Optimization Strategy of Expression Vectors and Regulatory Elements for Enhanced Protein Production in Bacillus subtilis
Abstract
1. Introduction
2. Optimization Strategies for Vector Stability and Copy Number in B. subtilis
2.1. Enhancing Vector Stability in B. subtilis
2.2. Increasing Vector Copy Number in B. subtilis
2.3. Current Vectors Optimization Strategies Promises and Challenges
3. Optimization Strategies for Regulatory Systems in B. subtilis
3.1. Enhancing the Utilization Efficiency of Inducers in B. subtilis
3.2. Implementing Alternative Regulatory Systems to Reduce Inducer Toxicity and Costs
3.3. Employing a Dual Transcription-Translation Strategy to Minimize Leaky Expression
3.4. Current Regulatory Systems Promises and Challenges
4. Optimization Strategies for Expression Elements in B. subtilis
4.1. Enhancement of Transcriptional Level via Promoters in B. subtilis
4.2. Augmentation of Translational Rate Through RBS in B. subtilis
4.3. Improvement of Secretion Efficiency by Signal Peptides in B. subtilis
4.4. Optimization of Termination Efficiency with Terminators in B. subtilis
4.5. Current Expression Elements Optimization Strategies Promises and Challenges
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koopman, N.; van Leeuwen, P.; Brul, S.; Seppen, J. History of Fecal Transplantation; Camel Feces Contains Limited Amounts of Bacillus subtilis Spores and Likely Has No Traditional Role in the Treatment of Dysentery. PLoS ONE 2022, 17, e0272607. [Google Scholar] [CrossRef] [PubMed]
- Sudan, S.; Zhan, X.; Li, J. A Novel Probiotic Bacillus subtilis Strain Confers Cytoprotection to Host Pig Intestinal Epithelial Cells during Enterotoxic Escherichia coli Infection. Microbiol. Spectr. 2022, 10, e0125721. [Google Scholar] [CrossRef]
- Li, K.; Guo, Y.; Sun, X.; Xi, X.; Wang, L.; Ren, X.; Wang, C.; Liu, X. Whole-Cell Biocatalysis for ε-Poly-l-Lysine Production by a Food-Grade Recombinant Bacillus subtilis. Enzym. Microb. Technol. 2024, 179, 110467. [Google Scholar] [CrossRef] [PubMed]
- Elmnasser, N.; Hassen, W.; Zmantar, T.; Ashraf, S.A.; Lajimi, R.H.; Humaidi, J.R.; Alreshidi, M.; Hamadou, W.S.; Emira, N.; Snoussi, M. Antagonistic and Enzymatic Activities of Bacillus Species Isolated from the Fish Gastrointestinal Tract as Potential Probiotics Use in Artemia Culture. Cell. Mol. Biol. 2024, 70, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Andleeb, S.; Shafique, I.; Naseer, A.; Abbasi, W.A.; Ejaz, S.; Liaqat, I.; Ali, S.; Khan, M.F.; Ahmed, F.; Ali, N.M. Molecular Characterization of Plant Growth-Promoting Vermi-Bacteria Associated with Eisenia Fetida Gastrointestinal Tract. PLoS ONE 2022, 17, e0269946. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, J.; Wang, B.; Yu, Z. Unraveling Aerobic Cultivable Cellulolytic Microorganisms within the Gastrointestinal Tract of Sheep (Ovis aries) and Their Evaluation for Cellulose Biodegradation. Can. J. Microbiol. 2022, 68, 237–248. [Google Scholar] [CrossRef]
- Pławińska-Czarnak, J.; Wódz, K.; Strzałkowska, Z.; Żychska, M.; Nowak, T.; Kwieciński, A.; Kwieciński, P.; Bielecki, W.; Rodo, A.; Rzewuska, M.; et al. Comparison of Automatic Methods MALDI-TOF, VITEK2 and Manual Methods for the Identification of Intestinal Microbial Communities on the Example of Samples from Alpacas (Vicugna pacos). J. Vet. Res. 2023, 67, 361–372. [Google Scholar] [CrossRef]
- Liu, Y.; Yue, Z.; Sun, Z.; Li, C. Harnessing Native Bacillus Spp. for Sustainable Wheat Production. Appl. Environ. Microbiol. 2023, 89, e0124722. [Google Scholar] [CrossRef]
- Rathod, K.; Rana, S.; Dhandhukia, P.; Thakker, J.N. From Sea to Soil: Marine Bacillus subtilis Enhancing Chickpea Production through in Vitro and in Vivo Plant Growth Promoting Traits. Braz. J. Microbiol. 2024, 55, 823–836. [Google Scholar] [CrossRef]
- Delbrück, A.I.; Zhang, Y.; Hug, V.; Trunet, C.; Mathys, A. Isolation, Stability, and Characteristics of High-Pressure Superdormant Bacillus subtilis Spores. Int. J. Food Microbiol. 2021, 343, 109088. [Google Scholar] [CrossRef]
- Isticato, R.; Lanzilli, M.; Petrillo, C.; Donadio, G.; Baccigalupi, L.; Ricca, E. Bacillus subtilis Builds Structurally and Functionally Different Spores in Response to the Temperature of Growth. Environ. Microbiol. 2020, 22, 170–182. [Google Scholar] [CrossRef]
- Huang, B.; Liu, N.; Huang, Y.; Chen, J. Coculture of Actinomycetes with Bacillus subtilis and Its Effect on the Bioactivesecondary Metabolites. Chin. J. Biotechnol. 2009, 25, 932–940. [Google Scholar]
- Liu, H.; Ji, H.; Wang, S.; Zhang, D.; Wang, J.; Shan, D.; Wang, Y. Research Progress on High-Density Culture of Feed Lactic Acid Bacteria. China Feed 2016, 4, 11–14. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Q.; Qi, X.; Gao, H.; Wang, M.; Guan, H.; Yu, B. Metabolic Engineering of Bacillus subtilis for Riboflavin Production: A Review. Microorganisms 2023, 11, 164. [Google Scholar] [CrossRef]
- Galinier, A.; Delan-Forino, C.; Foulquier, E.; Lakhal, H.; Pompeo, F. Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria. Biomolecules 2023, 13, 720. [Google Scholar] [CrossRef]
- Yang, H.; Qu, J.; Zou, W.; Shen, W.; Chen, X. An Overview and Future Prospects of Recombinant Protein Production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2021, 105, 6607–6626. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Gong, M.; Zhang, H.; Liu, Y.; Lv, X.; Li, J.; Du, G.; Liu, L. Production of Proteins and Commodity Chemicals Using Engineered Bacillus subtilis Platform Strain. Essays Biochem. 2021, 65, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Su, L.; Wu, J. Recent Advances in Recombinant Protein Production by Bacillus subtilis. Annu. Rev. Food Sci. Technol. 2020, 11, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A Universal Cell Factory for Industry, Agriculture, Biomaterials and Medicine. Microb. Cell Fact. 2020, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, Y.S.; More, S.S.; Keerthana, R.; Shaikh, I.A.; J., A.K.; More, V.S.; Niyonzima, F.N.; Muddapur, U.M.; Khan, A.A. Production and Purification of Pectinase from Bacillus subtilis 15A-B92 and Its Biotechnological Applications. Molecules 2022, 27, 4195. [Google Scholar] [CrossRef]
