Impact of the Technical Snow Production Process on Bacterial Community Composition, Antibacterial Resistance Genes, and Antibiotic Input—A Dual Effect of the Inevitable
Abstract
1. Introduction
2. Results and Discussion
2.1. Culture-Based Assessment of Bacteriological Contamination
2.2. Concentration of Antimicrobial Agents
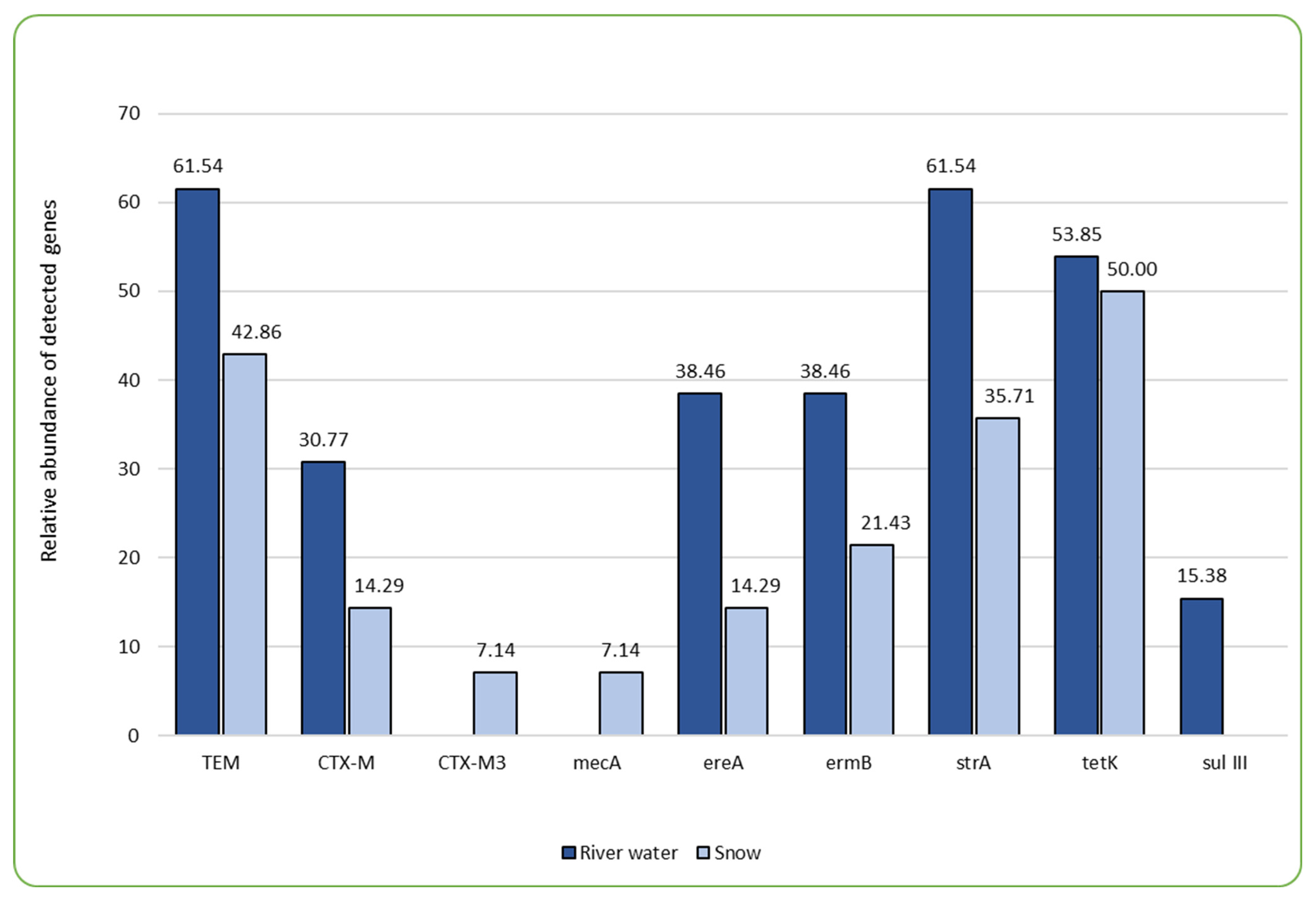
2.3. Detection and Prevalence of Antibiotic Resistance Genes (ARGs)
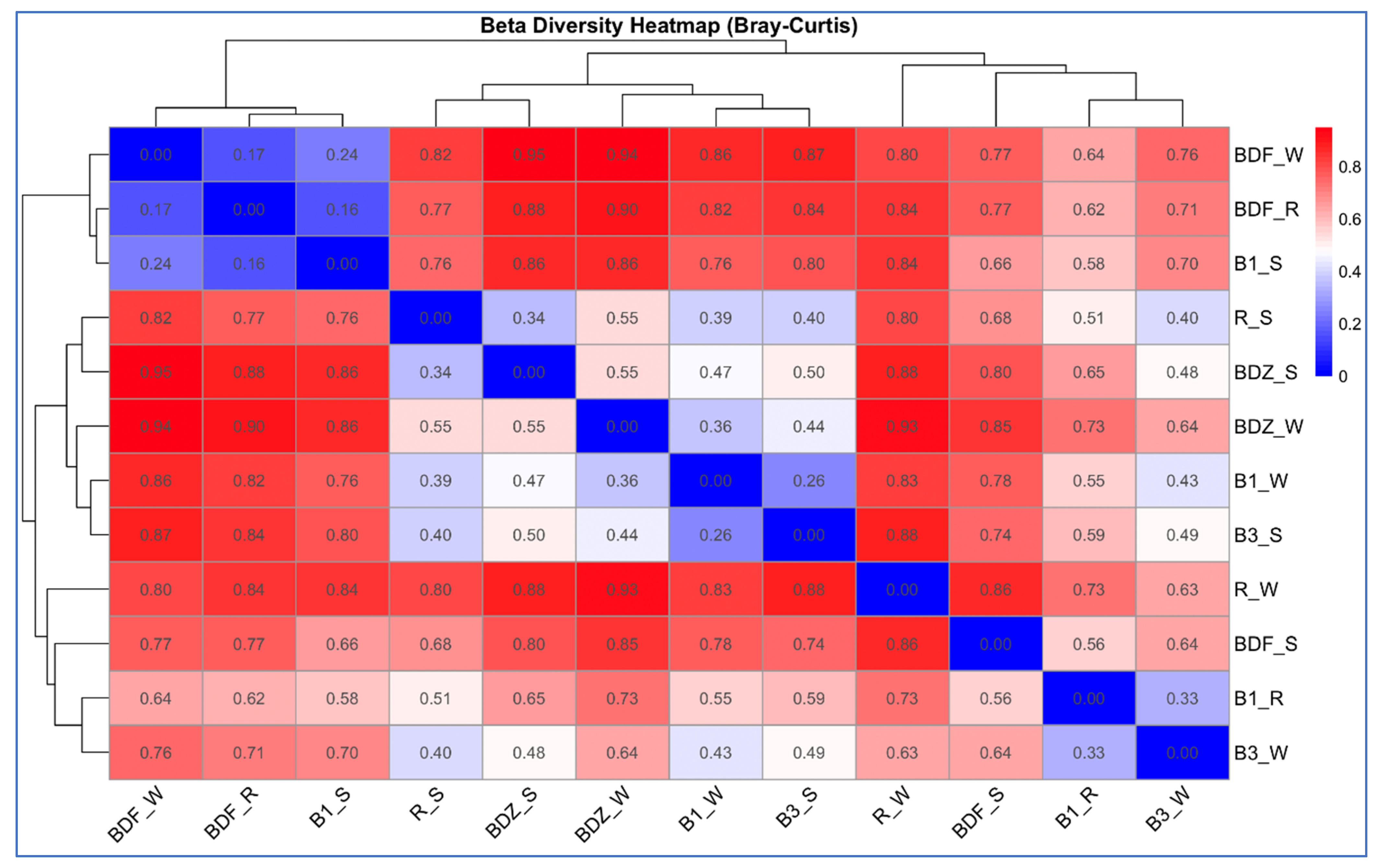
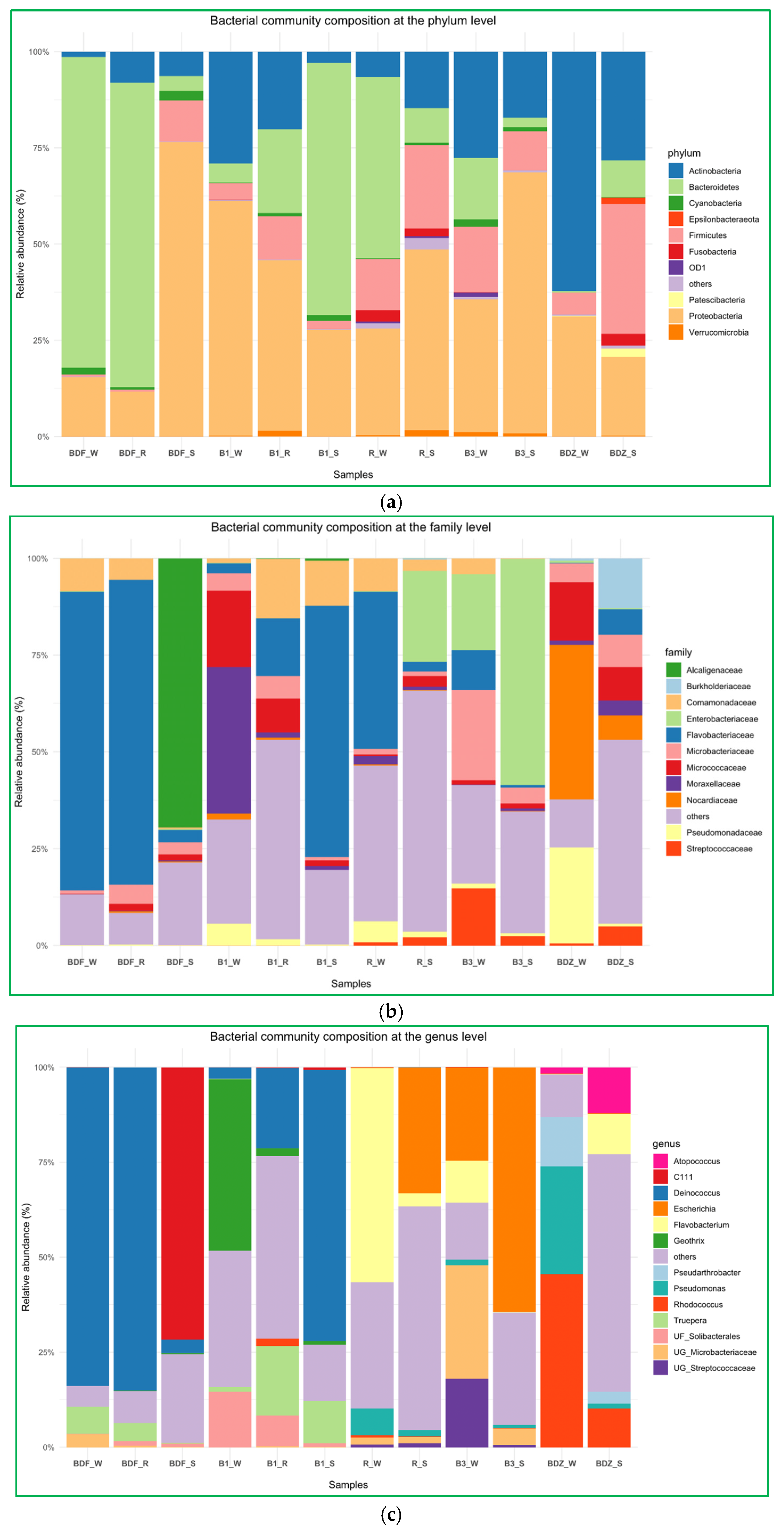
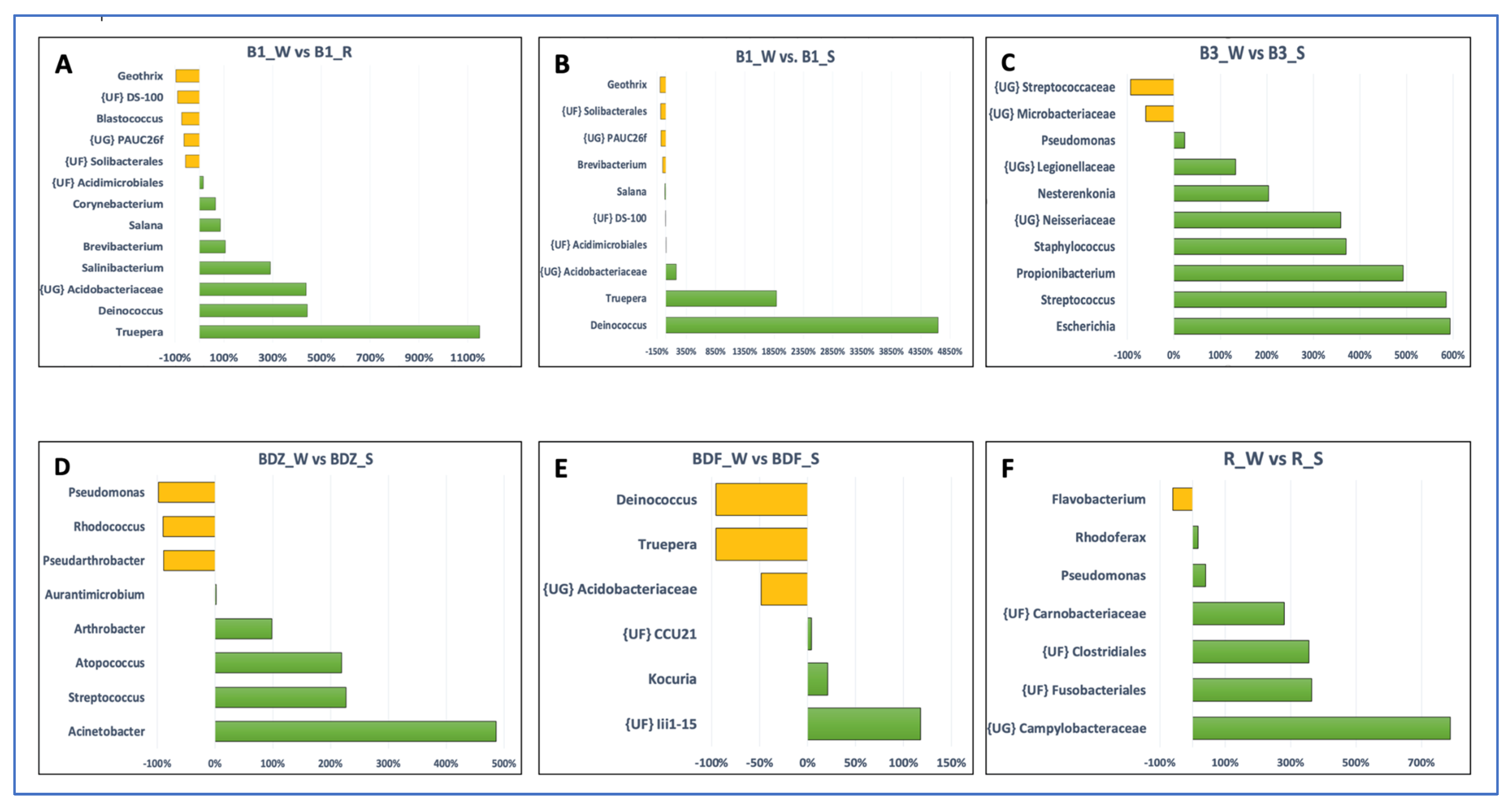
2.4. Bacterial Community Diversity and Composition
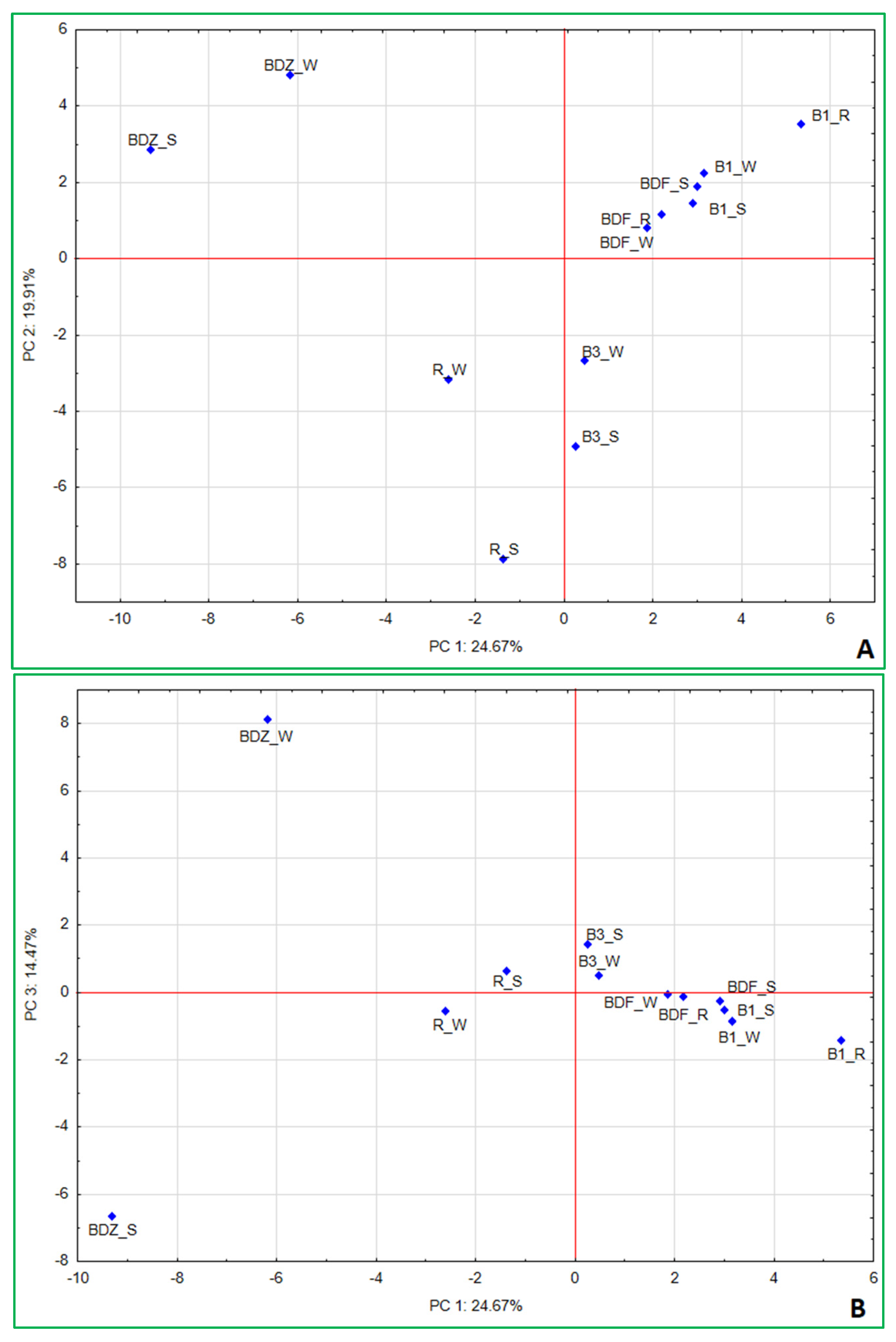
2.5. Multivariate Analysis of Data
3. Materials and Methods
3.1. Sampling Sites
| Code | Height Above Sea Level [m a.s.l] | Number of Inhabitants | Anthropogenic Pressure Description | Technical Reservoir [yes/no] | Sample Description and Code |
|---|---|---|---|---|---|
