus (H), Leucocytozoon (L) and Plasmodium (P).
Some host species showed a high number of infection occurrences: Sylvia atricapilla with 23 (17 Haemoproteus and six Leucocytozoon); Acrocephalus scirpaceus with 22 (Haemoproteus); Passer hispaniolensis with 21 and Parus major with 20 (Table S1).
Among the haemosporidian lineages detected in this study and recorded in MalAvi, some are relatable to morphologically described species. Plasmodium lineages SGS1 (Plasmodium relictum) and SYAT05 (Plasmodium vaughani) were the most frequently found: 10 times each in six species (from five families) and two species (from two families) respectively (Table 1). Another frequently detected haplotype, ARW1 (Haemoproteus belopolskyi), was found nine times but only in Acrocephalus scirpaceus. We also detected another seven lineages attributable to a described parasite species: MW1 that still pertains to Haemoproteus belopolskyi; PARUS1 to Haemoproteus majoris; ROBIN1 to Haemoproteus attenuates; SFC1 to Haemoproteus balmorali, SYAT01 and SYAT02 to Haemoproteus parabelopolskyi and TURDUS2 to Haemoproteus minutus.
We also identified 13 new host-parasite associations compared to data published on MalAvi (Table 1).
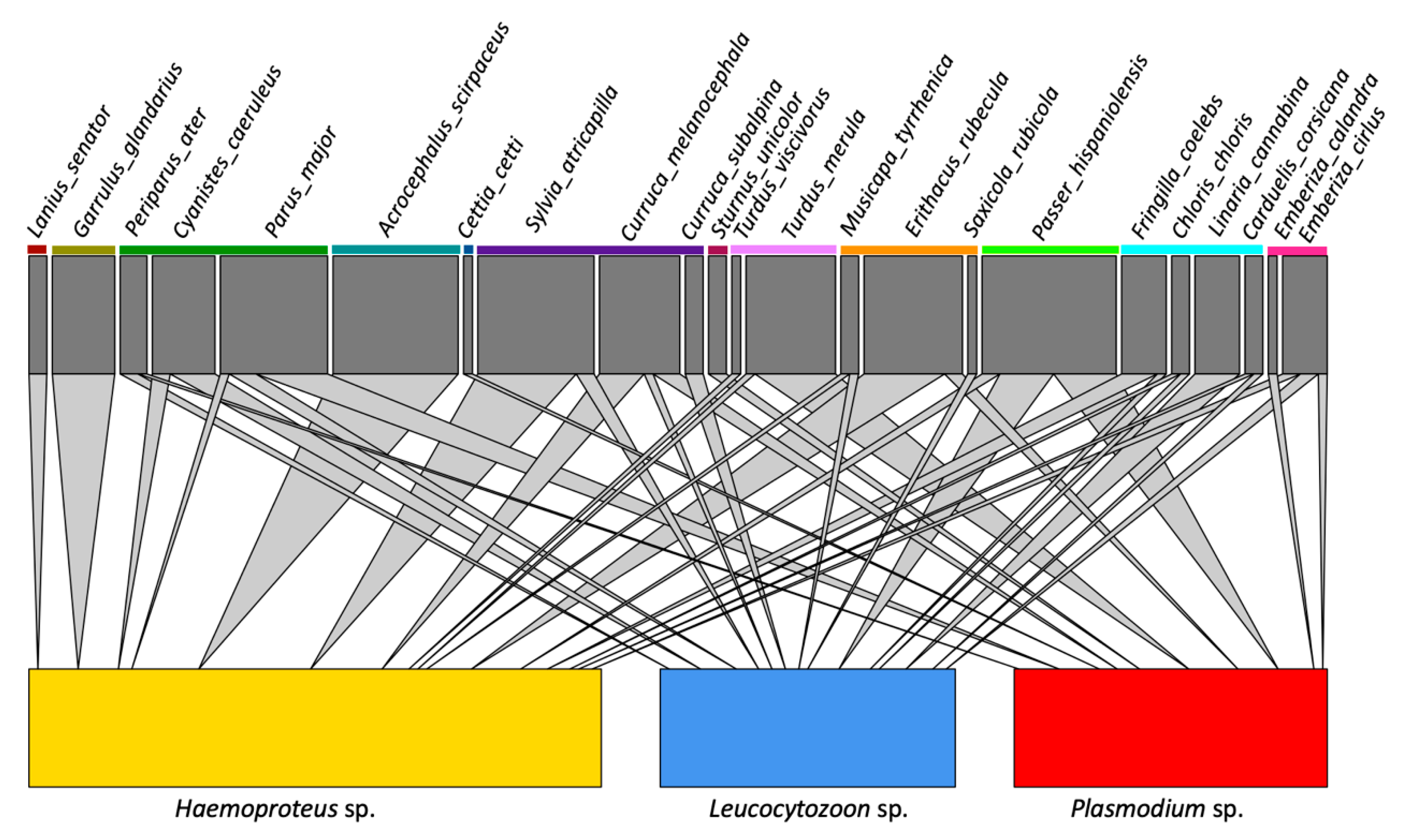
The host-parasite network is reported in Figure 2 (and see Figure S1). The host-parasite network-level specificity index in Sardinia showed a high value (H2′ = 0.756), significantly higher than the null model value (0.141; p ≤ 0.001). The species-level specificity index (d′) calculated on our dataset indicated a high specialization (≥0.70, null model value = 0.455; p < 0.001) for 12 haemosporidian lineages (Supporting information Table 2), three which are new lineages from Sardinia and nine having been previously described. Of the nine previously described, five were associated with a single species: CIRCUM05 found exclusively in Garrulus glandarius (five occurrences), RBS3 in Lanius senator (two occurrences), ROBIN1 in Erithacus rubecula (seven occurrences), ARW1 in Acrocephalus scirpaceus (nine occurrences), CUKI1 in Turdus viscivorus (two occurrences) (Table S1).
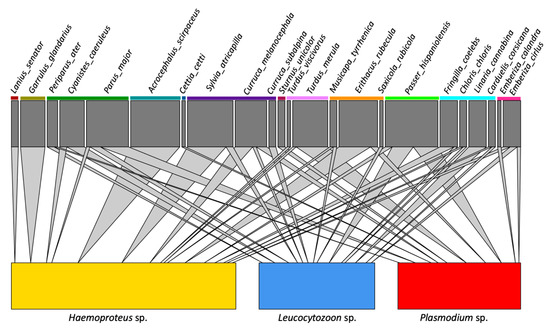
Figure 2.
Host-parasite networks of avian and haemosporidian Sardinian communities; parasites were grouped by genus. Interactions between a given host species and a haemosporidian genus is denoted with a cone; the size of the cone is proportional to the number of infections detected in each species. Colored lines under host species names indicate taxonomic family groupings.
3.2. Genetic Diversity and Phylogeny of Haemosporidian Lineages
p-distances calculated for each of the three haemosporidian genera were 0.054 (±0.006) in Haemoproteus, 0.062 (±0.006) in Leucocytozoon and 0.070 (±0.007) in Plasmodium. Between group mean distance across genera was 0.114 (±0.010) for Haemoproteus and Plasmodium, 0.160 (±0.014) between Haemoproteus and Leucocytozoon, and 0.166 (±0.014) between Plasmodium and Leucocytozoon.
