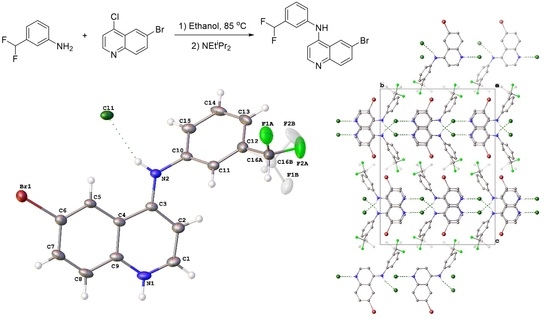
6-Bromo-N-(3-(difluoromethyl)phenyl)quinolin-4-amine
Abstract
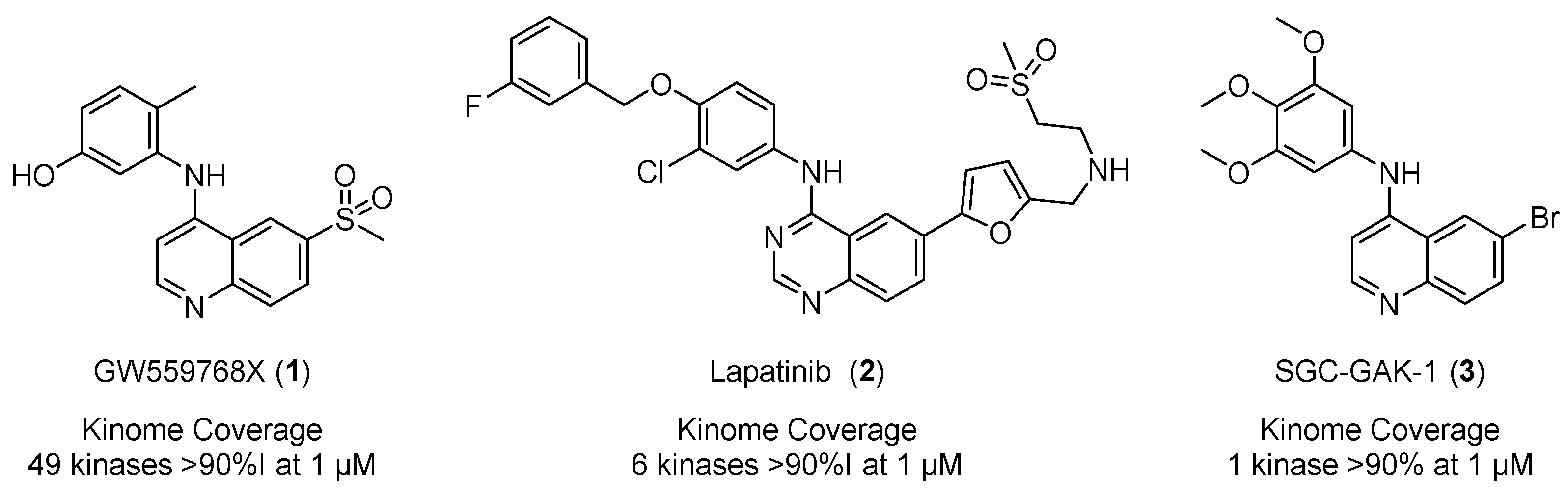
1. Introduction
2. Results
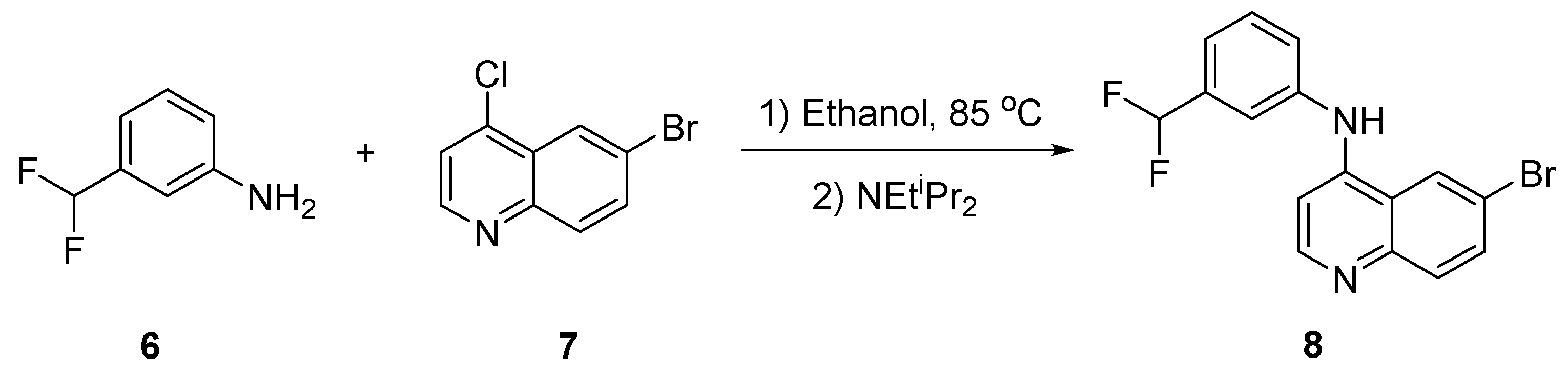
2.1. Synthesis of 8
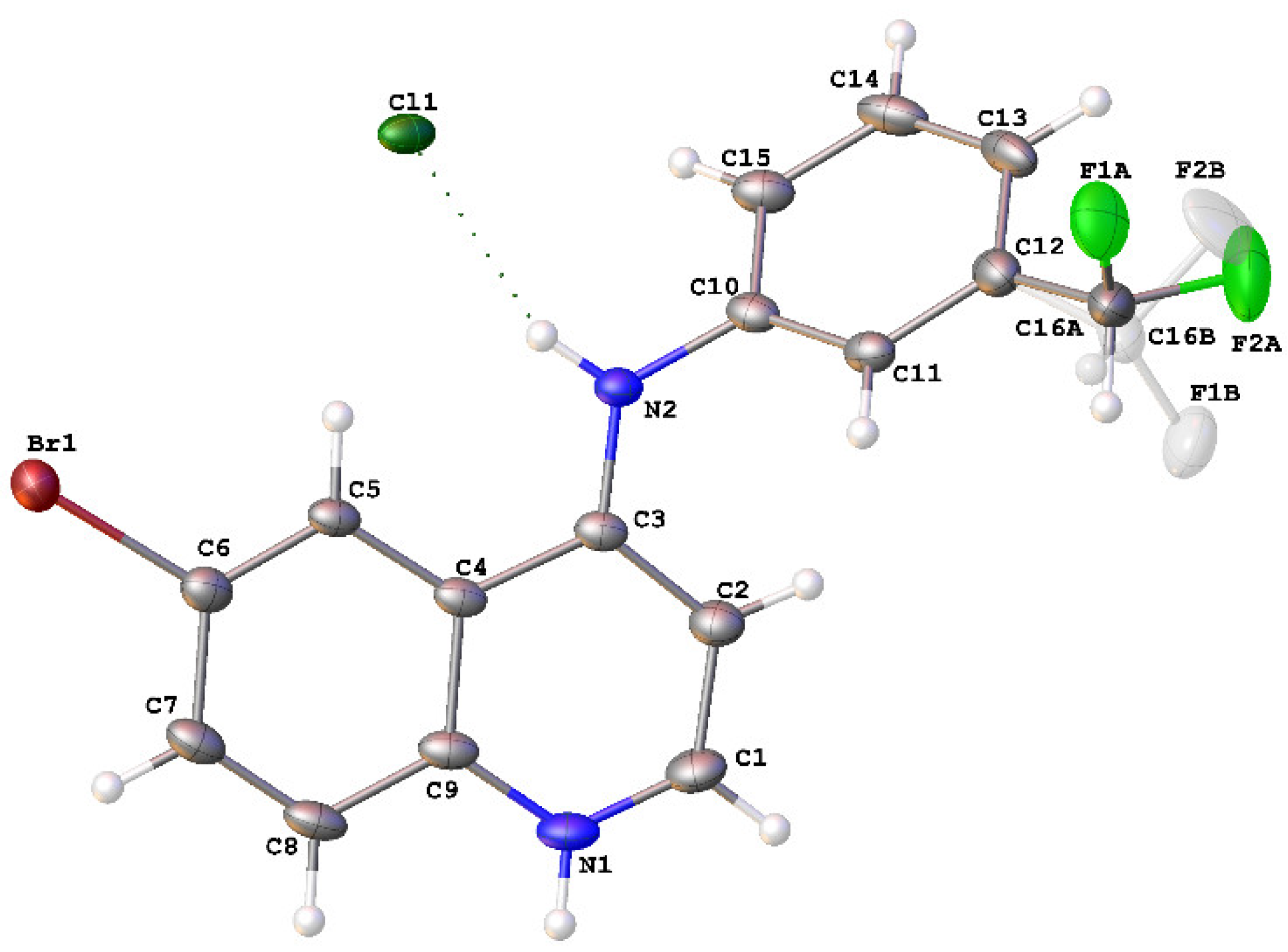
2.2. Crystal Structure of 8
3. Discussion
4. Material and Methods
4.1. Chemistry
4.2. Crystallography
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat. Rev. Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Alessi, D.R. Kinase drug discovery—What’s next in the field? ACS Chem. Biol. 2013, 8, 96–104. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Laitinen, T.; Bennett, J.M.; Godoi, P.H.; East, M.P.; Tizzard, G.J.; Graves, L.M.; Johnson, G.L.; Dornsife, R.E.; Wells, C.I.; et al. Identification and Optimization of 4-Anilinoquinolines as Inhibitors of Cyclin G Associated Kinase. ChemMedChem 2018, 13, 48–66. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Berger, B.T.; Wan, J.; Bennett, J.M.; Capuzzi, S.J.; Crona, D.J.; Drewry, D.H.; East, M.P.; Elkins, J.M.; Fedorov, O.; et al. SGC-GAK-1: A Chemical Probe for Cyclin G Associated Kinase (GAK). J. Med. Chem. 2019, 62, 2830–2836. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Naegeli, N.; East, M.P.; Laitinen, T.; Havener, T.M.; Wells, C.I.; Johnson, G.L.; Drewry, D.H.; Zuercher, W.J.; Morris, D.C. Design of a cyclin G associated kinase (GAK)/epidermal growth factor receptor (EGFR) inhibitor set to interrogate the relationship of EGFR and GAK in chordoma J. Med. Chem. 2019, 62, 4772–4778. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Treiber, D.K.; Zuercher, W.J. Utilizing comprehensive and mini-kinome panels to optimize the selectivity of quinoline inhibitors for cyclin G associated kinase (GAK). Bioorg. Med. Chem. Lett. 2019, 29, 1727–1731. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Bennett, J.M.; Su, L.; Laitinen, T.; Elkins, J.M.; Pickett, J.E.; Wells, C.I.; Li, Z.; Willson, T.M.; Zuercher, W.J. Development of SGC-GAK-1 as an orally active in vivo probe for cyclin G associated kinase through cytochrome P450 inhibition. bioRxiv 2019, 629220. [Google Scholar] [CrossRef]