- Cui, J.-N.; Hu, W.; Liu, Y.-X.; Li, Y.-L.; Hu, J.-H.; Liu, Z.-Y.; Chen, J.-H. Isolation and Screening of High-Yielding α-Amylase Mutants of Bacillus subtilis by Heavy Ion Mutagenesis. Appl. Biochem. Biotechnol. 2023, 195, 68–85. [Google Scholar] [CrossRef]
- Baradia, H.; Kumar, S.M.; Chattopadhyay, S. Techno-Economic Analysis of Production and Purification of Lipase from Bacillus subtilis (NCIM 2193). Prep. Biochem. Biotechnol. 2023, 53, 1237–1242. [Google Scholar] [CrossRef]
- Saravanakumar, S.; Prabakaran, N.N.; Ashokkumar, R.; Jamuna, S. Unlocking the Gut’s Treasure: Lipase-Producing Bacillus subtilis Probiotic from the Intestine of Microstomus kitt (Lemon Sole). Appl. Biochem. Biotechnol. 2023, 196, 4273–4286. [Google Scholar] [CrossRef]
- Zhou, M.-J.; Jing Wu, J.W.; Hu, L.-X.; Hu, W.-S.; Huang, J.-B.; Huang, X.-L.; Gao, X.-L.; Luo, Y.-N.; Xue, Z.-L.; Liu, Y. Enhanced Vitamin K2 Production by Engineered Bacillus subtilis during Leakage Fermentation. World J. Microbiol. Biotechnol. 2023, 39, 224. [Google Scholar] [CrossRef]
- Xu, Y.; Song, Y.; Ning, Y.; Li, S.; Qu, Y.; Jiao, B.; Lu, X. Macrolactin XY, a Macrolactin Antibiotic from Marine-Derived Bacillus subtilis Sp. 18. Mar. Drugs 2024, 22, 331. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, S.; Jiang, J.-Y.; Yao, Z.-Y.; Li, Q.; Lian, S.-J.; Liu, Q.; Shi, J.-S.; Xu, Z.-H.; Gong, J.-S. High-Level Extracellular Expression of Hyaluronate Lyase HylP in Bacillus subtilis for Hyaluronan Degradation. Appl. Biochem. Biotechnol. 2024, 196, 6782–6801. [Google Scholar] [CrossRef]
- Akinsemolu, A.A.; Onyeaka, H.; Odion, S.; Adebanjo, I. Exploring Bacillus subtilis: Ecology, Biotechnological Applications, and Future Prospects. J. Basic Microbiol. 2024, 64, e2300614. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Q.; Zhao, J.; Xia, J.-Y. γ-PGA Fermentation by Bacillus subtilis PG-001 with Glucose Feedback Control PH-Stat Strategy. Appl. Biochem. Biotechnol. 2022, 194, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhao, Y.; Dai, L.; Xu, G. Bacillus subtilis and Bifidobacteria Bifidum Fermentation Effects on Various Active Ingredient Contents in Cornus Officinalis Fruit. Molecules 2023, 28, 1032. [Google Scholar] [CrossRef]
- de O Nunes, P.S.; de Medeiros, F.H.; de Oliveira, T.S.; de Almeida Zago, J.R.; Bettiol, W. Bacillus subtilis and Bacillus licheniformis Promote Tomato Growth. Braz. J. Microbiol. 2023, 54, 397–406. [Google Scholar] [CrossRef]
- Mahapatra, S.; Yadav, R.; Ramakrishna, W. Bacillus subtilis Impact on Plant Growth, Soil Health and Environment: Dr. Jekyll and Mr. Hyde. J. Appl. Microbiol. 2022, 132, 3543–3562. [Google Scholar] [CrossRef]
- Goya, M.E.; Xue, F.; Sampedro-Torres-Quevedo, C.; Arnaouteli, S.; Riquelme-Dominguez, L.; Romanowski, A.; Brydon, J.; Ball, K.L.; Stanley-Wall, N.R.; Doitsidou, M. Probiotic Bacillus subtilis Protects against α-Synuclein Aggregation in C. Elegans. Cell Rep. 2020, 30, 367–380.e7. [Google Scholar] [CrossRef]
- Yang, M.; Hutchinson, N.; Ye, N.; Yin, J.; Guan, M.; Wang, Z.; Chen, P.; Yang, S.; Crane, J.D.; Zhang, K.; et al. Engineered Bacillus subtilis as Oral Probiotics to Enhance Clearance of Blood Lactate. bioRxiv 2024. [Google Scholar] [CrossRef]
- Nihorimbere, G.; Korangi Alleluya, V.; Nimbeshaho, F.; Nihorimbere, V.; Legrève, A.; Ongena, M. Bacillus-Based Biocontrol beyond Chemical Control in Central Africa: The Challenge of Turning Myth into Reality. Front. Plant Sci. 2024, 15, 1349357. [Google Scholar] [CrossRef]
- Payne, J.; Bellmer, D.; Jadeja, R.; Muriana, P. The Potential of Bacillus Species as Probiotics in the Food Industry: A Review. Foods 2024, 13, 2444. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, A.; Farinas, E.T. Applications of Bacillus subtilis Protein Display for Medicine, Catalysis, Environmental Remediation, and Protein Engineering. Microorganisms 2024, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Z.; Gao, C.; Jiang, Z.; Huang, S.; Li, X.; Yang, H. Bacillus subtilis as an Excellent Microbial Treatment Agent for Environmental Pollution: A Review. Biotechnol. J. 2025, 20, e70026. [Google Scholar] [CrossRef]
- Chen, Y.; Li, M.; Yan, M.; Chen, Y.; Saeed, M.; Ni, Z.; Fang, Z.; Chen, H. Bacillus subtilis: Current and Future Modification Strategies as a Protein Secreting Factory. World J. Microbiol. Biotechnol. 2024, 40, 195. [Google Scholar] [CrossRef]
- Kaspar, F.; Neubauer, P.; Gimpel, M. Bioactive Secondary Metabolites from Bacillus subtilis: A Comprehensive Review. J. Nat. Prod. 2019, 82, 2038–2053. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.; Song, Y.; Liu, C.; Yu, L.; Shang, Y.; Tang, H.; Sun, S.; Wang, F. Application of Bacillus subtilis as a Live Vaccine Vector: A Review. J. Vet. Med. Sci. 2020, 82, 1693–1699. [Google Scholar] [CrossRef]
- Falkenberg, K.B.; Mol, V.; de la Maza Larrea, A.S.; Pogrebnyakov, I.; Nørholm, M.H.H.; Nielsen, A.T.; Jensen, S.I. The ProUSER2.0 Toolbox: Genetic Parts and Highly Customizable Plasmids for Synthetic Biology in Bacillus subtilis. ACS Synth. Biol. 2021, 10, 3278–3289. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, Z.; Duan, X.; Li, H. The Research Progress on Bacillus Expression System. Genom. Appl. Biol. 2020, 39, 9. [Google Scholar]
- Chen, Z.; Huang, X.; Jiang, H.; Huang, Z.; Yin, G. Research Progress of Bacillus subtilis as an Exogenous Gene Presentation Vector. Ind. Sci. Trib. 2017, 16, 2. [Google Scholar]
- Yu, X.; Tian, J.; Liu, X.; Wu, N. Research Progress of Bacillus subtilis Expression System and Its Promoter Regulatory Elements. Biotechnol. Bull. 2015, 31, 10. [Google Scholar]
- Santos, K.O.; Costa-Filho, J.; Spagnol, K.L.; Marins, L.F. Comparing Methods of Genetic Manipulation in Bacillus subtilis for Expression of Recombinant Enzyme: Replicative or Integrative (CRISPR-Cas9) Plasmid? J. Microbiol. Methods 2019, 164, 105667. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Wang, H.; Pan, X. Present Status of Integration Vectors of Bacillus subtilis. Lett. Biotechnol. 2003, 14, 3. [Google Scholar]