| BDF | 850 | 540 | upstream of a small village next to the Tatra National Park, upstream of wastewater discharge sites | yes | river water (BDF_W) |
| storage reservoir (BDF_R) | |||||
| snowmelt water (BDF_S) | |||||
| B3 | 760 | 950 | upstream of a small village next to the Tatra National Park and the Polish/Slovakian border | no | river water (B3_W) |
| snowmelt water (B3_S) | |||||
| B1 | 700 | 2300 | center of a popular tourist resort, ca. 3 km downstream of a WWTP | yes | river water (B1_W) |
| storage reservoir (B1_R) | |||||
| snowmelt water (B1_S) | |||||
| R | 315 | 17,500 | center of a medium-sized town, downstream of several ski resorts, ca. 5 km downstream of a hospital, ca. 10 km of a WWTP | no | river water (R_W) |
| snowmelt water (R_S) | |||||
| BDZ | 750 | 25,000 | center of a popular tourist resort, ca. 3 km downstream of a WWTP, ca. 2 km downstream of a hospital | no | river water (BDZ_W) snowmelt water (BDZ_S) |
3.2. Sample Collection
3.3. Culture-Based Microbiological Analysis of Samples
3.4. Determining the Presence and Concentration of Antimicrobial Agents in Water and Snowmelt Samples
3.5. PCR Determination of Genetic Antimicrobial Resistance Determinants in Total Genomic DNA
3.6. Illumina Sequencing of V3-V4 16S rRNA Amplicon
3.7. Statistical Analyses
4. Conclusions
- ○
- The numbers of culturable bacteria drop sharply during the technical snowmaking process; thus, even if the snow is produced from water severely contaminated by wastewater, the resulting snow does not seem to pose a serious threat to human health.
- ○
- The presence and concentration of antimicrobial agents in water and the produced snow is strongly affected by the proximity of point sources of pollution, such as WWTPs and hospitals.
- ○
- As many as nine antibiotic resistance genes (ARGs) were detected at the examined sites, and their prevalence was strongly affected by the close vicinity of wastewater discharge sites and hospitals. The presence and concentration of antimicrobial agents is also associated with the occurrence of ARGs.