- Drewry, D.H.; Wells, C.I.; Andrews, D.M.; Angell, R.; Al-Ali, H.; Axtman, A.D.; Capuzzi, S.J.; Elkins, J.M.; Ettmayer, P.; Frederiksen, M.; et al. Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS ONE 2017, 12, e0181585. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.A.; Biggs, W.H., III; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Karaman, M.W.; Herrgard, S.; Treiber, D.K.; Gallant, P.; Atteridge, C.E.; Campbell, B.T.; Chan, K.W.; Ciceri, P.; Davis, M.I.; Edeen, P.T.; et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008, 26, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Westwell, A.D. The role of fluorine in medicinal chemistry. J. Enzyme Inhib. Med. Chem. 2007, 22, 527–540. [Google Scholar] [CrossRef]
- Hagmann, W.K. The Many Roles for Fluorine in Medicinal Chemistry. J. Med. Chem. 2008, 51, 4359–4369. [Google Scholar] [CrossRef]
- Swallow, S. Chapter Two-Fluorine in Medicinal Chemistry. Prog. Med. Chem. 2015, 54, 65–133. [Google Scholar] [CrossRef]
- Wang, Y.; Callejo, R.; Slawin, A.M.Z.; O’Hagan, D. The difluoromethylene (CF2) group in aliphatic chains: Synthesis and conformational preference of palmitic acids and nonadecane containing CF2 groups Beilstein. J. Org. Chem. 2014, 10, 18–25. [Google Scholar] [CrossRef]
- Corr, M.J.; Cormanich, R.A.; von Hahmann, C.N.; Bühl, M.; Cordes, D.B.; Slawin, A.M.Z.; O’Hagan, D. Fluorine in fragrances: Exploring the difluoromethylene (CF2) group as a conformational constraint in macrocyclic musk lactones. Org. Biomol. Chem. 2016, 14, 211. [Google Scholar] [CrossRef]