- Zhao, P.; Liu, J. Research Progress on the Expression System of Bacillus subtilis. BeiFang MuYe Chin. 2024, 12, 27. [Google Scholar]
- Zhenhua, W.; Jianzhen, L.; Kangcheng, P. The Research Progress on Probiotic Bacillus Expression System. Anim. Husb. Vet. Med. Chin. 2018, 50, 5. [Google Scholar]
- Far, B.E.; Ragheb, M.; Rahbar, R.; Mafakher, L.; Nojookambari, N.Y.; Achinas, S.; Yazdansetad, S. Cloning and Expression of Staphylococcus Simulans Lysostaphin Enzyme Gene in Bacillus subtilis WB600. AIMS Microbiol. 2021, 7, 271–283. [Google Scholar] [CrossRef]
- Song, W.; Zhang, M.; Li, X.; Zhang, Y.; Zheng, J. Heterologous Expression of Cyclodextrin Glycosyltransferase from Bacillus stearothermophilus in Bacillus subtilis and Its Application in Glycosyl Rutin Production. 3 Biotech 2023, 13, 84. [Google Scholar] [CrossRef]
- Shafaati, M.; Ghorbani, M.; Mahmoodi, M.; Ebadi, M.; Jalalirad, R. Expression and Characterization of Hemagglutinin-Neuraminidase Protein from Newcastle Disease Virus in Bacillus subtilis WB800. J. Genet. Eng. Biotechnol. 2022, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; You, Q.; Hao, W.; Liang, X.; Yuan, J.; Li, Q. Study on the Stability of Growth Differentiation Factor (BMP11) Recombinant Plasmid in Escherichia coli. Sci. Technol. Food Ind. Chin. 2017, 38, 5. [Google Scholar]
- Huang, K.; Zhang, T.; Jiang, B.; Yan, X.; Mu, W.; Miao, M. Overproduction of Rummeliibacillus Pycnus Arginase with Multi-Copy Insertion of the Arg R.Pyc Cassette into the Bacillus subtilis Chromosome. Appl. Microbiol. Biotechnol. 2017, 101, 6039–6048. [Google Scholar] [CrossRef]
- Ferrando, J.; Miñana-Galbis, D.; Picart, P. The Construction of an Environmentally Friendly Super-Secreting Strain of Bacillus subtilis through Systematic Modulation of Its Secretory Pathway Using the CRISPR-Cas9 System. Int. J. Mol. Sci. 2024, 25, 6957. [Google Scholar] [CrossRef]
- Itaya, M.; Kaneko, S. Integration of Stable Extracellular DNA Released from Escherichia coli into the Bacillus subtilis Genome Vector by Culture Mix Method. Nucleic Acids Res. 2010, 38, 2551–2557. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, C.; Li, Y.; Zheng, Y.; Liu, Y. Expression of Zearalenone Degrading Enzyme Gene Zlhy-6 in Bacillus subtilis. J. Nucl. Agric. Sci. 2022, 36, 885–894. [Google Scholar]
- Ogawa, T.; Iwata, T.; Kaneko, S.; Itaya, M.; Hirota, J. An Inducible RecA Expression Bacillus subtilis Genome Vector for Stable Manipulation of Large DNA Fragments. BMC Genom. 2015, 16, 209. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Jianghua, L.; Long, L.; Guocheng, D.; Yanfeng, L. Regulating the Synthesis of N-Acetylneuraminic Acid Based on Adaptive Evolution and Plasmid Stability Modification in Bacillus subtilis. Food Ferment. Ind. Chin. 2021, 47, 1–6. [Google Scholar]
- Tian, H.; Liu, B.; Yang, J.; Zhou, C.; Xu, X.; Zhang, Y.; Lu, Z.; Zhang, W. Genetic Transformation System for Bacillus velezensis NSZ-YBGJ001 and Curing of the Endogenous Plasmid PBV01. Biotechnol. Lett. 2021, 43, 1595–1605. [Google Scholar] [CrossRef]
- Lv, X.; Wu, Y.; Lin, L.; Xu, X.; Yu, W.; Cui, S.; Li, J.; Su, G.; Liu, L. Strategies and Tools for Metabolic Engineering in Bacillus subtilis. Chin. J. Biotechnol. 2021, 37, 18. [Google Scholar]
- Guo, Y.; Xia, Y.; Liang, Z.; Yang, S.; Guo, S.; Sun, L.; Huo, Y.-X. Plasmid-Stabilizing Strains for Antibiotic-Free Chemical Fermentation. ACS Synth. Biol. 2024, 13, 2820–2832. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, Y.; Lv, X.; Li, J.; Liu, L.; Du, G.; Chen, J.; Liu, Y. Reduced Genetic Heterogeneity for Stable Bioproduction by Harnessing the Bias and Mechanism of Mutation. Microb. Biotechnol. 2025, 18, e70162. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, J.; Tan, M.; Zhen, J.; Shu, W.; Yang, S.; Ma, Y.; Zheng, H.; Song, H. High Copy Number and Highly Stable Escherichia coli-Bacillus subtilis Shuttle Plasmids Based on PWB980. Microb. Cell Fact. 2020, 19, 25. [Google Scholar] [CrossRef]
- Boros, I.; Pósfai, G.; Venetianer, P. High-Copy-Number Derivatives of the Plasmid Cloning Vector PBR322. Gene 1984, 30, 257–260. [Google Scholar] [CrossRef]
- Bert, A.G.; Burrows, J.; Osborne, C.S.; Cockerill, P.N. Generation of an Improved Luciferase Reporter Gene Plasmid That Employs a Novel Mechanism for High-Copy Replication. Plasmid 2000, 44, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.J.; Ryu, A.J.; Li, L.; Han, N.S.; Jeong, K.J. Development of a High-Copy Plasmid for Enhanced Production of Recombinant Proteins in Leuconostoc citreum. Microb. Cell Fact. 2016, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Mu, W.; Jiang, B.; Yan, X.; Zhang, T. Food-Grade Expression of d-Psicose 3-Epimerase with Tandem Repeat Genes in Bacillus subtilis. J. Agric. Food Chem. 2016, 64, 5701–5707. [Google Scholar] [CrossRef]
- Xiong, H.; Wei, Y. Research Progress of Bacillus subtilis Expression System and Its Promoter Regulatory Elements. Guangxi Sci. Chin. 2018, 25, 9. [Google Scholar]
- Zhao, S.; Zhang, M.; Gao, X. Progress of Homologous Recombinant Expression of Exogenous Genes of Bacillus subtilis for Animal Husbandry Applications. Jilin Anim. Husb. Vet. Med. 2024, 45, 7–9. [Google Scholar]
- Yan, J.; Zhang, Z.; Feng, L.; Tang, H.; Yang, K.; Wei, G.; Zhang, X. Cloning and Expression of Trichoderma Viride Eg VII. J. Cent. South Univ. For. Technol. 2022, 42, 9. [Google Scholar]
- Tian, Y.; Song, Z.; Ma, Z.; Wang, C.; Xu, J.; Zhou, C. Expression and Fermentation Condition Optimization of Xylanase XynZF-318 in Bacillus subtilis WB600. Sci. Technol. Food Ind. 2020, 41, 7. [Google Scholar]
- Yao, D.; Han, X.; Gao, H.; Wang, B.; Fang, Z.; Li, H.; Fang, W.; Xiao, Y. Enhanced Extracellular Production of Raw Starch-Degrading α-Amylase in Bacillus subtilis through Expression Regulatory Element Modification and Fermentation Optimization. Microb. Cell Fact. 2023, 22, 118. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Li, D.; Cao, X.; Yuan, H.; Zhang, Y.; Yu, J.; Lu, F.; Li, Y. The Effect on Heterologous Expression of Alkaline Protease AprE by Two Different Promoter and Combinatorial. China Biotechnol. 2019, 39, 7. [Google Scholar]