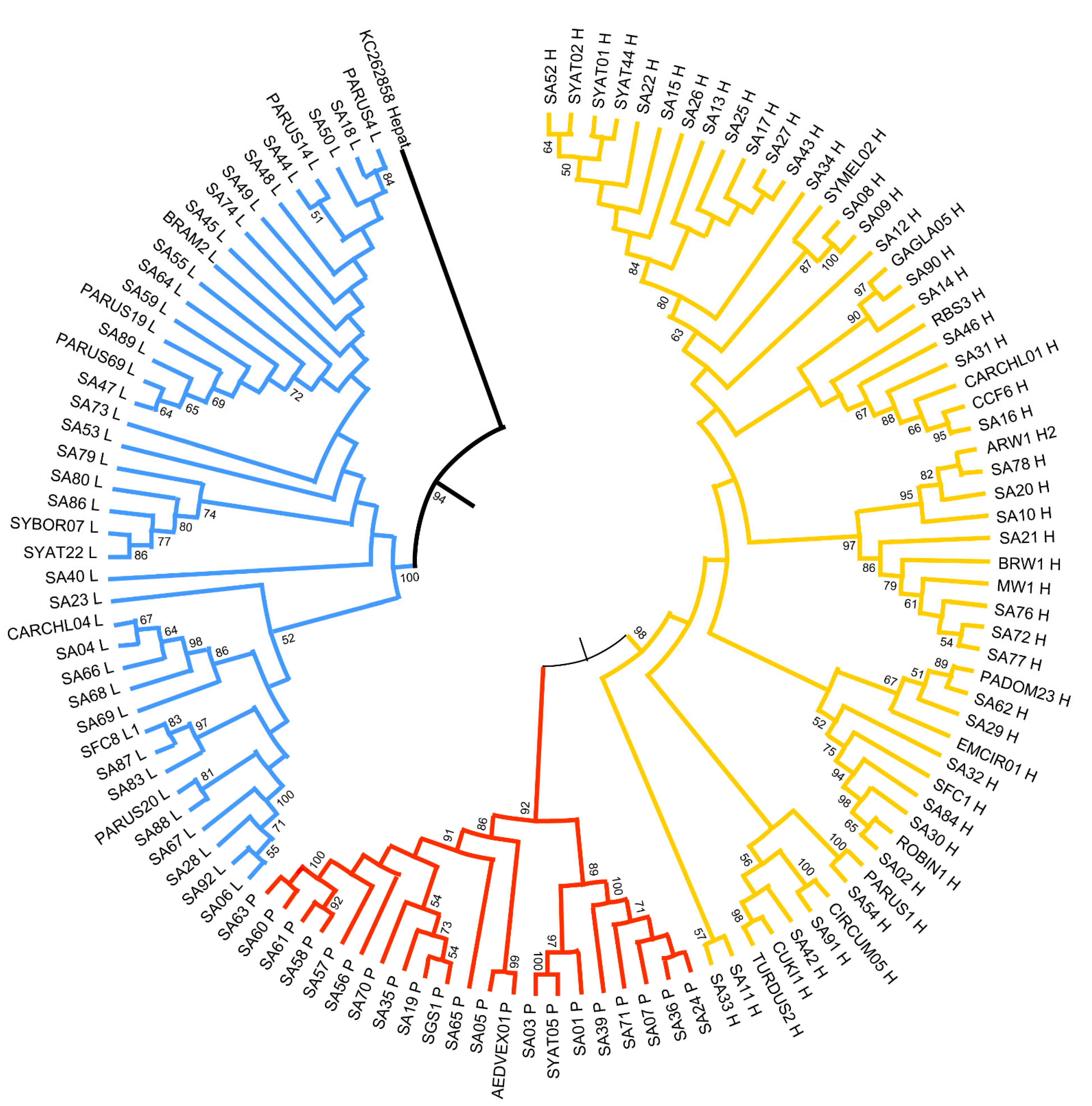
The Neighbor-joining tree (Figure 3) and Bayesian trees (GTR+G+I model) showed a similar topology. The phylogenetic reconstruction of avian haemosporidian parasite lineages clustered with strong support in three major clades representing the three genera: Plasmodium, Haemoproteus, and Leucocytozoon. Notably, most of the novel Sardinian lineages do not form monophyletic clusters; instead, many are more closely related to more widespread lineages than they are to other Sardinian-only lineages.
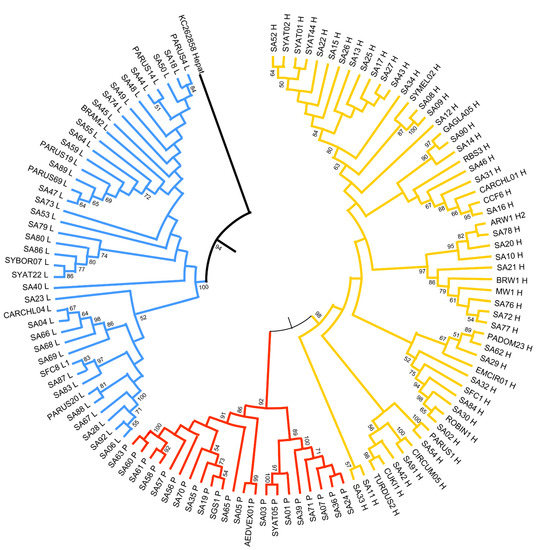
Figure 3.
Neighbor-joining tree of the partial cytb gene (470 bp) of Plasmodium (red), Haemoproteus (yellow), and Leucocytozoon (blue) sequenced from Sardinian birds. Bootstrap values (≥50%) are provided. “SA” precedes novel lineages found in this study; other lineages are named based on their match to MalAvi lineages.