- ○
- Importantly, the prevalence of most ARGs decreased during technical snowmaking. This was more evident within ski stations where water is stored in technical reservoirs prior to snowmaking. On the other hand, biofilm formation and its further detachment may contribute to reduced removal efficiency of ARGs.
- ○
- Finally, the NGS-based analysis of bacterial community composition evidently indicates its changes through the process of technical snowmaking, which may be due to the survivability of certain strains in freezing temperatures or the inhibitory effect of antimicrobial agents that enter the technical snow.
- ○
- The presence and concentration of antimicrobials in water seem to be the most significant factors affecting the changes in bacterial community composition. Therefore, measures should be taken to reduce their spread within the aquatic environment.
- ○
- Among the measures that a ski station management could implement, one can mention the construction of technical reservoirs. These appear to effectively eliminate pollutants and micropollutants from the resource water. In addition, ensuring the regular and frequent cleaning and maintenance of snowmaking devices is advisable.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Joemann, M.; Völkel, R.; Pollerberg, C.; Podesta, L.; Besana, F. All-Weather Snow Machine Driven by Solar Energy. In Proceedings of the ISES Solar World Congress 2017—IEA SHC International Conference on Solar Heating and Cooling for Buildings and Industry 2017, Abu Dhabi, United Arab Emirates, 29 October–2 November 2017; pp. 996–1007. [Google Scholar] [CrossRef]
- Krąż, P. DOLINY BIAŁKI Paweł Krąż Anthropogenic Hazards to the Białka Valley Natural Environment. Pract. Geogr. 2012, 128, 45–54. [Google Scholar] [CrossRef]
- Krzesiwo, K. Ocena Sytuacji Rozwojowej i Funkcjonalnej Stacji Narciarskich—Przykład Polskich Karpat. Stud. Ind. Geogr. Comm. Pol. Geogr. Soc. 2021, 35, 259–276. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Wolanin, A.; Jelonkiewicz, Ł.; Chmielewska-Błotnicka, D.; Zelazny, M. Spatiotemporal Variability in Microbiological Water Quality of the Białka River and Its Relation to the Selected Physicochemical Parameters of Water. Water Air Soil Pollut. 2016, 227, 22. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Prajsnar, J.; Guzik, M.; Boroń, P.; Chmiel, M. How Much of Antibiotics Can Enter Surface Water with Treated Wastewater and How It Affects the Resistance of Waterborne Bacteria: A Case Study of the Białka River Sewage Treatment Plant. Environ. Res. 2020, 191, 110037. [Google Scholar] [CrossRef]
- de Jong, C. Artificial Production of Snow. Encycl. Earth Sci. Ser. 2011, 3, 61–66. [Google Scholar] [CrossRef]
- Rixen, C.; Stoeckli, V.; Ammann, W. Does Artificial Snow Production Affect Soil and Vegetation of Ski Pistes? A Review. Perspect. Plant Ecol. Evol. Syst. 2003, 5, 219–230. [Google Scholar] [CrossRef]
- Faria, C.; Vaz-Moreira, I.; Serapicos, E.; Nunes, O.C.; Manaia, C.M. Antibiotic Resistance in Coagulase Negative Staphylococci Isolated from Wastewater and Drinking Water. Sci. Total Environ. 2009, 407, 3876–3882. [Google Scholar] [CrossRef] [PubMed]
- Lagriffoul, A.; Boudenne, J.-L.; Absi, R.; Ballet, J.-J.; Berjeaud, J.-M.; Chevalier, S.; Creppy, E.E.; Gilli, E.; Gadonna, J.-P.; Gadonna-Widehem, P.; et al. Bacterial-Based Additives for the Production of Artificial Snow: What Are the Risks to Human Health? Sci. Total Environ. 2010, 408, 1659–1666. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Prajsnar, J.; Boroń, P. Survival and Antibiotic Resistance of Bacteria in Artificial Snow Produced from Contaminated Water. Water Environ. Res. 2017, 89, 2059–2069. [Google Scholar] [CrossRef]
- Hyer, K.E. Escherichia coli Concentrations in Recreational Streams and Backcountry Drinking-Water Supplies in Shenandoah National Park, Virginia, 2005–2006; U.S. Geological Survey: Reston, VA, USA, 2007; 18p. [Google Scholar]
- Lu, Q.; Mao, J.; Xia, H.; Song, S.; Chen, W.; Zhao, D. Effect of Wastewater Treatment Plant Discharge on the Bacterial Community in a Receiving River. Ecotoxicol. Environ. Saf. 2022, 239, 113641. [Google Scholar] [CrossRef]
- Mackuľak, T.; Cverenkárová, K.; Vojs Staňová, A.; Fehér, M.; Tamáš, M.; Škulcová, A.B.; Gál, M.; Naumowicz, M.; Špalková, V.; Bírošová, L. Hospital Wastewater—Source of Specific Micropollutants, Antibiotic-Resistant Microorganisms, Viruses, and Their Elimination. Antibiotics 2021, 10, 1070. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, B.; Apori, S.O.; Giltrap, M.; Bhat, A.; Curtin, J.; Tian, F. Hospital Effluents and Wastewater Treatment Plants: A Source of Oxytetracycline and Antimicrobial-Resistant Bacteria in Seafood. Sustainability 2021, 13, 13967. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The Impact of On-Site Hospital Wastewater Treatment on the Downstream Communal Wastewater System in Terms of Antibiotics and Antibiotic Resistance Genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Maes, S.; Odlare, M.; Jonsson, A. Fecal Indicator Organisms in Northern Oligotrophic Rivers: An Explorative Study on Escherichia coli Prevalence in a Mountain Region with Intense Tourism and Reindeer Herding. Environ. Monit. Assess. 2022, 194, 264. [Google Scholar] [CrossRef]
- Staley, Z.R.; He, D.D.; Edge, T.A. Persistence of Fecal Contamination and Pathogenic Escherichia coli O157:H7 in Snow and Snowmelt. J. Great Lakes Res. 2017, 43, 248–254. [Google Scholar] [CrossRef]
- Vázquez-Sánchez, D.; Rodríguez-López, P. Chapter 5—Biofilm Formation of Staphylococcus aureus. In Staphylococcus aureus; Fetsch, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 87–103. ISBN 978-0-12-809671-0. [Google Scholar]