- Zhou, Z.; Wang, X. Rational Design and Structure-Based Engineering of Alkaline Pectate Lyase from Paenibacillus Sp. 0602 to Improve Thermostability. BMC Biotechnol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Li, X.; Wang, H.; Zhou, C.; Ma, Y.; Li, J.; Song, J. Cloning, Expression and Characterization of a Pectate Lyase from Paenibacillus sp. 0602 in Recombinant Escherichia coli. BMC Biotechnol. 2014, 14, 18. [Google Scholar] [CrossRef]
- Zhang, K.; Su, L.; Wu, J. Enhancing Extracellular Pullulanase Production in Bacillus subtilis Through DltB Disruption and Signal Peptide Optimization. Appl. Biochem. Biotechnol. 2022, 194, 1206–1220. [Google Scholar] [CrossRef]
- Han, L.; Chen, Q.; Luo, J.; Cui, W.; Zhou, Z. Development of a Glycerol-Inducible Expression System for High-Yield Heterologous Protein Production in Bacillus subtilis. Microbiol. Spectr. 2022, 10, e0132222. [Google Scholar] [CrossRef]
- Chu, P.T.B.; Phan, T.T.P.; Nguyen, T.T.T.; Truong, T.T.T.; Schumann, W.; Nguyen, H.D. Potent IPTG-Inducible Integrative Expression Vectors for Production of Recombinant Proteins in Bacillus subtilis. World J. Microbiol. Biotechnol. 2023, 39, 143. [Google Scholar] [CrossRef]
- Wu, J.; Liang, C.; Li, Y.; Zeng, Y.; Sun, X.; Jiang, P.; Chen, W.; Xiong, D.; Jin, J.-M.; Tang, S.-Y. Engineering and Application of LacI Mutants with Stringent Expressions. Microb. Biotechnol. 2024, 17, e14427. [Google Scholar] [CrossRef]
- Marklund, E.; van Oosten, B.; Mao, G.; Amselem, E.; Kipper, K.; Sabantsev, A.; Emmerich, A.; Globisch, D.; Zheng, X.; Lehmann, L.C.; et al. DNA Surface Exploration and Operator Bypassing during Target Search. Nature 2020, 583, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Marklund, E.; Mao, G.; Yuan, J.; Zikrin, S.; Abdurakhmanov, E.; Deindl, S.; Elf, J. Sequence Specificity in DNA Binding Is Mainly Governed by Association. Science 2022, 375, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhu, Y.; Fu, G.; Song, Y.; Jin, Z.; Sun, Y.; Zhang, D. High-Level Intra- and Extra-Cellular Production of D-Psicose 3-Epimerase via a Modified Xylose-Inducible Expression System in Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1577–1591. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fang, Y.; Shi, Y.; Xin, Y.; Gu, Z.; Yang, T.; Li, Y.; Ding, Z.; Shi, G.; Zhang, L. Analysis of Xylose Operon from Paenibacillus polymyxa ATCC842 and Development of Tools for Gene Expression. Int. J. Mol. Sci. 2022, 23, 5024. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-C. Xylose Metabolism and Transport in Bacillus subtilis and Its Application to D-Ribose Production. J. Microbiol. Biotechnol. 2025, 35, e2504021. [Google Scholar] [CrossRef]
- Heravi, K.M.; Altenbuchner, J. Regulation of the Bacillus subtilis Mannitol Utilization Genes: Promoter Structure and Transcriptional Activation by the Wild-Type Regulator (MtlR) and Its Mutants. Microbiology 2014, 160, 91–101. [Google Scholar] [CrossRef]
- Liu, H.; Liu, H.; Yang, S.; Wang, R.; Wang, T. Improved Expression and Optimization of Trehalose Synthase by Regulation of Pglv in Bacillus subtilis. Sci. Rep. 2019, 9, 6585. [Google Scholar] [CrossRef]
- Clermont, L.; Macha, A.; Müller, L.M.; Derya, S.M.; von Zaluskowski, P.; Eck, A.; Eikmanns, B.J.; Seibold, G.M. The α-Glucan Phosphorylase MalP of Corynebacterium Glutamicum Is Subject to Transcriptional Regulation and Competitive Inhibition by ADP-Glucose. J. Bacteriol. 2015, 197, 1394–1407. [Google Scholar] [CrossRef]
- Chen, S.; Tong, Q.; Guo, X.; Cong, H.; Zhao, Z.; Liang, W.; Li, J.; Zhu, P.; Yang, H. Complete Secretion of Recombinant Bacillus subtilis Levansucrase in Pichia Pastoris for Production of High Molecular Weight Levan. Int. J. Biol. Macromol. 2022, 214, 203–211. [Google Scholar] [CrossRef]
- Débarbouillé, M.; Martin-Verstraete, I.; Arnaud, M.; Klier, A.; Rapoport, G. Positive and Negative Regulation Controlling Expression of the Sac Genes in Bacillus subtilis. Res. Microbiol. 1991, 142, 757–764. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, P.; Wang, X.; Wen, J. Construction of Bacillus subtilis for Efficient Production of Fengycin from Xylose through CRISPR-Cas9. Front. Microbiol. 2023, 14, 1342199. [Google Scholar] [CrossRef]
- Park, Y.-C.; Jun, S.Y.; Seo, J.-H. Construction and Characterization of Recombinant Bacillus subtilis JY123 Able to Transport Xylose Efficiently. J. Biotechnol. 2012, 161, 402–406. [Google Scholar] [CrossRef]
- Gao, W.; Yin, Y.; Wang, P.; Tan, W.; He, M.; Wen, J. Production of Fengycin from D-Xylose through the Expression and Metabolic Regulation of the Dahms Pathway. Appl. Microbiol. Biotechnol. 2022, 106, 2557–2567. [Google Scholar] [CrossRef]
- Choi, J.W.; Song, N.-E.; Hong, S.-P.; Rhee, Y.K.; Hong, H.-D.; Cho, C.-W. Engineering Bacillus subtilis J46 for Efficient Utilization of Galactose through Adaptive Laboratory Evolution. AMB Express 2024, 14, 14. [Google Scholar] [CrossRef]
- Yan, P.; Wu, Y.; Yang, L.; Wang, Z.; Chen, T. Engineering Genome-Reduced Bacillus subtilis for Acetoin Production from Xylose. Biotechnol. Lett. 2018, 40, 393–398. [Google Scholar] [CrossRef]
- Yue, J.; Fu, G.; Zhang, D.; Wen, J. A New Maltose-Inducible High-Performance Heterologous Expression System in Bacillus subtilis. Biotechnol. Lett. 2017, 39, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Neira, J.L.; Cámara-Artigas, A.; Hernández-Cifre, J.G.; Ortore, M.G. The Histidine Phosphocarrier Kinase/Phosphorylase from Bacillus subtilis Is an Oligomer in Solution with a High Thermal Stability. Int. J. Mol. Sci. 2021, 22, 3231. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gu, Y.; Ren, C.; Yang, S.; Jiang, W. Recent Research on Catabolite Control Protein A in Microorganisms. Chin. Bull. Life Sci. 2011, 23, 9. [Google Scholar]
- Liu, Z.; Wang, Y.; Liu, S.; Guo, X.; Zhao, T.; Wu, J.; Chen, S. Boosting the Heterologous Expression of D-Allulose 3-Epimerase in Bacillus subtilis through Protein Engineering and Catabolite-Responsive Element Box Engineering. J. Agric. Food Chem. 2022, 70, 12128–12134. [Google Scholar] [CrossRef]