3.3. Haemosporidian Occurrence Predictors
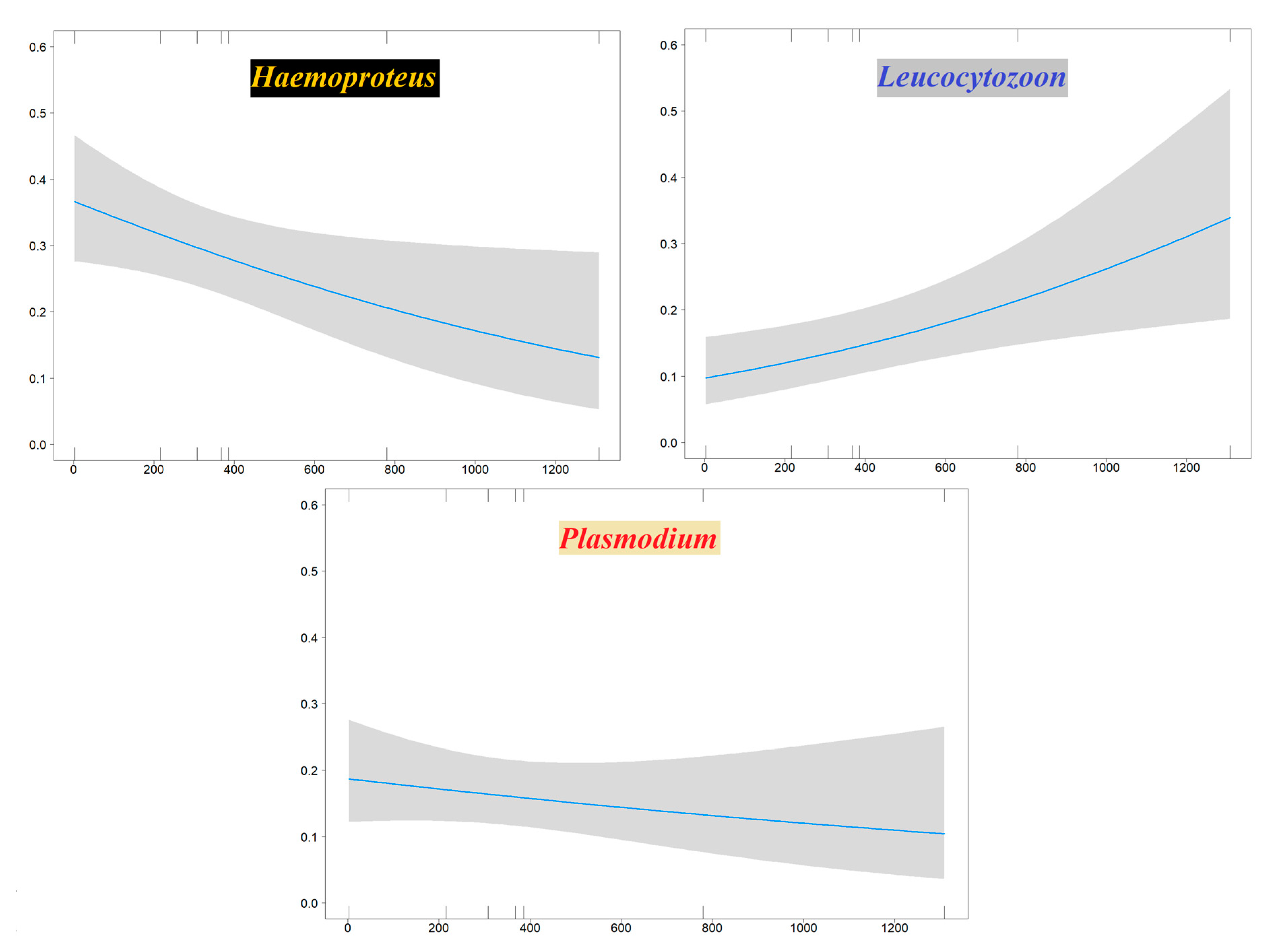
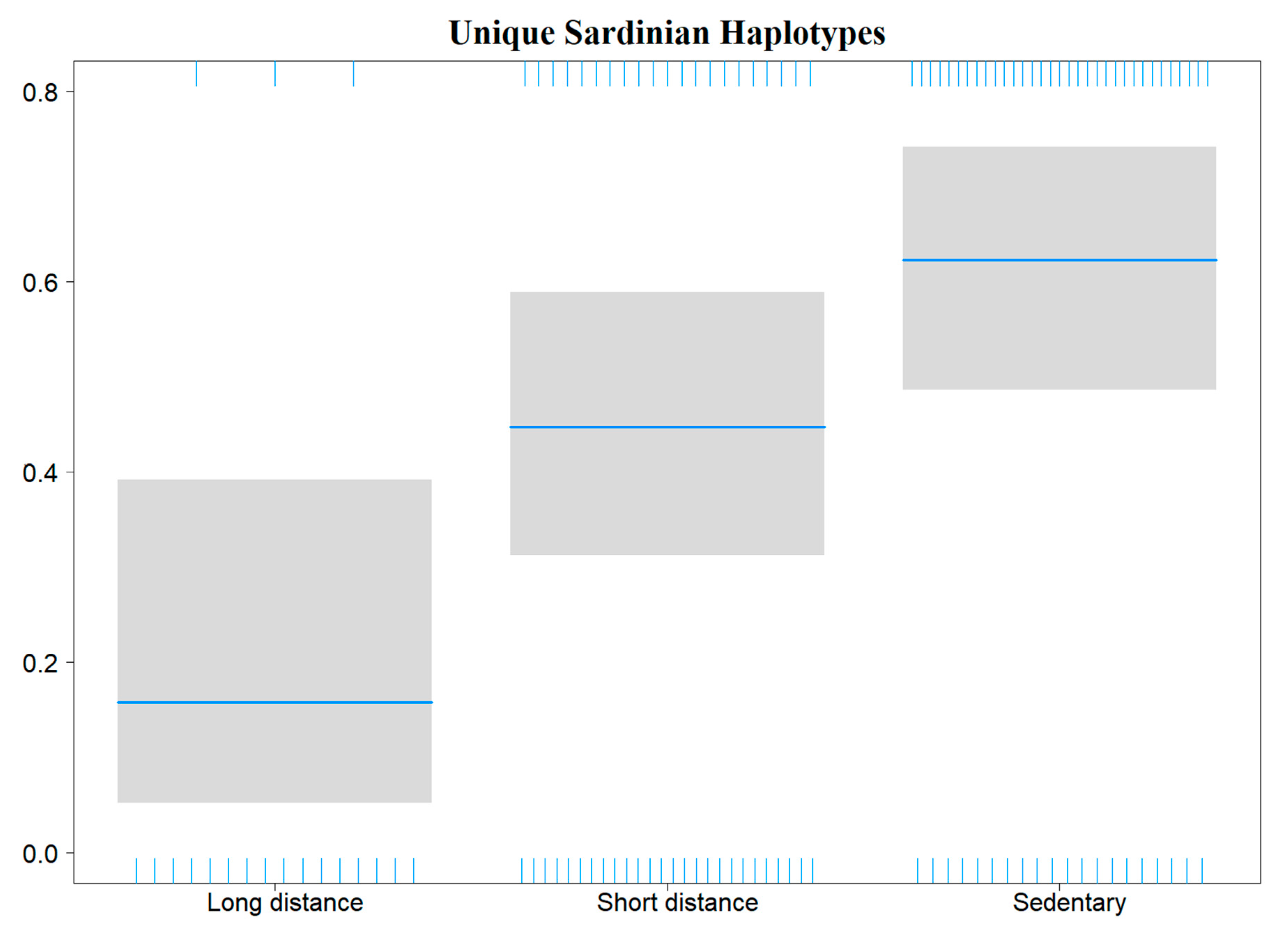
Binomial generalized linear models (GLMs) performed to evaluate haemosporidian genus occurrence probability in the Sardinian bird community showed a significant influence of sampling site elevation. This relationship was negative for Haemoproteus (decreasing occurrence with increasing elevation) and positive for Leucocytozoon; no effect of elevation was evident for Plasmodium (Table 3, Figure 4). A model assessing novel Sardinian haplotypes occurrence probability showed an effect related to host species migratory patterns, where the three migratory categories (sedentary, short-distance migrants and long-distance migrants) respectively display higher, intermediate and lower infection probabilities (Table 3, Figure 5).

Table 3.
Binomial generalized linear models (GLMs) results. The first three (each parasite genus) assess the effects of sample site elevation on haemosporidian occurrence probability, across the entire Sardinian bird community that we sampled. The last result shows the influence of species migratory pattern in affecting novel Sardinian haplotype occurrence probability in infected individuals.