- Rageot, D.; Bohnacker, T.; Keles, E.; McPhail, J.A.; Hoffmann, R.M.; Melone, A.; Borsari, C.; Sriramaratnam, R.; Sele, A.M.; Beaufils, F.; et al. (S)-4-(Difluoromethyl)-5-(4-(3-methylmorpholino)-6-morpholino-1,3,5-triazin-2-yl)pyridin-2-amine (PQR530), a potent, orally bioavailable, and brain-penetrable dual inhibitor of class I PI3K and mTOR kinase. J. Med. Chem. 2019, 62, 6241–6261. [Google Scholar] [CrossRef]
- Braun, M.-G.; Hanan, E.; Staben, S.T.; Heald, R.A.; MacLeod, C.; Elliott, R. Benzoxazepin Oxazolidinone Compounds and Methods of Use. U.S. Patent Application 20,170,210,733, 16 May 2017. [Google Scholar]
- Han, C.; Kelly, S.M.; Cravillion, T.; Savage, S.J.; Nguyen, T.; Gosselin, F. Synthesis of PI3K inhibitor GDC-0077 via a stereocontrolled N-arylation of α-amino acids. Tetrahedron 2019, 75, 4351–4357. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Tizzard, G.J.; Bennett, J.M.; Wells, C.I.; Elkins, J.M.; Willson, T.; Poso, A.; Laitinen, T. Targeting the water network in cyclin G associated kinase (GAK) with 4-anilino-quin(az)oline inhibitors. ChemMedChem 2020, 15, 1200–1215. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Laitinen, T.; Bennett, J.M.; Wells, C.I.; Elkins, J.M.; Zuercher, W.J.; Tizzard, G.J.; Poso, A. Design and analysis of the 4-anilino-quin(az)oline kinase inhibition profiles of GAK/SLK/STK10 using quantitative structure activity relationships. ChemMedChem 2020, 15, 26–49. [Google Scholar] [CrossRef] [PubMed]
- Saul, S.; Pu, S.; Zuercher, W.J.; Einav, S.; Asquith, C.R.M. Potent antiviral activity of novel multi-substituted 4-anilinoquin(az)olines. Bioorg. Med. Chem. Lett. 2020, 30, 127284. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Laitinen, T.; Wells, C.I.; Tizzard, G.J.; Zuercher, W.J. New insights into 4-anilinoquinazolines as inhibitors of cardiac troponin I–interacting kinase (TNNi3K). Molecules 2020, 25, 1697. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Tizzard, G.J. 6-Bromo-N-(2-methyl-2H-benzo[d][1,2,3]triazol-5-yl)quinolin-4-amine. Molbank 2019, 2019, 1087. [Google Scholar] [CrossRef]