- Li, D.; Guo, J.; Zhang, Z.; Liu, Y.; Lu, F.; Li, Q.; Liu, Y.; Li, Y. Sequence Composition and Location of CRE Motifs Affect the Binding Ability of CcpA Protein. Int. J. Biol. Macromol. 2023, 253, 126407. [Google Scholar] [CrossRef]
- Sprehe, M.; Seidel, G.; Diel, M.; Hillen, W. CcpA Mutants with Differential Activities in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2007, 12, 96–105. [Google Scholar] [CrossRef]
- Chen, B.; Wen, J.; Zhao, X.; Ding, J.; Qi, G. Surfactin: A Quorum-Sensing Signal Molecule to Relieve CCR in Bacillus amyloliquefaciens. Front. Microbiol. 2020, 11, 631. [Google Scholar] [CrossRef]
- Haller, D.J.; Castillo-Hair, S.M.; Tabor, J.J. Optogenetic Control of B. subtilis Gene Expression Using the CcaSR System. Methods Mol. Biol. 2025, 2840, 1–17. [Google Scholar] [CrossRef]
- Khani, M.-H.; Bagheri, M. Skimmed Milk as an Alternative for IPTG in Induction of Recombinant Protein Expression. Protein Expr. Purif. 2020, 170, 105593. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ding, D.; Wang, H.; Ren, X.; Lee, S.Y.; Zhang, D. Balancing Cell Growth and Product Synthesis for Efficient Microbial Cell Factories. Adv. Sci. 2025, 12, e10649. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Tong, Y.; Li, Y.; Tao, J.; Rao, S.; Li, J.; Zhou, J.; Liu, S. Efficient, Flexible Autoinduction Expression Systems with Broad Initiation in Bacillus subtilis. ACS Synth. Biol. 2021, 10, 3084–3093. [Google Scholar] [CrossRef]
- Xu, K.; Tong, Y.; Li, Y.; Tao, J.; Rao, S.; Li, J.; Zhou, J.; Liu, S. Autoinduction Expression Modules for Regulating Gene Expression in Bacillus subtilis. ACS Synth. Biol. 2022, 11, 4220–4225. [Google Scholar] [CrossRef]
- Zeng, M.; Sarker, B.; Howitz, N.; Shah, I.; Andrews, L.B. Synthetic Homoserine Lactone Sensors for Gram-Positive Bacillus subtilis Using LuxR-Type Regulators. ACS Synth. Biol. 2024, 13, 282–299. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, G.G.; Lins, M.R.d.C.R.; Silva, B.F.; de Paiva, G.B.; Zocca, V.F.B.; Ribeiro, N.V.; Picheli, F.P.; Mack, M.; Pedrolli, D.B. A Modular Autoinduction Device for Control of Gene Expression in Bacillus subtilis. Metab. Eng. 2020, 61, 326–334. [Google Scholar] [CrossRef]
- Xu, K.; Tong, Y.; Li, Y.; Tao, J.; Rao, S.; Li, J.; Zhou, J.; Liu, S. Autoinduction AND Gate Inhibits Cell Lysis to Enhance Protein Production in Bacillus subtilis Controlled by Population Density and Cell Physiological State. ACS Synth. Biol. 2023, 12, 842–851. [Google Scholar] [CrossRef]
- Wu, G.; Yin, C.; Zheng, J.; Wang, M.; Abdalmegeed, D.; Zhang, F.; Sun, S.; Sun, S.; Shao, Y.; Xin, Z. Dynamic Regulation of Iturin Production via Reconstructing the Quorum-Sensing System ComQXPA in Bacillus subtilis. World J. Microbiol. Biotechnol. 2025, 41, 173. [Google Scholar] [CrossRef]
- Wang, B.; Wang, K.; Zhao, X.; Fang, Z.; Zhao, Y.; Fang, Y.; Xiao, Y.; Yao, D. Development and Construction of a Novel Bacillus subtilis Autoinducible Extracellular Expression System Based on a LuxI/R Device. Microb. Cell Fact. 2025, 24, 86. [Google Scholar] [CrossRef]
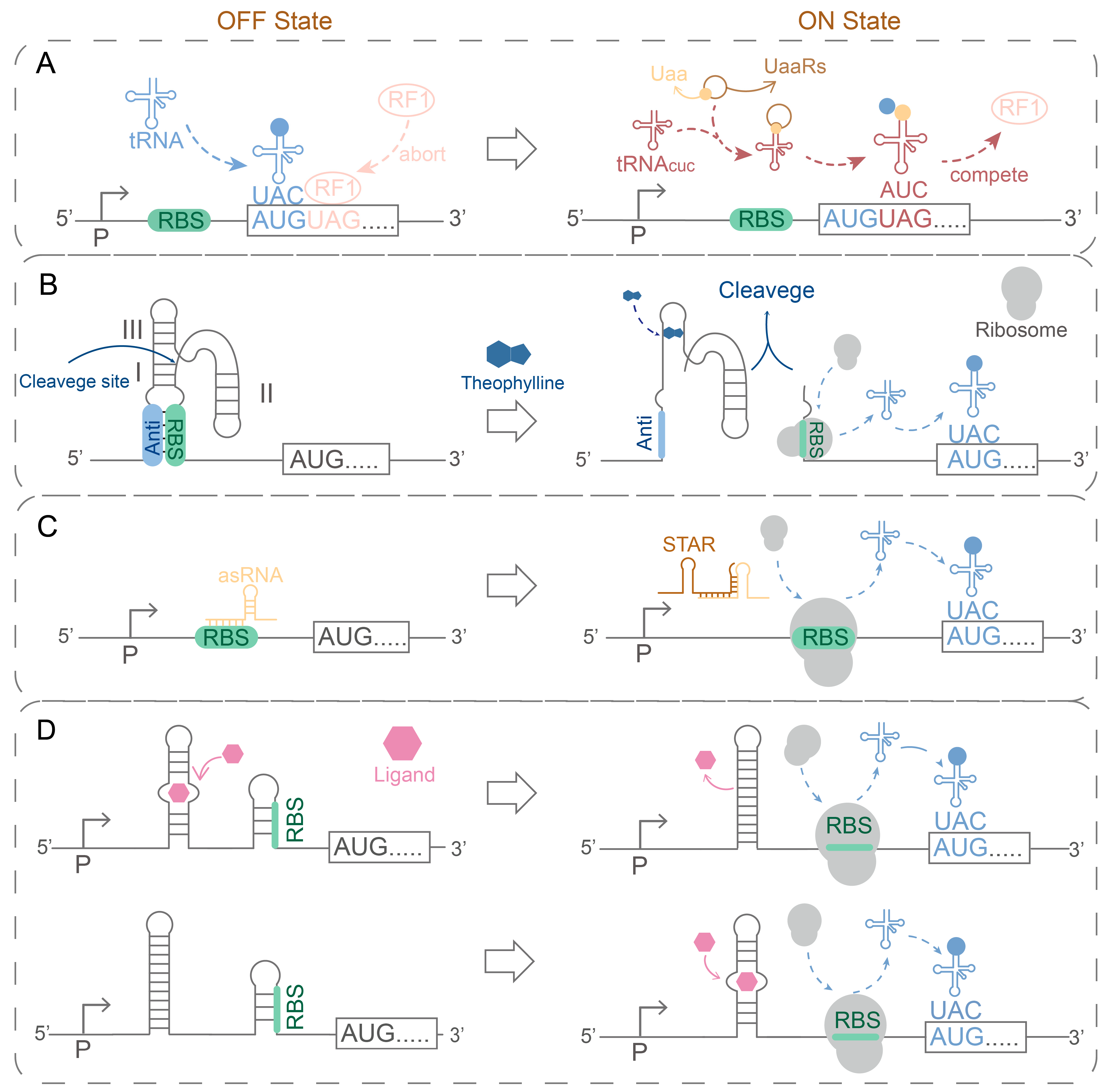
- Kato, Y. Extremely Low Leakage Expression Systems Using Dual Transcriptional-Translational Control for Toxic Protein Production. Int. J. Mol. Sci. 2020, 21, 705. [Google Scholar] [CrossRef]
- Kato, Y. Tunable Translational Control Using Site-Specific Unnatural Amino Acid Incorporation in Escherichia coli. PeerJ 2015, 3, e904. [Google Scholar] [CrossRef]
- Kato, Y. Tight Translational Control Using Site-Specific Unnatural Amino Acid Incorporation with Positive Feedback Gene Circuits. ACS Synth. Biol. 2018, 7, 1956–1963. [Google Scholar] [CrossRef]
- Peng, A.; Yin, G.; Zuo, W.; Zhang, L.; Du, G.; Chen, J.; Wang, Y.; Kang, Z. Regulatory RNAs in Bacillus subtilis: A Review on Regulatory Mechanism and Applications in Synthetic Biology. Synth. Syst. Biotechnol. 2024, 9, 223–233. [Google Scholar] [CrossRef]
- Chen, L.; Xin, X.; Zhang, Y.; Li, S.; Zhao, X.; Li, S.; Xu, Z. Advances in Biosynthesis of Non-Canonical Amino Acids (NcAAs) and the Methods of NcAAs Incorporation into Proteins. Molecules 2023, 28, 6745. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Tian, R.; Wu, Y.; Lv, X.; Li, J.; Liu, L.; Du, G.; Chen, J.; Liu, Y. Facilitating Stable Gene Integration Expression and Copy Number Amplification in Bacillus subtilis through a Reversible Homologous Recombination Switch. Synth. Syst. Biotechnol. 2024, 9, 577–585. [Google Scholar] [CrossRef]
- Miao, S.; Yang, T.; Cui, W.; Zhou, Z. Construction and Application of Theophylline-Activated RNA Switches in the Regulation of Expression of Recombinant Proteins in Bacillus subtilis. Sheng Wu Gong Cheng Xue Bao 2019, 35, 1478–1490. [Google Scholar] [CrossRef]
- Niu, T.; Liu, Y.; Li, J.; Koffas, M.; Du, G.; Alper, H.S.; Liu, L. Engineering a Glucosamine-6-Phosphate Responsive GlmS Ribozyme Switch Enables Dynamic Control of Metabolic Flux in Bacillus subtilis for Overproduction of N-Acetylglucosamine. ACS Synth. Biol. 2018, 7, 2423–2435. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Iordanescu, S.; Projan, S.J.; Kornblum, J.; Edelman, I. PT181 Plasmid Replication Is Regulated by a Countertranscript-Driven Transcriptional Attenuator. Cell 1989, 59, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Han, L.; Cheng, J.; Liu, Z.; Zhou, L.; Guo, J.; Zhou, Z. Engineering an Inducible Gene Expression System for Bacillus subtilis from a Strong Constitutive Promoter and a Theophylline-Activated Synthetic Riboswitch. Microb. Cell Fact. 2016, 15, 199. [Google Scholar] [CrossRef]
- Gatti-Lafranconi, P.; Dijkman, W.P.; Devenish, S.R.A.; Hollfelder, F. A Single Mutation in the Core Domain of the Lac Repressor Reduces Leakiness. Microb. Cell Fact. 2013, 12, 67. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, X.; Zhang, W.; Huang, Z.; Wu, Y.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. De Novo 2’-Fucosyllactose Biosynthesis Using Glucose as the Sole Carbon Source by Multiple Engineered Bacillus subtilis. Metab. Eng. 2024, 88, 85–93. [Google Scholar] [CrossRef]