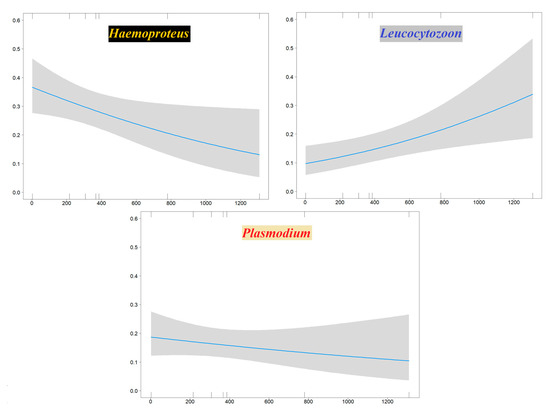
Figure 4.
Plots showing the effects of the sampling site elevation (X axis: meters asl) on the occurrence probability (Y axis) of Haemoproteus (left), Leucocytozoon (right), Plasmodium (bottom) on Sardinian birds.
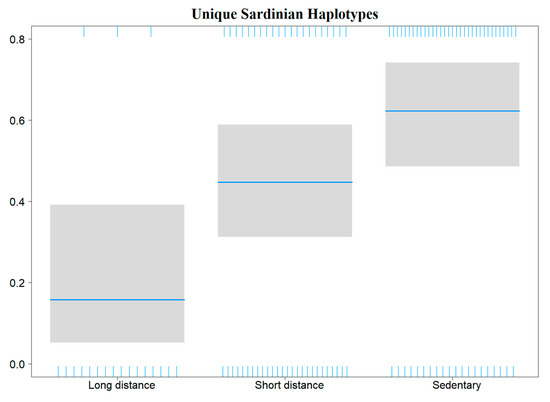
Figure 5.
Plot showing the effects of species migratory patterns (X axis) on the occurrence probability of novel haplotypes found in Sardinia (Y axis) in those birds infected by haemosporidians.
4. Discussion
4.1. Sardinian Haemosporidian Diversity
The present study provides the first molecular screening of avian malaria parasites in the bird breeding community from Sardinia, and in general the first from Mediterranean islands.
Our analyses detected haemosporidian infections in 119 individuals from 23 species (of 32 bird species analyzed), and recovered 116 haplotypes, 84 of which are new to MalAvi. In nine host species, we did not observe any infections, and, although the number of birds sampled in some species was low, the negative results from Caprimulgus europaeus, Dendrocopos major, Lullula arborea, Regulus ignicapilla, and Curruca sarda is consistent with there being no molecular evidence of infections recorded to date (see MalAvi).We identified for the first time a haemosporidian parasite infection in four species: Muscicapa tyrrhenica, Saxicola rubicola, Carduelis corsicana and Lanius senator. For the latter species, a previous association was indirectly related to louse flies [49].
In most of the examined host species, we detected prevalence levels comparable to those previously published, such as high prevalence of infections of Plasmodium in Turdus merula [50] and of Haemoproteus in Garrulus glandarius [51].
In other species, we detected an intermediate level of prevalence, which is comparable to other studies. For example, the prevalence level of 63.6% in Acrocephalus scirpaceus, a species typical of marshes dominated by common reed (Phragmites australis) and sampled only in Marceddì (reed-dominated wetlands), is somewhat lower than that 84.6% detected in Spain by Fernandez et al. [52], but it is higher than the 33.1% detected in Portuguese wetlands by Ventim et al. [53]. The most highly sampled host species in our study was Passer hispaniolensis, which showed a prevalence of 44.4%. This level of prevalence is generally similar to the value (55.6%) reported for Turkish populations of this species [54]. However, interestingly, the infection rates between that study and ours differed with respect to levels of parasite genus infections: our Passer hispaniolensis were infected by, in ascending order, Haemoproteus (13.3%), Leucocytozoon (40%) and Plasmodium (46.7%), while in Turkey Plasmodium infections were lacking and Haemoproteus was the main infecting genus.
We found substantial differences in infection rate across bird families (Table 1) suggesting that certain families experience a higher amount of parasite infections than others do. This result is in line with findings reported by for example, Harvey and Voelker [17] in Benin, Clark et al. [18] in the Western Palearctic, and Bodawatta et al. [19] in both New Guinea and the Canary Islands. Despite these differences being observed across diverse regions, the factors that may affect the haemosporidian prevalence at the host family level have not yet been identified [20].
The host-parasite network in Sardinia indicated a high value of the specificity H2′ index (0.756), which suggests that the Sardinian host-parasite community is specialized, and this result differs from the more generalized host-parasite community observed in the Canary and New Guinea Islands [19]. These different community structures could be due to the differences in bird species and environmental diversity: i.e., the Canary Islands are smaller and contain fewer breeding species than Sardinia, while the more diverse bird community in New Guinea was sampled over a long elevational gradient (from 200 to 3700 m asl).
When considering the specialization of individual haemosporidian lineages, we found some lineages that showed a high d’ species-level specificity index [46]. In particular, a high value (d’ ≥ 0.70) was found for three new Sardinian lineages (SA11_H, SA03_P, SA05_P). Another eight cases of specialization pertain to previously described lineages showing a high specificity, that was confirmed in two cases (CUKI1 and SYATT44), while high specificity was not a general character in five lineages that were also found in other host species sampled elsewhere ([50]; MalAvi database). These values could indicate an artifact due to sampling or a trend to specialization in an insular context. Overall, the number of sampled Sardinian birds per species in some cases are low and there is evidence to suggest that wider sampling, on both islands and in mainland areas, could increase the chance of detecting these apparently novel or rare haplotypes [55].
4.2. Lineage Distributions
While we detected 84 haemosporidian lineages from Sardinia that are novel to MalAvi, we also detected 32 lineages previously described in other studies. Of these 32, only four were shared with the Italian peninsula and only two with Tunisia; these are the closest mainland countries to Sardinia. The most observed haplotypes in our samples are SGS1 and SYAT05. The former is also the most widespread haplotype in the world, is ascribable to Plasmodium relictum, and has been reported in 127 different host species, encompassing 73 genera and 32 families (MalAvi). SYAT05 (Plasmodium vaughani), that we detected in two species (with 10 occurrences), is another common lineage reported from 19 different countries (including Italy) for 33 host species, encompassing 23 genera and 16 families (MalAvi). We identified the presence of Leucocytozoon sp. SYAT22 in three different bird species on Sardinia; this lineage has been detected from the Canary Islands to Sweden [56,57,58].
Our results also provide new insights regarding specific host-parasite relationships. For example, Haemoproteus haplotypes CIRCUM05 and GAGLA05 were found only in Garrulus glandarius (a corvid species) in this study, which is consistent with them having only been recorded from Corvidae species to date (MalAvi). This may indicate a close relationship between these haplotypes and Corvidae, indicative of some level of host-parasite specificity, and by extension parasite specialization. Similar host-specificity appears evident for SYAT44, which has been only recorded from Sylvia atricapilla, both in Sardinia and Europe, for RBS3 which appears to parasitize only Lanius host species [59,60], for SYLMEL02 [52] and SYBOR07 [51,57,60,61,62] which have only been found in Sylvia hosts, for PADOM23 found only in Passer hosts [51,53,63], for AEDVEX1 found only in Emberiza hosts [24] and for BRAM2 found only in Fringilla hosts [51,57,60]. Conversely, some haemosporidian lineages are less specific, and include SFC1 (Haemoproteus balmorali), which we only detected in Muscicapa tyrrhennica (a Muscicapidae), but which has also been observed in Turdidae and Paridae (MalAvi).