- Williams, M.M.; Domingo, J.W.S.; Meckes, M.C.; Kelty, C.A.; Rochon, H.S. Phylogenetic Diversity of Drinking Water Bacteria in a Distribution System Simulator. J. Appl. Microbiol. 2004, 96, 954–964. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Moles, S.; Gozzo, S.; Ormad, M.P.; Mosteo, R.; Gómez, J.; Laborda, F.; Szpunar, J. Long-Term Study of Antibiotic Presence in Ebro River Basin (Spain): Identification of the Emission Sources. Water 2022, 14, 1033. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial Pharmaceuticals in the Aquatic Environment—Occurrence and Environmental Implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Iglesias, A.; Nebot, C.; Miranda, J.M.; Vázquez, B.I.; Abuín, C.M.F.; Cepeda, A. Determination of the Presence of Three Antimicrobials in Surface Water Collected from Urban and Rural Areas. Antibiotics 2013, 2, 46–57. [Google Scholar] [CrossRef]
- Sims, N.; Kannan, A.; Holton, E.; Jagadeesan, K.; Mageiros, L.; Standerwick, R.; Craft, T.; Barden, R.; Feil, E.J.; Kasprzyk-Hordern, B. Antimicrobials and Antimicrobial Resistance Genes in a One-Year City Metabolism Longitudinal Study Using Wastewater-Based Epidemiology. Environ. Pollut. 2023, 333, 122020. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tan, L.; Zhang, L.; Tian, W.; Ma, L. A Review of the Distribution of Antibiotics in Water in Different Regions of China and Current Antibiotic Degradation Pathways. Front. Environ. Sci. 2021, 9, 692298. [Google Scholar] [CrossRef]
- Daghrir, R.; Drogui, P. Tetracycline Antibiotics in the Environment: A Review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Osorio, V.; Marcé, R.; Pérez, S.; Ginebreda, A.; Cortina, J.L.; Barceló, D. Occurrence and Modeling of Pharmaceuticals on a Sewage-Impacted Mediterranean River and Their Dynamics under Different Hydrological Conditions. Sci. Total Environ. 2012, 440, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Mutuku, C.; Gazdag, Z.; Melegh, S. Occurrence of Antibiotics and Bacterial Resistance Genes in Wastewater: Resistance Mechanisms and Antimicrobial Resistance Control Approaches. World J. Microbiol. Biotechnol. 2022, 38, 152. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, T.; Wolfsperger, F. Water Losses During Technical Snow Production: Results from Field Experiments. Front. Earth Sci. 2019, 7, 78. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, S.Y.; Kim, G.E.; Jho, E.H. Effect of PH and Temperature on the Biodegradation of Oxytetracycline, Streptomycin, and Validamycin A in Soil. Appl. Biol. Chem. 2023, 66, 63. [Google Scholar] [CrossRef]
- Loftin, K.A.; Adams, C.D.; Meyer, M.T.; Surampalli, R. Effects of Ionic Strength, Temperature, and PH on Degradation of Selected Antibiotics. J. Environ. Qual. 2008, 37, 378–386. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the Aquatic Environment—A Review—Part II. Chemosphere 2009, 75, 435–441. [Google Scholar] [CrossRef]
- Cortes, L.G.; Marinov, D.; Sanseverino, I.; Cuenca, A.N.; Niegowska, M.; Rodriguez, E.P.; Lettieri, T. Selection of Substances for the 4th Watch List Under the Water Framework Directive; Publications Office of the European Union: Luxembourg, 2020; ISBN 9789276194262. [Google Scholar]
- Arsand, J.B.; Hoff, R.B.; Jank, L.; Bussamara, R.; Dallegrave, A.; Bento, F.M.; Kmetzsch, L.; Falção, D.A.; do Carmo Ruaro Peralba, M.; de Araujo Gomes, A.; et al. Presence of Antibiotic Resistance Genes and Its Association with Antibiotic Occurrence in Dilúvio River in Southern Brazil. Sci. Total Environ. 2020, 738, 139781. [Google Scholar] [CrossRef]
- Takawira, H.; Mbanga, J. Occurrence of Multidrug-Resistant Escherichia coli and Antibiotic Resistance Genes in a Wastewater Treatment Plant and Its Associated River Water in Harare, Zimbabwe. Water SA 2023, 49, 396–403. [Google Scholar] [CrossRef]
- Wu, Y.; Li, S.; Yu, K.; Hu, J.; Chen, Q.; Sun, W. Wastewater Treatment Plant Effluents Exert Different Impacts on Antibiotic Resistome in Water and Sediment of the Receiving River: Metagenomic Analysis and Risk Assessment. J. Hazard. Mater. 2023, 460, 132528. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.J.; Chen, H. Antibiotic-Resistant Genes and Antibiotic-Resistant Bacteria in the Effluent of Urban Residential Areas, Hospitals, and a Municipal Wastewater Treatment Plant System. Environ. Sci. Pollut. Res. Int. 2015, 22, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of Antibiotics and Antibiotic Resistance Genes in Hospital and Urban Wastewaters and Their Impact on the Receiving River. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Wilson, G.J.L.; Perez-Zabaleta, M.; Owusu-Agyeman, I.; Kumar, A.; Ghosh, A.; Polya, D.A.; Gooddy, D.C.; Cetecioglu, Z.; Richards, L.A. Discovery of Sulfonamide Resistance Genes in Deep Groundwater below Patna, India. Environ. Pollut. 2024, 356, 124205. [Google Scholar] [CrossRef]
- Pazda, M.; Rybicka, M.; Stolte, S.; Piotr Bielawski, K.; Stepnowski, P.; Kumirska, J.; Wolecki, D.; Mulkiewicz, E. Identification of Selected Antibiotic Resistance Genes in Two Different Wastewater Treatment Plant Systems in Poland: A Preliminary Study. Molecules 2020, 25, 2851. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, X.; Huang, H.; Zhang, J. Prevalence of Antibiotic Resistance Genes and Their Association with Antibiotics in a Wastewater Treatment Plant: Process Distribution and Analysis. Water 2019, 11, 2495. [Google Scholar] [CrossRef]
- Su, H.; Li, W.; Okumura, S.; Wei, Y.; Deng, Z.; Li, F. Transfer, Elimination and Accumulation of Antibiotic Resistance Genes in Decentralized Household Wastewater Treatment Facility Treating Total Wastewater from Residential Complex. Sci. Total Environ. 2024, 912, 169144. [Google Scholar] [CrossRef]
- Kulik, K.; Lenart-Boroń, A.; Wyrzykowska, K. Impact of Antibiotic Pollution on the Bacterial Population within Surface Water with Special Focus on Mountain Rivers. Water 2023, 15, 975. [Google Scholar] [CrossRef]
- Pei, R.; Cha, J.; Carlson, K.H.; Pruden, A. Response of Antibiotic Resistance Genes (ARG) to Biological Treatment in Dairy Lagoon Water. Environ. Sci. Technol. 2007, 41, 5108–5113. [Google Scholar] [CrossRef]