- Tran, D.T.M.; Phan, T.T.P.; Huynh, T.K.; Dang, N.T.K.; Huynh, P.T.K.; Nguyen, T.M.; Truong, T.T.T.; Tran, T.L.; Schumann, W.; Nguyen, H.D. Development of Inducer-Free Expression Plasmids Based on IPTG-Inducible Promoters for Bacillus subtilis. Microb. Cell Fact. 2017, 16, 130. [Google Scholar] [CrossRef]
- Zhang, G.; An, Y.; Zabed, H.M.; Yun, J.; Parvez, A.; Zhao, M.; Zhang, C.; Ravikumar, Y.; Li, J.; Qi, X. Rewiring Bacillus subtilis and Bioprocess Optimization for Oxidoreductive Reaction-Mediated Biosynthesis of D-Tagatose. Bioresour. Technol. 2023, 389, 129843. [Google Scholar] [CrossRef]
- Cong, G.; Li, M.; Jiang, M.; Guo, D.; Liu, Y.; Wang, C.; Li, X. Heterologous Expression and Characterization of Maltotetraose Amylase in Bacillus subtilis Based on Maltose-Inducible Promoter. J. Microbiol. 2024, 44, 23–31. [Google Scholar]
- Xu, Y.; Tang, L.; Zhang, B.; Liu, J. Application of Promoter Optimization Strategies in Microbial Metabolic Engineering. Chin. Bull. Life Sci. 2022, 34, 871–879. [Google Scholar]
- Ling, M.; Liu, Y.; Li, J.; Shin, H.D.; Chen, J.; Du, G.; Liu, L. Combinatorial Promoter Engineering of Glucokinase and Phosphoglucoisomerase for Improved N-Acetylglucosamine Production in Bacillus subtilis. Bioresour. Technol. 2017, 245, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yao, D.; Pan, Y.; Chen, X.; Fang, Z.; Xiao, Y. Enhanced Extracellular Raw Starch-Degrading α-Amylase Production in Bacillus subtilis by Promoter Engineering and Translation Initiation Efficiency Optimization. Microb. Cell Fact. 2022, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Yan, R.; Shen, J.; Zhu, X.; Meng, F.; Lu, Z.; Lu, F. Cis-Element Engineering Promotes the Expression of Bacillus subtilis Type I L-Asparaginase and Its Application in Food. Int. J. Mol. Sci. 2022, 23, 6588. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, B.; Cheng, S.; Zhao, L.; Liu, Y.; Jiang, J.; Yan, X. Promoter Engineering Enables Overproduction of Foreign Proteins from a Single Copy Expression Cassette in Bacillus subtilis. Microb. Cell Fact. 2019, 18, 111. [Google Scholar] [CrossRef]
- Phan, T.T.P.; Tran, L.T.; Schumann, W.; Nguyen, H.D. Development of Pgrac100-Based Expression Vectors Allowing High Protein Production Levels in Bacillus subtilis and Relatively Low Basal Expression in Escherichia coli. Microb. Cell Fact. 2015, 14, 72. [Google Scholar] [CrossRef]
- Harms, M.; Wolfgramm, H.; Schedlowski, M.; Michalik, S.; Hildebrandt, P.; Schaffer, M.; Völker, U.; Reder, A. Characterization of the MgsR-Dependent Promoter Structure in Bacillus subtilis-Application of a Novel PHIS Plasmid-Based Screening System for Promoter Element Analysis. Nucleic Acids Res. 2025, 53, gkaf636. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Arsalan, A.; Zhang, G.; Yun, J.; Zhang, C.; Qi, X. Coexpression of D-Allulose 3-Epimerase and L-Rhamnose Isomerase in Bacillus subtilis through a Dual Promoter Enables High-Level Biosynthesis of D-Allose from D-Fructose in One Pot. J. Agric. Food Chem. 2025, 73, 2056–2067. [Google Scholar] [CrossRef]
- Lu, J.; Zhao, Y.; Cheng, Y.; Hu, R.; Fang, Y.; Lyu, M.; Wang, S.; Lu, Z. Optimal Secretory Expression of Acetaldehyde Dehydrogenase from Issatchenkia Terricola in Bacillus subtilis through a Combined Strategy. Molecules 2022, 27, 747. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, T.; Liu, Y.; Tian, R.; Lv, X.; Li, J.; Du, G.; Chen, J.; Ledesma-Amaro, R.; Liu, L. Design of a Programmable Biosensor-CRISPRi Genetic Circuits for Dynamic and Autonomous Dual-Control of Metabolic Flux in Bacillus subtilis. Nucleic Acids Res. 2020, 48, 996–1009. [Google Scholar] [CrossRef]
- Li, X.; Xu, S.; Zhang, X.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Yang, S.; et al. Design of a High-Efficiency Synthetic System for l-Asparaginase Production in Bacillus subtilis. Eng. Life Sci. 2019, 19, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Abdulmalek, H.W.; Yazgan-Karataş, A. Improvement of Bacilysin Production in Bacillus subtilis by CRISPR/Cas9-Mediated Editing of the 5’-Untranslated Region of the Bac Operon. J. Microbiol. Biotechnol. 2023, 33, 410–418. [Google Scholar] [CrossRef]
- Volkenborn, K.; Kuschmierz, L.; Benz, N.; Lenz, P.; Knapp, A.; Jaeger, K.-E. The Length of Ribosomal Binding Site Spacer Sequence Controls the Production Yield for Intracellular and Secreted Proteins by Bacillus subtilis. Microb. Cell Fact. 2020, 19, 154. [Google Scholar] [CrossRef]
- Tan, M.; Chen, X.; Peng, Z.; Zhang, J.; Zhang, G. Enhancing the Extracellular Expression of Bacillus licheniformis Keratinase in Bacillus subtilis by Combinatorial Strategies of Signal Peptides Screening and Ribosome Binding Site Optimization. Food Ferment. Ind. 2023, 49, 8–14. [Google Scholar]
- Zhou, C.; Xue, Y.; Ma, Y. Characterization and High-Efficiency Secreted Expression in Bacillus subtilis of a Thermo-Alkaline β-Mannanase from an Alkaliphilic Bacillus clausii Strain S10. Microb. Cell Fact. 2018, 17, 124. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Zhang, Z.; Chen, M.; Wang, S.; Niu, S.; Yu, H.; Yan, Z.; Zheng, X. Promoter and Signal Peptide Optimization Increases the Secretory Expression of Glutaminase in Bacillus subtilis. Microbiol. China 2024, 51, 1536–1549. [Google Scholar]
- Licheng, Z.; Ran, Z.; Bo, J.; Qing, M.; Jingjing, C.; Xiaoyong, L. Efficient Production of an Alginate Lyase in Bacillus subtilis with Combined Strategy: Vector and Host Selection, Promoter and Signal Peptide Screening, and Modification of a Translation Initiation Region. J. Agric. Food Chem. 2024, 72, 19403–19412. [Google Scholar] [CrossRef]
- Qiao, L.; Zhemin, Z.; Wenjing, C. Gene Tandem Strategy Strengthens the Function of Terminators and Its Application in Gene Expression in Bacillus subtilis. Acta Microbiol. Sin. 2021, 61, 13. [Google Scholar]
- Gan, T.; Liu, Y.; Qiao, Y.; Dong, Y.; Feng, J.; Chen, X.; Zhu, L. Translation Regulation in Bacillus subtilis and Its Applications in Heterologous Protein Expression: A Review. Int. J. Biol. Macromol. 2025, 311, 143653. [Google Scholar] [CrossRef] [PubMed]
- Dahlgren, A.; Rydén-Aulin, M. Effects of Two Cis-Acting Mutations on the Regulation and Expression of Release Factor One in Escherichia coli. Biochimie 2004, 86, 431–438. [Google Scholar] [CrossRef]