4.3. Genetic Diversity
While phylogenetic reconstruction readily identified the three parasite genera and their relationships to one another, the relationship of haplotypes in each genus is generally unresolved. Therefore, while cytb is a good molecular barcode for Haemosporidian identification and characterization, it is unable to reliably disentangle phylogenetic structure within genera. This is likely attributable to both the short portion of the gene that is used as the barcode standard (479 base pairs), and the high substitution rate possible at third codon positions [64].
Haemosporidian genetic diversity observed in Sardinia is high as shown by the large number of new (84) and previously described (32) lineages detected. In this regard, the comparatively high numbers of new lineages have only been found in a few other studies, e.g., one from Benin in western Africa [17], with 85 new lineages and an infection rate of 50.9%, and another from northwestern Iberia and northwestern Africa with 111 new lineages and 56.5% of infection rate [52].
The high genetic diversity shown in the present study could reflect the complexity and the considerable molecular rate variation among haemosporidians [18,65]. Moreover, it could be explained by Sardinia as a general source of biodiversity [66], with co-evolution between host species and haemosporidian lineages being driven by isolation [67]. In this context, the last connections between Sardinia and continental land masses are dated to at least 5 mya, during the Messinian salinity crisis events [68]. Another possible factor contributing to the high number of novel lineages could be that a burst of diversification may have occurred following a mosquito vector eradication program that was enacted on Sardinia from 1946–1950 [69]. During this period, 267 metric tons of DDT (dichloro-diphenyl-trichloroethane), a wide range insecticide, were spread across the island, vegetation was cut back, and 100,000 acres of swampland were drained; the presumed reduction of mosquito breeding habitat was 99.93%, and zero cases of human malaria were reported in 1950 (the disease has been considered endemic to Sardinia since 502 B.C., with significant annual human mortality [69]). While targeting human malaria mosquito vectors (Anopheles sp. that breed in open water, in marshes and mountain streams), DDT would obviously have impacted avian haemosporidian vectors including not only mosquitos, but also black flies [70] and likely midges as well. Thus, this decrease of vectors could also explain the high number of novel haplotypes relative to previously known haplotypes across all three genera in our study. Indeed, Imamura et al. [71] recorded a severe bottleneck in another protozoan parasite, Leishmania donovani, coincident with DDT spraying campaigns for malaria eradication on the Indian subcontinent in the 1960′s. Shortly after this eradication, Leishmania donovani populations not only increased, but they underwent significant bursts of diversification that generated new lineages (strains) of the disease agent. Finally, at least some of lineages that we have called novel might simply be an artifact of incomplete sampling on nearby continents. A new study of the avian haemosporidian community on São Tomé [72] also found high numbers of novel lineages that they termed “endemics”. In justifying the naming of endemic lineages, they referred to the nearly 650 described sub-Saharan African haemosporidian lineages in MalAvi, only some of which were present in their study. By comparison, sampling in the Western Palearctic has been far more extensive in terms of both geographic coverage and avian host diversity, and as such, we are confident that a considerable number of our novel lineages will remain so as new continental areas are sampled.
4.4. Effect of Elevation and Host Species Migratory Pattern
Two of our GLMs analyses support an effect of elevation on infection occurrence probability: a positive effect for Leucocytozoon and a negative one for Haemoproteus, while no significant effect was recovered for Plasmodium. The Leucocytozoon pattern is likely due to distributional constraints affecting its insect vectors. Indeed, the occurrence of this haemosporidian genus is positively related to elevation since in its main vector species, black flies (Simuliidae), larvae often reach high-density values in streams with high flow speed and low water temperature [73]. These stream features both tend to increase with elevation, and black fly species richness is highest near mid-level elevations (e.g., 1985 m asl in the Italian Alps [74]; 1405 m asl in Malaysia, [75]). The positive relationship between Leucocytozoon and elevation we found on Sardinia is consistent with other studies carried out in other geographic regions [15,76]. The decreasing occurrence of Haemoproteus with elevation on Sardinia is a pattern consistent with several studies which also found a negative relationship between elevation and the distribution of this genus (e.g., [77]; also see Doussang et al. [78] for a converse relationship). Haemoproteus is vectored mainly by biting midges (Ceratopogonidae) and louse flies (Hippoboscidae). Biting midges should be negatively affected by low temperatures and as such should decrease with increasing elevation [79], particularly above mid-level elevations [80]. Although there is a general decrease of Plasmodium infections at high elevation [76], in our study a negative effect of elevation on Plasmodium occurrence was not statistically significant.
Finally, we found a relationship between avian host species migratory patterns and the occurrence probability of haemosporidian haplotypes found exclusively on Sardinia. Infected individuals belonging to sedentary host species, for which we would assume each one is part of an endemic population, show a higher probability of carrying haplotypes (so far) unique to Sardinia, as compared to-short-distance (i.e., intra-Palaearctic) and long-distance (i.e., trans-Saharan) migrants. While this relationship might be due at least in part to the DDT eradication program mentioned above, it might also be due to the fact that long-distance migratory species spend a considerable part of the year in tropical latitudes, where haemosporidian transmission is likely not constrained by climatic conditions (i.e., temperature) for both vector and protozoan reproductive cycles [6,27]. Conversely, haemosporidian infection in sedentary species and short-distance migrants (restricted by the temperate climate) occurs mostly during spring and summer seasons [81]. This also likely reduces the possibility of the latter two categories becoming infected by parasite lineages not occurring in Sardinia (e.g., during overwintering of northern bird populations in Sardinia, or long-distance migrants passing through).
In conclusion, while the Western Palearctic is one of the most studied regions with respect to avian haemosporidian parasites, our results indicate that there is much yet to learn about their distributions, associations, and means of transmission via habitat, elevation and host life-history variables. While we have discussed 84 Sardinia-novel lineages here, we fully expect that some may prove to have wider distributions as other insular (e.g., Corsica, Sicily) or under-sampled (e.g., Italy) avian host populations are sampled. Furthermore, a focus on the insect vectors of avian haemosporidians is required to better assess haemosporidian ecological determinants, such as elevation and other environmental features to include available water sources and types for insect vector breeding. Indeed, studies focused on avian haemosporidians from other Mediterranean islands would not only complete the knowledge gap in this geographic context but would also provide for robust assessments of the comparative role of islands in their diversification.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/13/2/75/s1, Figure S1: Host-parasite network of bird species and haemosporidian lineages. Table S1: Table of lineage, host, locality and GenBank accession numbers Table S2: Value of d’ species-level specificity index calculated for each lineage.
Author Contributions
G.B., I.P., M.P. and G.V. conceived of and designed the study. G.B., M.P., H.L.P. and G.V. collected the samples in the field. I.P. performed the molecular and data analyses. M.C. and L.I. performed statistical analyses. I.P. and L.I. prepared the manuscripts with contributions from all other authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ricerca Locale FAR 2019-2021 University of Eastern Piedmont.