- Li, Z.; Yu, E.; Zhang, K.; Gong, W.; Xia, Y.; Tian, J.; Wang, G.; Xie, J. Water Treatment Effect, Microbial Community Structure, and Metabolic Characteristics in a Field-Scale Aquaculture Wastewater Treatment System. Front. Microbiol. 2020, 11, 930. [Google Scholar] [CrossRef]
- Ji, B.; Liang, J.; Ma, Y.; Zhu, L.; Liu, Y. Bacterial Community and Eutrophic Index Analysis of the East Lake. Environ. Pollut. 2019, 252, 682–688. [Google Scholar] [CrossRef]
- Wu, D.; Zou, Y.; Xiao, J.; Mo, L.; Lek, S.; Chen, B.; Fu, Q.; Guo, Z. The Spatiotemporal Variations of Microbial Community in Relation to Water Quality in a Tropical Drinking Water Reservoir, Southmost China. Front. Microbiol. 2024, 15, 1354784. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Nakagawa, Y.; Holmes, B.; Subcommittee on the taxonomy of Flavobacterium and Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes. Proposed Minimal Standards for Describing New Taxa of the Family Flavobacteriaceae and Emended Description of the Family. Int. J. Syst. Evol. Microbiol. 2002, 52, 1049–1070. [Google Scholar] [CrossRef]
- Smith, J.J.; Howington, J.P.; McFeters, G.A. Survival, Physiological Response and Recovery of Enteric Bacteria Exposed to a Polar Marine Environment. Appl. Environ. Microbiol. 1994, 60, 2977–2984. [Google Scholar] [CrossRef]
- Gerber, E.; Bernard, R.; Castang, S.; Chabot, N.; Coze, F.; Dreux-Zigha, A.; Hauser, E.; Hivin, P.; Joseph, P.; Lazarelli, C.; et al. Deinococcus as New Chassis for Industrial Biotechnology: Biology, Physiology and Tools. J. Appl. Microbiol. 2015, 119, 1–10. [Google Scholar] [CrossRef]
- Jin, M.; Xiao, A.; Zhu, L.; Zhang, Z.; Huang, H.; Jiang, L. The Diversity and Commonalities of the Radiation-Resistance Mechanisms of Deinococcus and Its up-to-Date Applications. AMB Express 2019, 9, 138. [Google Scholar] [CrossRef]
- Pan, J.; Wang, J.; Zhou, Z.; Yan, Y.; Zhang, W.; Lu, W.; Ping, S.; Dai, Q.; Yuan, M.; Feng, B.; et al. IrrE, a Global Regulator of Extreme Radiation Resistance in Deinococcus Radiodurans, Enhances Salt Tolerance in Escherichia coli and Brassica napus. PLoS ONE 2009, 4, e4422. [Google Scholar] [CrossRef]
- van Overbeek, L.; Duhamel, M.; Aanstoot, S.; van der Plas, C.L.; Nijhuis, E.; Poleij, L.; Russ, L.; van der Zouwen, P.; Andreo-Jimenez, B. Transmission of Escherichia coli from Manure to Root Zones of Field-Grown Lettuce and Leek Plants. Microorganisms 2021, 9, 2289. [Google Scholar] [CrossRef] [PubMed]
- Banach, J.L.; van Bokhorst-van de Veen, H.; van Overbeek, L.S.; van der Zouwen, P.S.; van der Fels-Klerx, H.J.; Groot, M.N.N. The Efficacy of Chemical Sanitizers on the Reduction of Salmonella Typhimurium and Escherichia coli Affected by Bacterial Cell History and Water Quality. Food Control 2017, 81, 137–146. [Google Scholar] [CrossRef]
- Seo, H.; Kim, J.H.; Lee, S.-M.; Lee, S.-W. The Plant-Associated Flavobacterium: A Hidden Helper for Improving Plant Health. Plant Pathol. J. 2024, 40, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.A.; Aman, S.; Singh, B. Unveiling the Positive Impacts of the Genus Rhodococcus on Plant and Environmental Health. J. Exp. Biol. Agric. Sci. 2024, 12, 557–572. [Google Scholar] [CrossRef]
- Huerta, B.; Marti, E.; Gros, M.; López, P.; Pompêo, M.; Armengol, J.; Barceló, D.; Balcázar, J.L.; Rodríguez-Mozaz, S.; Marcé, R. Exploring the Links between Antibiotic Occurrence, Antibiotic Resistance, and Bacterial Communities in Water Supply Reservoirs. Sci. Total Environ. 2013, 456–457, 161–170. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Cheng, W.; Huang, C.; Ren, J.; Wan, T.; Gao, K. Effects of Water Environmental Factors and Antibiotics on Bacterial Community in Urban Landscape Lakes. Aquat. Toxicol. 2023, 265, 106740. [Google Scholar] [CrossRef]
- Mu, X.; Huang, Z.; Ohore, O.E.; Yang, J.; Peng, K.; Li, S.; Li, X. Impact of Antibiotics on Microbial Community in Aquatic Environment and Biodegradation Mechanism: A Review and Bibliometric Analysis. Environ. Sci. Pollut. Res. 2023, 30, 66431–66444. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, Y.; Te, S.H.; He, Y.; Gin, K.Y. The Effects of Antibiotics on Microbial Community Composition in an Estuary Reservoir during Spring and Summer Seasons. Water 2018, 10, 154. [Google Scholar] [CrossRef]
- Santos, R.G.; Hurtado, R.; Gomes, L.G.R.; Profeta, R.; Rifici, C.; Attili, A.R.; Spier, S.J.; Mazzullo, G.; Morais-Rodrigues, F.; Gomide, A.C.P.; et al. Complete Genome Analysis of Glutamicibacter creatinolyticus from Mare Abscess and Comparative Genomics Provide Insight of Diversity and Adaptation for Glutamicibacter. Gene 2020, 741, 144566. [Google Scholar] [CrossRef] [PubMed]
- Ham, S.H.; Yoon, A.R.; Oh, H.E.; Park, Y.G. Plant Growth-Promoting Microorganism Pseudarthrobacter sp. NIBRBAC000502770 Enhances the Growth and Flavonoid Content of Geum aleppicum. Microorganisms 2022, 10, 1241. [Google Scholar] [CrossRef]
- Issifu, M.; Songoro, E.K.; Onguso, J.; Ateka, E.M.; Ngumi, V.W. Potential of Pseudarthrobacter chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants. Bacteria 2022, 1, 191–206. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Consumption in the EU/EEA (ESAC-Net)—Annual Epidemiological Report 2020; European Centre for Disease Prevention and Control: Solna, Sweden, 2021. [Google Scholar]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2019 and 2020 (EMA/294674/2019); European Medicines Agency: Amsterdam, The Netherlands, 2021; ISBN 9789291550685. [Google Scholar]