- Kozak, M. Point Mutations Define a Sequence Flanking the AUG Initiator Codon That Modulates Translation by Eukaryotic Ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Zhang, M.; Song, J.; Xiao, J.; Jin, J.; Nomura, C.T.; Chen, S.; Wang, Q. Engineered Multiple Translation Initiation Sites: A Novel Tool to Enhance Protein Production in Bacillus licheniformis and Other Industrially Relevant Bacteria. Nucleic Acids Res. 2022, 50, 11979–11990. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, B.; Pan, L. Efficient Expression of γ-Glutamyl Transpeptidase in Bacillus subtilis via CRISPR/Cas9n and Its Immobilization. Appl. Microbiol. Biotechnol. 2024, 108, 149. [Google Scholar] [CrossRef]
- Chen, H.; Wu, J.; Huang, X.; Feng, X.; Ji, H.; Zhao, L.; Wang, J. Overexpression of Bacillus circulans Alkaline Protease in Bacillus subtilis and Its Potential Application for Recovery of Protein from Soybean Dregs. Front. Microbiol. 2022, 13, 968439. [Google Scholar] [CrossRef]
- Caspers, M.; Brockmeier, U.; Degering, C.; Eggert, T.; Freudl, R. Improvement of Sec-Dependent Secretion of a Heterologous Model Protein in Bacillus subtilis by Saturation Mutagenesis of the N-Domain of the AmyE Signal Peptide. Appl. Microbiol. Biotechnol. 2010, 86, 1877–1885. [Google Scholar] [CrossRef]
- Zhang, M.; Zhen, J.; Teng, J.; Zhao, X.; Fu, X.; Song, H.; Zhang, Y.; Zheng, H.; Bai, W. N-Terminal Sequences of Signal Peptides Assuming Critical Roles in Expression of Heterologous Proteins in Bacillus subtilis. Microorganisms 2024, 12, 1275. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, H.; Zhang, K.; Ma, Y.; Shen, W.; Xia, Y.; Chen, L. Signal Peptide Mutant and Its Application to Enhance Extracellular Production of Recombinant Bacillus subtilis Protein. Chinese Patent CN202011322536.7, 22 April 2022. [Google Scholar]
- Zhang, Z.; Li, Y.; Zheng, L.; Jin, M.; Wu, Y.; Xu, R.; Luo, Y.; Wu, J.; Su, W.; Luo, S.; et al. A Novel Method for High Level Production of Protein Glutaminase by SfGFP Tag in Bacillus subtilis. Int. J. Biol. Macromol. 2024, 262, 130092. [Google Scholar] [CrossRef]
- Le, N.T.P.; Phan, T.T.P.; Phan, H.T.T.; Truong, T.T.T.; Schumann, W.; Nguyen, H.D. Influence of N-Terminal His-Tags on the Production of Recombinant Proteins in the Cytoplasm of Bacillus subtilis. Biotechnol. Rep. 2022, 35, e00754. [Google Scholar] [CrossRef]
- Heinrich, J.; Drewniok, C.; Neugebauer, E.; Kellner, H.; Wiegert, T. The YoaW Signal Peptide Directs Efficient Secretion of Different Heterologous Proteins Fused to a StrepII-SUMO Tag in Bacillus subtilis. Microb. Cell Fact. 2019, 18, 31. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Kang, C. Lysis Delay and Burst Shrinkage of Coliphage T7 by Deletion of Terminator Tφ Reversed by Deletion of Early Genes. J. Virol. 2014, 88, 2107–2115. [Google Scholar] [CrossRef]
- McGary, K.; Nudler, E. RNA Polymerase and the Ribosome: The Close Relationship. Curr. Opin. Microbiol. 2013, 16, 112–117. [Google Scholar] [CrossRef]
- Mandell, Z.F.; Vishwakarma, R.K.; Yakhnin, H.; Murakami, K.S.; Kashlev, M.; Babitzke, P. Comprehensive Transcription Terminator Atlas for Bacillus subtilis. Nat. Microbiol. 2022, 7, 1918–1931. [Google Scholar] [CrossRef] [PubMed]
- Bidnenko, V.; Nicolas, P.; Grylak-Mielnicka, A.; Delumeau, O.; Auger, S.; Aucouturier, A.; Guerin, C.; Repoila, F.; Bardowski, J.; Aymerich, S.; et al. Termination Factor Rho: From the Control of Pervasive Transcription to Cell Fate Determination in Bacillus subtilis. PLoS Genet. 2017, 13, e1006909. [Google Scholar] [CrossRef] [PubMed]
- Bidnenko, V.; Nicolas, P.; Guérin, C.; Dérozier, S.; Chastanet, A.; Dairou, J.; Redko-Hamel, Y.; Jules, M.; Bidnenko, E. Termination Factor Rho Mediates Transcriptional Reprogramming of Bacillus subtilis Stationary Phase. PLoS Genet. 2023, 19, e1010618. [Google Scholar] [CrossRef]
- Mandell, Z.F.; Zemba, D.; Babitzke, P. Factor-Stimulated Intrinsic Termination: Getting by with a Little Help from Some Friends. Transcription 2022, 13, 96–108. [Google Scholar] [CrossRef]
- Yakhnin, A.V.; Babitzke, P. Mechanism of NusG-Stimulated Pausing, Hairpin-Dependent Pause Site Selection and Intrinsic Termination at Overlapping Pause and Termination Sites in the Bacillus subtilis Trp Leader. Mol. Microbiol. 2010, 76, 690–705. [Google Scholar] [CrossRef]
- Feng, C.-Q.; Zhang, Z.-Y.; Zhu, X.-J.; Lin, Y.; Chen, W.; Tang, H.; Lin, H. ITerm-PseKNC: A Sequence-Based Tool for Predicting Bacterial Transcriptional Terminators. Bioinformatics 2019, 35, 1469–1477. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, W.; Zhu, Q. Iterb-PPse: Identification of Transcriptional Terminators in Bacterial by Incorporating Nucleotide Properties into PseKNC. PLoS ONE 2020, 15, e0228479. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, H.; Li, H.; Zhang, Y.; Wang, M. A Programmable CRISPR/Cas9 Toolkit Improves Lycopene Production in Bacillus subtilis. Appl. Environ. Microbiol. 2023, 89, e0023023. [Google Scholar] [CrossRef] [PubMed]
- Jun, J.-S.; Jeong, H.-E.; Hong, K.-W. Exploring and Engineering Novel Strong Promoters for High-Level Protein Expression in Bacillus subtilis DB104 through Transcriptome Analysis. Microorganisms 2023, 11, 2929. [Google Scholar] [CrossRef]
- Sinumvayo, J.P.; Yang, S.; Chen, J.; Du, G.; Kang, Z. Engineering and Characterization of New Intrinsic Transcriptional Terminators in Bacillus subtilis 168. Sheng Wu Gong Cheng Xue Bao 2017, 33, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Welsch, N.; Homuth, G.; Schweder, T. Stepwise Optimization of a Low-Temperature Bacillus subtilis Expression System for “Difficult to Express” Proteins. Appl. Microbiol. Biotechnol. 2015, 99, 6363–6376. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Lin, Q.; Hu, R.; Han, L.; Cheng, Z.; Zhang, L.; Zhou, Z. Data-Driven and in Silico-Assisted Design of Broad Host-Range Minimal Intrinsic Terminators Adapted for Bacteria. ACS Synth. Biol. 2021, 10, 1438–1450. [Google Scholar] [CrossRef] [PubMed]
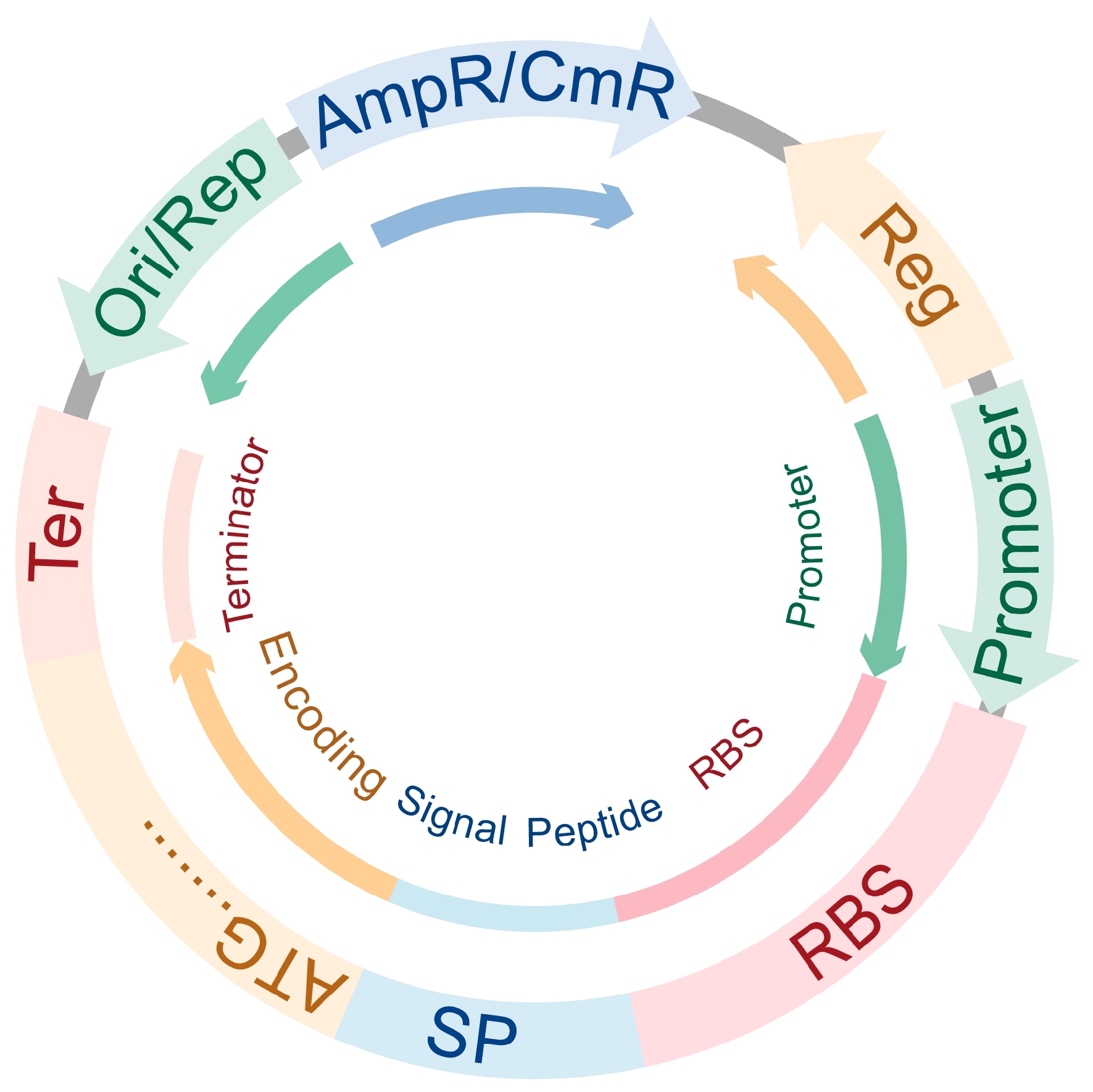