Institutional Review Board Statement
The study was approved by the Assessorato della Difesa dell’Ambiente of the Regione Autonoma della Sardegna (Prot. 822/18 of 15 January 2018, Det. 10616/288 of 11 May 2018) and of the Italian Istituto Superiore per la Protezione e la Ricerca Ambientale (Prot. 9447 of 26 Aprch 2018).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Marcello Grussu for his help during fieldwork and Danilo Pisu for the suggestions about Monte Limbara, we also thank Valeria Benotti who made with us with the lab analyses and Johanna Harvey for the useful suggestions about lab protocol. We also would like to thank Paolo Onida (Regione Sardegna) and Lorenzo Serra (ISPRA) for the sampling and ringing authorizations. Finally, we would thank three anonymous reviewers for their comments and suggestions that improved the whole manuscript. This is publication number 1633 of the Biodiversity Research and Teaching Collections at Texas A&M University and publication number 10 of the W.I.N.E.S. collaborative research group.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fletcher, K.; Träff, J.; Gustafsson, L. Importance of infection of haemosporidia blood parasites during different life history stages for long-term reproductive fitness of collared flycatchers. J. Avian Biol. 2019, 50, 1–11. [Google Scholar] [CrossRef]
- Schoener, E.R.; Banda, M.; Howe, L.; Castro, I.C.; Alley, M.R. Avian malaria in New Zealand. N. Z. Vet. J. 2014, 62, 189–198. [Google Scholar] [CrossRef]
- Van Riper, C., III; van Riper, S.G.; Lee Goff, M.; Laird, M. The Epizootiology and Ecological Significance of Malaria in Hawaiian Land Birds. Ecol. Monogr. 1986, 56, 327–344. [Google Scholar] [CrossRef]
- Magallanes, S.; García-Longoria, L.; López-Calderón, C.; Reviriego, M.; de Lope, F.; Møller, A.P.; Marzal, A. Uropygial gland volume and malaria infection are related to survival in migratory house martins. J. Avian Biol. 2017, 48, 1355–1359. [Google Scholar] [CrossRef]
- Bensch, S.; Hellgren, O.; PÉrez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef]
- Valkiūnas, G. Avian Malaria Parasites and Other Haemosporidia; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Santiago-Alarcon, D.; Palinauskas, V.; Schaefer, H.M. Diptera vectors of avian Haemosporidian parasites: Untangling parasite life cycles and their taxonomy. Biol. Rev. 2012, 87, 928–964. [Google Scholar] [CrossRef] [PubMed]
- Drovetski, S.V.; Aghayan, S.A.; Mata, V.A.; Lopes, R.J.; Mode, N.A.; Harvey, J.A.; Voelker, G. Does the niche breadth or trade-off hypothesis explain the abundance-occupancy relationship in avian Haemosporidia? Mol. Ecol. 2014, 23, 3322–3329. [Google Scholar] [CrossRef]
- Harrigan, R.J.; Sedano, R.; Chasar, A.C.; Chaves, J.A.; Nguyen, J.T.; Whitaker, A.; Smith, T.B. New host and lineage diversity of avian haemosporidia in the northern Andes. Evol. Appl. 2014, 7, 799–811. [Google Scholar] [CrossRef]
- Zehtindjiev, P.; Križanauskienė, A.; Bensch, S.; Palinauskas, V.; Asghar, M.; Dimitrov, D.; Scebba, S.; Valkiūnas, G. A New Morphologically Distinct Avian Malaria Parasite That Fails Detection by Established Polymerase Chain Reaction–Based Protocols for Amplification of the Cytochrome B Gene. J. Parasitol. 2012, 98, 657–665. [Google Scholar] [CrossRef]
- Scaglione, F.E.; Cannizzo, F.T.; Chiappino, L.; Sereno, A.; Ripepi, M.; Salamida, S.; Manuali, E.; Bollo, E. Plasmodium spp. In a captive raptor collection of a safaripark in northwest Italy. Res. Vet. Sci. 2016, 104, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Fecchio, A.; Pinheiro, R.; Felix, G.; Faria, I.P.; Pinho, J.B.; Lacorte, G.A.; Braga, E.M.; Farias, I.P.; Aleixo, A.; Tkach, V.V.; et al. Host community similarity and geography shape the diversity and distribution of haemosporidian parasites in Amazonian birds. Ecography 2018, 41, 505–515. [Google Scholar] [CrossRef]
- Ricklefs, R.E.; Swanson, B.L.; Fallon, S.M.; Martínez-Abraín, A.; Scheuerlein, A.; Gray, J.; Latta, S.C. Community relationships of avian malaria parasites in southern Missouri. Ecol. Monogr. 2005, 75, 543–559. [Google Scholar] [CrossRef]
- Liu, B.; Deng, Z.; Huang, W.; Dong, L.; Zhang, Y. High prevalence and narrow host range of haemosporidian parasites in Godlewski’s bunting (Emberiza godlewskii) in northern China. Parasitol. Int. 2019, 69, 121–125. [Google Scholar] [CrossRef]
- González, A.D.; Matta, N.E.; Ellis, V.A.; Miller, E.T.; Ricklefs, R.E.; Gutiérrez, H.R. Mixed species flock, nest height, and elevation partially explain avian haemoparasite prevalence in Colombia. PLoS ONE 2014, 9, e100695. [Google Scholar] [CrossRef]
- Fecchio, A.; Ellis, V.A.; Bell, J.A.; Andretti, C.B.; D’horta, F.M.; Silva, A.M.; Tkach, V.V.; Weckstein, J.D. Avian malaria, ecological host traits and mosquito abundance in southeastern Amazonia. Parasitology 2017, 144, 1117–1132. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.A.; Voelker, G. Host associations and climate influence avian haemosporidian distributions in Benin. Int. J. Parasitol. 2019, 49, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.J.; Drovetski, S.V.; Voelker, G. Robust geographical determinants of infection prevalence and a contrasting latitudinal diversity gradient for haemosporidian parasites in Western Palearctic birds. Mol. Ecol. 2020, 29, 3131–3143. [Google Scholar] [CrossRef] [PubMed]
- Bodawatta, K.H.; Synek, P.; Bos, N.; Garcia-del-Rey, E.; Koane, B.; Marki, P.Z.; Albrecht, T.; Lifjeld, J.; Poulsen, M.; Munclinger, P.; et al. Spatiotemporal patterns of avian host–parasite interactions in the face of biogeographical range expansions. Mol. Ecol. 2020, 29, 2431–2448. [Google Scholar] [CrossRef]
- Clark, N.J.; Clegg, S.M.; Lima, M.R. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int. J. Parasitol. 2014, 44, 329–338. [Google Scholar] [CrossRef]
- Illera, J.C.; López, G.; García-Padilla, L.; Moreno, Á. Factors governing the prevalence and richness of avian haemosporidian communities within and between temperate mountains. PLoS ONE 2017, 12, e0184587. [Google Scholar] [CrossRef]
- Ferraguti, M.; de la Puente, J.M.; Bensch, S.; Roiz, D.; Ruiz, S.; Viana, D.S.; Soriguer, R.C.; Figuerola, J. Ecological determinants of avian malaria infections: An integrative analysis at landscape, mosquito and vertebrate community levels. J. Anim. Ecol. 2018, 87. [Google Scholar] [CrossRef]