- Lenart-Boroń, A.M.; Boroń, P.M.; Prajsnar, J.A.; Guzik, M.W.; Żelazny, M.S.; Pufelska, M.D.; Chmiel, M.J. COVID-19 Lockdown Shows How Much Natural Mountain Regions Are Affected by Heavy Tourism. Sci. Total Environ. 2022, 806, 151355. [Google Scholar] [CrossRef]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia coli Strains of Human, Animal, and Food Origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, M.; Hopkins, K.; Threlfall, E.J.; Clifton-Hadley, F.A.; Stallwood, A.D.; Davies, R.H.; Liebana, E. BlaCTX-M Genes in Clinical Salmonella Isolates Recovered from Humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005, 49, 1319–1322. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Sáenz, Y.; Vinué, L.; Rojo-Bezares, B.; Jouini, A.; Zarazaga, M.; Rodrigues, J.; Torres, C. Detection of Escherichia coli Harbouring Extended-Spectrum β-Lactamases of the CTX-M, TEM and SHV Classes in Faecal Samples of Wild Animals in Portugal. J. Antimicrob. Chemother. 2006, 58, 1311–1312. [Google Scholar] [CrossRef]
- Bouallègue-Godet, O.; Ben Salem, Y.; Fabre, L.; Demartin, M.; Grimont, P.A.D.; Mzoughi, R.; Weill, F.-X. Nosocomial Outbreak Caused by Salmonella enterica Serotype Livingstone Producing CTX-M-27 Extended-Spectrum Beta-Lactamase in a Neonatal Unit in Sousse, Tunisia. J. Clin. Microbiol. 2005, 43, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Geha, D.J.; Uhl, J.R.; Gustaferro, C.A.; Persing, D.H. Multiplex PCR for Identification of Methicillin-Resistant Staphylococci in the Clinical Laboratory. J. Clin. Microbiol. 1994, 32, 1768–1772. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.; Grebe, T.; Tait-Kamradt, A.; Wondrack, L. Detection of Erythromycin-Resistant Determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef]
- Lina, G.; Quaglia, A.; Reverdy, M.E.; Leclercq, R.; Vandenesch, F.; Etienne, J. Distribution of Genes Encoding Resistance to Macrolides, Lincosamides, and Streptogramins among Staphylococci. Antimicrob. Agents Chemother. 1999, 43, 1062–1066. [Google Scholar] [CrossRef]
- Walsh, F.; Ingenfeld, A.; Zampicolli, M.; Hilber-Bodmer, M.; Frey, J.E.; Duffy, B. Real-Time PCR Methods for Quantitative Monitoring of Streptomycin and Tetracycline Resistance Genes in Agricultural Ecosystems. J. Microbiol. Methods 2011, 86, 150–155. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.-H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 Clinically Relevant Antibiotic-Resistance Genes in the Plasmid Metagenome of Wastewater Treatment Plant Bacteria Showing Reduced Susceptibility to Selected Antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, A.D.; Jenabi, A.; Montazeri, E.A. Distribution of Genes Encoding Resistance to Aminoglycoside Modifying Enzymes in Methicillin-Resistant Staphylococcus aureus (MRSA) Strains. Kaohsiung J. Med. Sci. 2017, 33, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Warsa, U.C.; Nonoyama, M.; Ida, T.; Okamoto, R.; Okubo, T.; Shimauchi, C.; Kuga, A.; Inoue, M. Detection of Tet(K) and Tet(M) in Staphylococcus aureus of Asian Countries by the Polymerase Chain Reaction. J. Antibiot. 1996, 49, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; Wagner, H.; Barbour, M.; Bedward, M.; Bolker, B.; et al. Package ‘Vegan’; The R Foundation: Vienna, Austria, 2025. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Kolde, R. Package “Pheatmap”: Pretty Heatmaps; Version 1.0.12; The R Foundation: Vienna, Austria, 2019; pp. 1–8. [Google Scholar]


| Code | Sample Description and Code | E. coli | E. faecalis/E. faecium | Salmonella | Coagulase-Positive Staphylococci |
|---|---|---|---|---|---|
| CFU/100 mL | CFU/mL | ||||
| BDF | river water (BDF_W) | 1 | 4 | 1 | 0 |
| storage reservoir (BDF_R) | 0 | 5 | 0 | 350 | |
| snowmelt water (BDF_S) | 1 | 7 | 1 | 0 | |
| B3 | river water (B3_W) | 119 | 45 | 0 | 2 |
| snowmelt water (B3_S) | 0 | 26 | 0 | 2 | |
| B1 | river water (B1_W) | 186 | 94 | 0 | 0 |
| storage reservoir (B1_R) | 14 | 13 | 0 | 0 | |
| snowmelt water (B1_S) | 2 | 7 | 0 | 12 | |
| R | river water (R_W) | 298 | 45 | 56 | 328 |
| snowmelt water (R_S) | 87 | 26 | 5 | 232 | |
| BDZ | river water (BDZ_W) | 224,067 | 273,641 | 10,785 | 8667 |
| snowmelt water (BDZ_S) | 153 | 286 | 4 | 190 | |
| Chemical Group | Antibiotic | BDF_W | BDF_R | BDF_S | B1_W | B1_R | B1_S | R_W | R_S | B3_W | B3_S | BDZ_W | BDZ_S | Frequency of Detection (% of All Samples) | Total Concentration of Antibiotics in All Samples (ng/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2nd gen. cephalosporins | cefoxitin | 0.00 | 0.00 | 112.59 | 0.00 | 0.00 | 73.47 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 9.7 | 519.05 |
| fluoroquinolones | ciprofloxacin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 65.20 | 24.31 | 0.00 | 0.00 | 0.00 | 15.37 | 9.7 | 314.65 |
| enrofloxacin | 0.00 | 0.00 | 0.00 | 1.67 | 6.09 | 3.04 | 626.28 | 34.95 | 3.38 | 0.95 | 2.94 | 0.00 | 3.2 | 204.78 | |
| ofloxacin | 0.00 | 0.00 | 0.00 | 0.49 | 0.00 | 0.00 | 95.64 | 28.39 | 0.00 | 0.00 | 7.00 | 6.29 | 29.0 | 2030.53 | |
| lincosamids | clindamycin | 0.51 | 0.00 | 0.00 | 1.83 | <LOQ ** | 2.47 | 37.66 | 9.96 | 5.80 | 0.00 | 15.59 | 11.18 | 29.0 | 3.71 |
| macrolides | erythromycin | 0.10 | 0.00 | 0.00 | 0.07 | 0.00 | 0.05 | 0.30 | 0.24 | 0.00 | 0.00 | 0.30 | 0.20 | 71.0 | 251.15 |
| tylosin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 56.59 | 10.23 | 0.00 | 0.00 | 0.00 | 0.00 | 22.6 | 64.45 | |
| tetracyclines | doxycycline | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 68.26 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 16.1 | 413.43 |
| oxytetracycline | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 13.47 | 0.00 | 0.00 | 0.00 | 0.00 | 61.36 | 6.5 | 224.46 | |
| tetracycline | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 9.23 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 35.5 | 214.61 | |
| sulphonamids | sulfamethoxazole | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.56 | 20.90 | 4.69 | 1.48 | 0.00 | 34.05 | 8.83 | 3.2 | 27.70 |
| antifolates | trimethoprim | 0.00 | 0.00 | 0.00 | 1.10 | 0.00 | 0.18 | 38.57 | 7.70 | 2.59 | 0.00 | 8.61 | 4.93 | 35.5 | 188.63 |
| glycopeptides | vancomycin | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 142.66 | 21.84 | 0.00 | 0.00 | 4.23 | 0.00 | 9.7 | 506.19 |
| oxazolidinones | linezolid | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.33 | 17.40 | 1.48 | 1.63 | 0.00 | 3.99 | 1.55 | 6.5 | 200.44 |