- Carabajal, M.A.; Asquith, C.R.M.; Laitinen, T.; Tizzard, G.J.; Yim, L.; Rial, A.; Chabalgoity, J.; Zuercher, W.J.; Véscovi, E.G. Quinazoline-based anti-virulence compounds that selectively target Salmonella PhoP/PhoQ signal transduction system. Antimicrob. Agents Chemother. 2019, 64, e01744-19. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Fleck, N.; Torrice, C.D.; Crona, D.J.; Grundner, C.; Zuercher, W.J. Anti-tubercular activity of novel 4-anilinoquinolines and 4-anilinoquinazolines. Bioorg. Med. Chem. Lett. 2019, 18, 2695–2699. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Maffuid, K.A.; Laitinen, T.; Torrice, C.D.; Tizzard, G.J.; Crona, D.J.; Zuercher, W.J. Targeting an EGFR water network using novel 4-anilinoquin(az)olines inhibitors for chordoma. ChemMedChem 2019. [Google Scholar] [CrossRef]
- Sheldrick, G.M. ShelXT-Integrated space-group and crystal-structure determination. Acta Cryst. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. Olex2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with ShelXL. Acta Cryst. 2015, C27, 3–8. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asquith, C.R.M.; Tizzard, G.J. 6-Bromo-N-(3-(difluoromethyl)phenyl)quinolin-4-amine. Molbank 2020, 2020, M1161. https://doi.org/10.3390/M1161
Asquith CRM, Tizzard GJ. 6-Bromo-N-(3-(difluoromethyl)phenyl)quinolin-4-amine. Molbank. 2020; 2020(4):M1161. https://doi.org/10.3390/M1161
Chicago/Turabian StyleAsquith, Christopher R. M., and Graham J. Tizzard. 2020. "6-Bromo-N-(3-(difluoromethyl)phenyl)quinolin-4-amine" Molbank 2020, no. 4: M1161. https://doi.org/10.3390/M1161
APA StyleAsquith, C. R. M., & Tizzard, G. J. (2020). 6-Bromo-N-(3-(difluoromethyl)phenyl)quinolin-4-amine. Molbank, 2020(4), M1161. https://doi.org/10.3390/M1161