- Garcia-Longoria, L.; Marzal, A.; De Lope, F.; Garamszegi, L. Host-parasite interaction explains variation in the prevalence of avian haemosporidians at the community level. PLoS ONE 2019, 14, e0205624. [Google Scholar] [CrossRef] [PubMed]
- Stanković, D.; Jönsson, J.; Raković, M. Diversity of avian blood parasites in wild passerines in Serbia with special reference to two new lineages. J. Ornithol. 2019, 160, 545–555. [Google Scholar] [CrossRef]
- De La Puente, J.M.; Muñoz, J.; Capelli, G.; Montarsi, F.; Soriguer, R.; Arnoldi, D.; Rizzoli, A.; Figuerola, J. Avian malaria parasites in the last supper: Identifying encounters between parasites and the invasive Asian mosquito tiger and native mosquito species in Italy. Malar. J. 2015, 14, 1–7. [Google Scholar] [CrossRef]
- Romano, A.; Nodari, R.; Bandi, C.; Caprioli, M.; Costanzo, A.; Ambrosini, R.; Rubolini, D.; Parolini, M.; Epis, S.; Saino, N. Haemosporidian parasites depress breeding success and plumage coloration in female barn swallows Hirundo rustica. J. Avian Biol. 2019, 50, 1–14. [Google Scholar] [CrossRef]
- Gutiérrez-López, R.; Gangoso, L.; De La Puente, J.M.; Fric, J.; López-López, P.; Mailleux, M.; Muñoz, J.; Touati, L.; Samraoui, B.; Figuerola, J. Low prevalence of blood parasites in a long-distance migratory raptor: The importance of host habitat. Parasites Vectors 2015, 8, 4–9. [Google Scholar] [CrossRef]
- Hellgren, O.; Wood, M.J.; Waldenström, J.; Hasselquist, D.; Ottosson, U.; Stervander, M.; Bensch, S. Circannual variation in blood parasitism in a sub-Saharan migrant passerine bird, the garden warbler. J. Evol. Biol. 2013, 26, 1047–1059. [Google Scholar] [CrossRef]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J. Biogeogr. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Medail, F.; Quezel, P. Biodiversity hotspots in the Mediterranean Basin: Setting global conservation priorities. Conserv. Biol. 1999, 13, 1510–1513. [Google Scholar] [CrossRef]
- Grussu, M. Checklist of the birds of Sardinia. Aves Ichnusae 2001, 4, 2–55. [Google Scholar]
- Pellegrino, I.; Negri, A.; Boano, G.; Cucco, M.; Kristensen, T.; Pertoldi, C.; Randi, E.; Salek, M.; Mucci, N. Evidence for strong genetic structure in European populations of the little owl Athene noctua. J. Avian Biol. 2015, 46, 462–1475. [Google Scholar] [CrossRef]
- Pellegrino, I.; Boatti, L.; Cucco, M.; Mignone, F.; Kristensen, T.N.; Mucci, N.; Randi, E.; Ruiz-Gonzalez, A.; Pertoldi, C. Development of SNP markers for population structure and phylogeography characterization in little owl (Athene noctua) using a genotyping- by-sequencing approach. Conserv. Genet. Resour. 2016, 8, 13. [Google Scholar] [CrossRef]
- Pons, J.-M.; Thibault, J.-C.; Aymí, R.; Grussu, M.; Muntaner, J.; Olioso, G.; Sunyer, J.R.; Touihri, M.; Fuchs, J. The role of western Mediterranean islands in the evolutionary diversification of the spotted flycatcher Muscicapa striata, a long-distance migratory passerine species. J. Avian Biol. 2016, 47, 386–398. [Google Scholar] [CrossRef]
- Zuccon, D.; Pons, J.; Boano, G.; Chiozzi, G.; Gamauf, A.; Mengoni, C.; Nespoli, D.; Olioso, G.; Pavia, M.; Pellegrino, I.; et al. Type specimens matter: New insights on the systematics, taxonomy and nomenclature of the subalpine warbler (Sylvia cantillans) complex. Zool. J. Linn. Soc. 2020, 169, 1–28. [Google Scholar] [CrossRef]
- Cucco, M.; Grenna, M.; Pellegrino, I.; Malacarne, G. Egg-sequence rather than mating preference influences female egg investment in the red-legged partridge. Ethol. Ecol. Evol. 2011, 23, 343–357. [Google Scholar] [CrossRef]
- Seutin, G.; White, B.N.; Boag, P.T. Preservation of avian blood and tissue samples for DNA analysis. Can. J. Zool. 1991, 69, 82–90. [Google Scholar] [CrossRef]
- Gill, F.; Donsker, D.; Rasmussen, P. (Eds.) IOC World Bird List (v11.1). 2021. Available online: https://www.worldbirdnames.org/ioc-lists/crossref/ (accessed on 29 September 2020). [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Haplotype phasing: Existing methods and new developments. Nat. Rev. Genet. 2011, 12, 703–714. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Gen. 2001, 68, 978-–989. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Blüthgen, N.; Menzel, F.; Blüthgen, N. Measuring specialization in species interaction networks. BMC Ecol. 2006, 6. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 29 October 2020).
- Breheny, P.; Burchett, W. Visualization of regression models using Visreg. R J. 2017, 9, 56–71. [Google Scholar] [CrossRef]
- Gangoso, L.; Gutiérrez-López, R.; Martínez-de la Puente, J.; Figuerola, J. Louse flies of Eleonora’s falcons that also feed on their prey are evolutionary dead-end hosts for blood parasites. Mol. Ecol. 2019, 1812–1825. [Google Scholar] [CrossRef]
- Himmel, T.; Harl, J.; Pfanner, S.; Nedorost, N.; Nowotny, N.; Weissenböck, H. Haemosporidioses in wild Eurasian blackbirds (Turdus merula) and song thrushes (T. philomelos): An in situ hybridization study with emphasis on exo-erythrocytic parasite burden. Malar. J. 2020, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mata, V.A.; da Silva, L.P.; Lopes, R.J.; Drovetski, S.V. The Strait of Gibraltar poses an effective barrier to host-specialised but not to host-generalised lineages of avian Haemosporidia. Int. J. Parasitol. 2015, 45, 711–719. [Google Scholar] [CrossRef]
- Fernández, M.; Rojo, M.Á.; Casanueva, P.; Carrión, S.; Hernández, M.Á.; Campos, F. High prevalence of haemosporidians in Reed Warbler Acrocephalus scirpaceus and Sedge Warbler Acrocephalus schoenobaenus in Spain. J. Ornithol. 2010, 151, 27–32. [Google Scholar] [CrossRef]
- Ventim, R.; Tenreiro, P.; Grade, N.; Encarnação, P.; Araújo, M.; Mendes, L.; Pérez-Tris, J.; Ramos, J.A. Characterization of haemosporidian infections in warblers and sparrows at south-western European reed beds. J. Ornithol. 2012, 153, 505–512. [Google Scholar] [CrossRef]
- Ciloglu, A.; Ergen, A.G.; Inci, A.; Dik, B.; Duzlu, O.; Onder, Z.; Yetismis, G.; Bensch, S.; Valkiūnas, G.; Yildirim, A. Prevalence and genetic diversity of avian haemosporidian parasites at an intersection point of bird migration routes: Sultan Marshes National Park, Turkey. Acta Trop. 2020, 210. [Google Scholar] [CrossRef] [PubMed]
- Jovani, R.; Tella, J.L. Parasite prevalence and sample size: Misconceptions and solutions. Trends Parasitol. 2006, 22, 214–218. [Google Scholar] [CrossRef]