| number of antibiotics detected | 2 | 0 | 1 | 5 | 2 | 7 | 13 | 10 | 5 | 1 | 8 | 8 | |||
| total concentration of antibiotics | 1.21 | 0 | 225.19 | 15.45 | 12.17 | 328.40 | 3559.06 | 431.37 | 29.77 | 1.90 | 230.14 | 329.10 | |||
| Site | Beta-Lactamases | Altered Penicillin-Binding Protein (PBP2a) | Erythromycin Esterase | Macrolide Ribosomal Methylase | Aminoglycoside 3’-phosphotransferase | Tetracycline Efflux Protein | Dihydropteroate Synthase | ||
|---|---|---|---|---|---|---|---|---|---|
| blaTEM | blaCTX-M | blaCTX-M3 | mecA | ereA | ermB | strA | tetK | sulIII | |
| BDF | 16.7 | 16.7 | 0 | 0 | 0 | 0 | 16.7 | 50.0 | 0 |
| B3 | 50.0 | 25.0 | 0 | 0 | 0 | 0 | 0 | 50.0 | 0 |
| B1 | 12.5 | 12.5 | 0 | 0 | 0 | 12.5 | 37.5 | 75 | 0 |
| R | 66.7 | 16.7 | 0 | 16.7 | 16.7 | 0 | 66.7 | 33.3 | 16.7 |
| BDZ | 100 | 33.3 | 16.7 | 0 | 83.3 | 100 | 100 | 66.7 | 16.7 |
| total share | 46.7 | 20.0 | 3.3 | 3.3 | 20.0 | 23.3 | 46.7 | 56.7 | 6.7 |
| No. | Resistance Mechanism | Gene | Primer | Sequence (5′-3′) | Annealing Temp. (°C) | Product Length (bp) | Reference |
|---|---|---|---|---|---|---|---|
| 1. | Extended-spectrum beta-lactamases (ESBL) | blaTEM | blaTEM-F | ATTCTTGAAGACGAAAGGGC ACGCTCAGTGGAACGAAAAC | 60 | 1150 | [67] |
| blaTEM-R | |||||||
| 2. | blaCTX-M | blaCTX-M-F | CGATGTGCAGTACCAGTAA TTAGTGACCAGAATCAGCGG | 55 | 585 | [68] | |
| blaCTX-M-R | |||||||
| 3. | blaCTX-M3 | blaCTX-M3-F | GTTACAATGTGTGAGAAGCAG CCGTTTCCGCTATTACAAAC | 50 | 1049 | [69] | |
| blaCTX-M3-R | |||||||
| 4. | blaCTX-M9 | blaCTX-M9-F | GTGACAAAGAGAGTGCAACGG ATGATTCTCGCCGCTGAAGCC | 54 | 856 | [70] | |
| blaCTX-M9-R | |||||||
| 5. | blaSHV | blaSHV-F | CACTCAAGGATGTATTGTG TTAGCGTTGCCAGTGCTCG | 52 | 885 | [67] | |
| blaSHV-R | |||||||
| 6. | blaOXA-1 | blaOXA-1-F | ACACAATACATATCAACTTCGC AGTGTGTTTAGAATGGTGATC | 61 | 813 | [67] | |
| blaOXA-1-R | |||||||
| 7. | Carbapenemases class D | blaOXA-48 | blaOXA-48-F | GCTTGATCGCCCTCGATT GATTTGCTCCGTGGCCGAAA | 60 | 281 | [71] |
| blaOXA-48-R | |||||||
| 8. | Carbapenemases class A | blaKPC | blaKPC-F | TGTTGCTGAAGGAGTTGGGC ACGACGGCATAGTCATTTGC | 57 | 340 | [71] |
| blaKPC-R | |||||||
| 9. | Carbapenemases class B | blaIMP | blaIMP-F | TTGACACTCCATTTACAG GATCGAGAATTAAGCCACCC | 56 | 139 | [71] |
| blaIMP-R | |||||||
| 10. | blaVIM | blaVIM-F | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG | 60 | 390 | [71] | |
| blaVIM-R | |||||||
| 11. | blaNDM | blaNDM-F | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC | 60 | 621 | [72] | |
| blaNDM-R | |||||||
| 12. | Methicillin-resistance | mecA | mecA-F | GTAGAAAATGACTGAACGTCCGATAA CCAATTCCACATTGTTTCGGTCTAA | 55 | 310 | [73] |
| mecA-R | |||||||
| 13. | Macrolide–lincosamide–streptogramin B (MLSb) resistance genes | ereA | ereA-F ereA-R | AACACCCTGAACCCAAGGGACG CTTCACATCCGGATTCGCTCGA | 57 | 420 | [74] |
| 14. | ereB | ereB-F ereB-R | AGAAATGGAGGTTCATACTTACCA CATATAATCATCACCAATGGCA | 52 | 546 | [74] | |
| 15. | ermA | ermA-F ermA-R | TCTAAAAAGCATGTAAAAGAA CTTCGATAGTTTATTAATATTAGT | 52 | 645 | [74] | |
| 16. | ermB | ermB-F ermB-R | GAAAAGGTACTCAACCAAATA AGTAACGGTACTTAAATTGTTTAC | 55 | 639 | [74] | |
| 17. | msrA | msrA-F msrA-R | GGCACAATAAGAGTGTTTAAAGG AAGTTATATCATGAATAGATTGTCCTGTT | 50 | 940 | [75] | |
| 18. | msrB | msrB-F msrB-R | TATGATATCCATAATAATTATCCAATC AAGTTATATCATGAATAGATTGTCCTGTT | 50 | 595 | [75] | |
| 19. | mphA | mphA-F mphA-R | AACTGTACGCACTTGC GGTACTCTTCGTTACC | 50 | 837 | [74] | |
| 20. | lnuA | lnuA-F lnuA-R | GGTGGCTGGGGGGTAGATGTATTAACTGG GCTTCTTTTGAAATACATGGTATTTTTCGATC | 57 | 323 | [75] | |
| 21. | vatA | vatA-F vatA-R | CAATGACCATGGACCTGATC CTTCAGCATTTCGATATCTC | 52 | 619 | [75] | |
| 22. | vatB | vatB-F vatB -R | CCCTGATCCAAATAGCATATATCC CTAAATCAGAGCTACAAAGT | 52 | 602 | [75] | |
| 23. | vga | vga-F vga-R | CCAGAACTGCTATTAGCAGATGAA AAGTTCGTTTCTCTTTTCGACG | 54 | 470 | [75] | |
| 24. | vgb | vgb-F vgb-R | ACTAACCAAGATACAGGACC TTATTGCTTGTCAGCCTTCC | 53 | 734 | [75] | |
| 25. | Streptomycin resistance | strA | strA-F strA-R | TCAATCCCGACTTCTTACCG CACCATGGCAAACAACCATA | 52 | 126 | [76] |
| 26. | Trimetophrim resistance | dfrA12 | dfrA12-F dfrA12-R | TTTATCTCGTTGCTGCGATG AGGCTTGCCGATAGACTCAA | 50 | 155 | [77] |
| 27. | Aminoglycoside resistance | aac(6′)/aph(2′) | aac(6′)/aph(2′′)-F aac(6′)/aph(2′′)-R | CAGAGCCTTGGGAAGATGAAG CCTCGTGTAATTCATGTTCTGGC | 55 | 348 | [78] |
| 28. | Tetracyclines resistance | tetK | tetK-F tetK-R | TCGATAGGAACAGCAGTA CAGCAGATCCTACTCCTT | 55 | 169 | [79] |
| 29. | Sulfonamides resistance | sulIII | sulIII-F sulIII-R | ACCACCGATAGTTTTTCCGA TGCCTTTTTCTTTTAAAGCC | 62 | 199 | [77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stankiewicz, K.; Bulanda, K.; Prajsnar, J.; Lenart-Boroń, A. Impact of the Technical Snow Production Process on Bacterial Community Composition, Antibacterial Resistance Genes, and Antibiotic Input—A Dual Effect of the Inevitable. Int. J. Mol. Sci. 2025, 26, 2771. https://doi.org/10.3390/ijms26062771
Stankiewicz K, Bulanda K, Prajsnar J, Lenart-Boroń A. Impact of the Technical Snow Production Process on Bacterial Community Composition, Antibacterial Resistance Genes, and Antibiotic Input—A Dual Effect of the Inevitable. International Journal of Molecular Sciences. 2025; 26(6):2771. https://doi.org/10.3390/ijms26062771
Chicago/Turabian StyleStankiewicz, Klaudia, Klaudia Bulanda, Justyna Prajsnar, and Anna Lenart-Boroń. 2025. "Impact of the Technical Snow Production Process on Bacterial Community Composition, Antibacterial Resistance Genes, and Antibiotic Input—A Dual Effect of the Inevitable" International Journal of Molecular Sciences 26, no. 6: 2771. https://doi.org/10.3390/ijms26062771
APA StyleStankiewicz, K., Bulanda, K., Prajsnar, J., & Lenart-Boroń, A. (2025). Impact of the Technical Snow Production Process on Bacterial Community Composition, Antibacterial Resistance Genes, and Antibiotic Input—A Dual Effect of the Inevitable. International Journal of Molecular Sciences, 26(6), 2771. https://doi.org/10.3390/ijms26062771