- Illera, J.C.; Fernández-Álvarez, Á.; Hernández-Flores, C.N.; Foronda, P. Unforeseen biogeographical patterns in a multiple parasite system in Macaronesia. J. Biogeogr. 2015, 42, 1858–1870. [Google Scholar] [CrossRef]
- Ellis, V.A.; Huang, X.; Westerdahl, H.; Jönsson, J.; Hasselquist, D.; Neto, J.M.; Nilsson, J.Å.; Nilsson, J.; Hegemann, A.; Hellgren, O.; et al. Explaining prevalence, diversity and host specificity in a community of avian haemosporidian parasites. Oikos 2020, 129, 1314–1329. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, A.; Ramírez, Á.; Richardson, D.S.; Pérez-Tris, J. Evolution of parasite island syndromes without long-term host population isolation: Parasite dynamics in Macaronesian blackcaps Sylvia atricapilla. Glob. Ecol. Biogeogr. 2013, 22, 1272–1281. [Google Scholar] [CrossRef]
- Dimitrov, D.; Zehtindjiev, P.; Bensch, S. Genetic diversity of avian blood parasites in SE Europe: Cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitol. 2010, 55, 201–209. [Google Scholar] [CrossRef]
- Hellgren, O.; Waldenström, J.; Peréz-Tris, J.; Szöll Ösi, E.; Hasselquist, D.; Krizanauskiene, A.; Ottosson, U.; Bensch, S. Detecting shifts of transmission areas in avian blood parasites—A phylogenetic approach. Mol. Ecol. 2007, 16, 1281–1290. [Google Scholar] [CrossRef]
- Hellgren, O.; Kutzer, M.; Bensch, S.; Valkiūnas, G.; Palinauskas, V. Identification and characterization of the merozoite surface protein 1 (msp1) gene in a host-generalist avian malaria parasite, Plasmodium relictum (lineages SGS1 and GRW4) with the use of blood transcriptome. Malar. J. 2013, 12, 381. [Google Scholar] [CrossRef]
- López, G.; Muñoz, J.; Soriguer, R.; Figuerola, J. Increased Endoparasite Infection in Late-Arriving Individuals of a Trans-Saharan Passerine Migrant Bird. PLoS ONE 2013, 8, e61236. [Google Scholar] [CrossRef]
- Ayadi, T.; Selmi, S.; Hammouda, A.; Reis, S.; Boulinier, T.; Loiseau, C. Diversity, prevalence and host specificity of avian parasites in southern Tunisian oases. Parasitology 2018, 145, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, M.A.; Matta, N.E.; Valkiünas, G.; Parker, P.G.; Mello, B.; Stanley, C.E.; Lentino, M.; Garcia-Amado, M.A.; Cranfield, M.; Pond, S.L.K.; et al. Mode and rate of evolution of haemosporidian mitochondrial genomes: Timing the radiation of avian parasites. Mol. Biol. Evol. 2018, 35, 383–403. [Google Scholar] [CrossRef]
- Outlaw, D.C.; Ricklefs, R.E. Comparative Gene Evolution in Haemosporidian (Apicomplexa) Parasites of Birds and Mammals. Mol. Biol. Evol. 2010, 27, 537–542. [Google Scholar] [CrossRef]
- Grill, A.; Casula, P.; Lecis, R.; Menken, S. Endemism in Sardinia. In Phylogeography of Southern European Refugia: Evolutionary Perspectives on the Origins and Conservation of European Biodiversity; Weiss, S., Ferrand, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 273–296. ISBN 978-1-4020-4904-0. [Google Scholar]
- Gupta, P.; Vishnudas, C.K.; Ramakrishnan, U.; Robin, V.V.; Dharmarajan, G. Geographical and host species barriers differentially affect generalist and specialist parasite community structure in a tropical sky-island archipelago. Proc. R. Soc. B Biol. Sci. 2019, 286. [Google Scholar] [CrossRef]
- Hsü, K.J.; Montadert, L.; Bernouilli, D.; Cita, M.B.; Erikson, A.; Garrison, R.E.; Kidd, R.B.; Melieres, F.; Muller, C.; Wright, R. History of the Mediterranean salinity crisis. Nature 1978, 267, 399–403. [Google Scholar] [CrossRef]
- Tognotti, E. Program to eradicate malaria in Sardinia, 1946–1950. Emerg. Infect. Dis. 2009, 15, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Hocking, B.; Richards, W. Biology and Control of Labrador Black Flies (Diptera: Simuliidae). Bull. Entomol. Res. 1952, 43, 237–257. [Google Scholar] [CrossRef]
- Imamura, H.; Downing, T.; van den Broeck, F.; Sanders, M.J.; Rijal, S.; Sundar, S.; Mannaert, A.; Vanaerschot, M.; Berg, M.; de Muylder, G.; et al. Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. Elife 2016, 5, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.; Melo, M.; Covas, R.; Doutrelant, C.; Pereira, H.; de Lima, R.; Loiseau, C. Influence of land use and host species on parasite richness, prevalence and co-infection patterns. Int. J. Parasitol. 2021, 51, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Talbalaghi, A.; Pessino, M.; Agosta, P.; Bo, T.; Ignjatović-Ćupina, A. Overview of Possible Simuliidae Related Problems in the Alessandria District (Piedmont, Italy). Suppl. Acta Entomol. Serbica 2006, 77–81. [Google Scholar]
- Margoni, S.; Maiolini, B. Distribuzione altitudinale di Simulidi in torrenti alpini d’alta quota (Trentino, Italia). Stud. Trent. Sci. Nat. Acta Biol. 2003, 80, 63–64. [Google Scholar]
- Ya’Cob, Z.; Takaoka, H.; Pramual, P.; Low, V.L.; Sofian-Azirun, M. Distribution pattern of black fly (Diptera: Simuliidae) assemblages along an altitudinal gradient in Peninsular Malaysia. Parasites Vectors 2016, 9, 1–16. [Google Scholar] [CrossRef]
- Van Rooyen, J.; Lalubin, F.; Glaizot, O.; Christe, P. Avian haemosporidian persistence and co-infection in great tits at the individual level. Malar. J. 2013, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Latta, S.C.; Ricklefs, R.E. Prevalence patterns of avian haemosporida on hispaniola. J. Avian Biol. 2010, 41, 25–33. [Google Scholar] [CrossRef]
- Doussang, D.; González-Acuña, D.; Torres-Fuentes, L.G.; Lougheed, S.C.; Clemente-Carvalho, R.B.; Greene, K.C.; Vianna, J.A. Spatial distribution, prevalence and diversity of haemosporidians in the rufous-collared sparrow, Zonotrichia capensis. Parasites Vectors 2019, 12, 1–12. [Google Scholar] [CrossRef]
- Mukhopadhyay, E.; Hazra, S.; Banerjee, D. Effect of Environment and Elevation on Seasonal Prevalence of Culicoides in West Bengal. Legum. Res. Int. J. 2020, 54, 1125–1135. [Google Scholar]
- Tschuor, A.C.; Kaufmann, C.; Schaffner, F.; Mathis, A. Vorkommen von gnitzen (Culicoides spp.) in drei höhenlagen einer alpinen region der schweiz. Schweiz. Arch. Tierheilkd. 2009, 151, 215–221. [Google Scholar] [CrossRef]
- Cosgrove, C.L.; Wood, M.J.; Day, K.P.; Sheldon, B.C. Seasonal variation in Plasmodium prevalence in a population of blue tits Cyanistes caeruleus. J. Anim. Ecol. 2008, 77, 540–548. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).