| Vector Type | Copy Method | Advantage | Disadvantage | Common Vectors | Reference |
|---|---|---|---|---|---|
| Plasmid vector | Self-reproduction | High copy. High expression. | Low stability. Antibiotic dependence. | pEB20, pUB18, pMA5, pHT43, pWB980 | [38,42,49,50,51,52] |
| Integration vector | Integrate into the host genome for replication | High stability. | Low copy. Low expression. | pDL, pDG1662, pAX01, pSG1151, pMutin-GFP, pHT01 | [42,53,54] |
| Thermosensitive phage DNA | High stability. No antibiotic dependence. | Low expression. Less research. | Φ105, Φ105J27, Φ105 dcM | [42,47,48,55] |
| Vector | Optimization Process | Conclusion | Reference |
|---|---|---|---|
| pMA5 | (1) Integrated expression in B. subtilis. (2) Build dual promoters PHapII-P43. (3) Knockout resistance gene. | (1) Construct a marker-free expression strain for zearalenone-degrading enzyme. (2) Vector stability increased to 98% by the 100th generation, up from 72%. | [56] |
| BGM | (1) Using plasmid DNA released by lysed E. coli. (2) Adopt the Culture mixed method. | (1) Transforming large DNA fragments (>100 kb). (2) Plasmid DNA requires no biochemical purification. (3) Suitable for other hosts. (4) DNA stability remains at 69%. | [55,57] |
| iREX | (1) Introducing xylite-induced recA expression cassette. (2) Deleting the endogenous recA. | (1) DNA stability increased from 69% to 73% under xylose induction. (2) DNA stability reaches 93% without xylose induction. | [57] |
| pl36 | (1) Construct plasmids with spc and erm resistance genes. (2) Use double-resistance screening. (3) Inserted floB into recombinant plasmid. (4) Deleting the endogenous gene floB. | (1) Couple growth with yield. (2) Yield increased 31.7%. (3) Plasmid loss rate dropped from 34.1% to 11.8%. | [58] |
| pUBC01 | (1) Construct pBV01-incompatible plasmid, pBV02. (2) pBV01 endogenous plasmid ori, pUC-ori, and kan genes formed pBV03 plasmid. | (1) Eliminate pBV01. (2) Plasmid stability reached 85% after 40 generations without antibiotic. (3) Efficiently express GFP. | [59] |
| pHT01 | Knock out the B. subtilis 168 yueB gene. | (1) Enhanced plasmid stability in BsΔyueB versus Bs168. (2) Acetoin titer increased by 61.99%. | [61] |
| pHT01 | (1) Construct SiteMuB. (2) Knockout genes yloD, yozK, yozL. (3) Enhanced uvrC expression. (4) Knockout genes mfd. (5) The ChassisLMR-SiteMuB combination. | (1) The mutation rate varied up to 110.63-fold across sites. (2) 41.8% decrease in spontaneous mutation rate. (3) 57.8% decrease in spontaneous mutation rate. (4) 89.1% decrease in spontaneous mutation rate. (5) The stable genetic generation increased 2.1-fold. | [62] |
| pWB980 | (1) Construct pWB980-DB by deleting bleoR gene. (2) ori from E. coli was inserted into the site upstream of the membrane binding region BA3-1. | (1) Copy numbers increased to 584. (2) Separation stability reached 98%. (3) The alkaline pectinate lyase and the alkaline protease reached 5200 U/mL and 21,537 U/mL. | [63] |
| pBR322 | DNA near the 3′ end of encoding RNA I gene sequence mutation G→T. | (1) RNAI is unable to bind RNAII. (2) Copy numbers increased to 1000. | [64] |
| pGL3 | ColE1 RNA II site-specific mutation C→A. | (1) RNAII promoter strength elevated. (2) RNAII concentration elevated. (3) Increased vector copy number. | [65] |
| pCB4170 | Replication IR III region site-specific mutation C→T. | (1) Initiation protein-replication locus affinity strengthens. (2) Increased vector copy number. | [66] |
| pDG1730 | (1) Targeted integration of P43-DPEase tandem repeats into the amyE locus. (2) Knock out the resistance gene spc. | (1) Copy number of 3. (2) 2.2-fold increase in DPEase activity. (3) Vector stability declined sharply beyond generations 3–4. | [67] |
| pJOE8999.1 | (1) Knockout of amyE generated the BS2 strain. (2) Build dual promoters PamyQ-Pcry3A. (3) PamyQ-Pcry3A-amyQ was iteratively integrated into B. subtilis BS2. (4) PrsA and SppA overexpression. | (1) Copy number of 6. (2) α-amylase activity increased 20.9-fold. (3) α-amylase production increased to 1439.2 U/mL. | [54] |
| System | Principle | Inducer/Promoter | Features | Reference |
|---|---|---|---|---|
| IPTG/Lactose | An inducer activates transcription by binding to the repressor protein, which alleviates its suppression of the promoter. | IPTG/Lactose Pgrac100 | (1) Widely used. (2) Suffering from induced toxicity and leaky expression. | [78,79,80,81] |
| Xylose | Xylose/PxylA/xylB | (1) Low-cost, readily available inducers. (2) Low utilization rate of inducers. (3) Affected by the CCR effect seriously. | [82,83,84] | |
| Mannitol | Mannitol/PmtlA | [85] | ||
| Maltose | The inducer combines with the regulatory protein, activating the regulatory protein to transform into a transcription activator, thereby activating transcription. | Maltose/PglvA/malA | [86,87] | |
| Methanol | Methanol/PAOX1 | (1) Low-cost, readily available inducers. (2) Inducer low toxicity. | [88] | |
| Glycerol | Inducer-antiterminator binding disrupts terminator structure, thereby activating transcription. | Glycerol/PglpD | (1) Realize self-induced expression. (2) Dependent on medium components. (3) No significant advantage over constitutive expression. | [77] |
| Fructose | Fructose/PsacA/sacB | (1) Inexpensive, non-toxic inducer. (2) Weak promoter and leaky expression. | [89] |
| Strain | Optimization Process | Conclusion | Reference |
|---|---|---|---|
| B. subtilis 168 | Integration of PxylA-araE-Tfba expression box into the amyE locus of B. subtilis 168. | (1) Strain JY123 was successfully constructed. (2) JY123 fully consumed xylose within 15 h; JY121 (cassette-negative) metabolized only 50% in 19 h. (3) AraE facilitates efficient xylose transport in B. subtilis. | [91] |
| BSUY00 | (1) Knock out araR gene. (2) Expressed araE gene. (3) Screen the optimal promoter Pveg. | (1) 6.25-fold increase in xylose consumption. (2) Fengycin yield of 376.58 mg/L. | [90] |
| B. subtilis J46 | (1) Screen galactose-adapted strains BSGA14. (2) Mutate the araRH226R gene. (3) Knockout of the glcR gene involved sugar phosphorylation. | (1) BSGA14 exhibits enhanced galactose consumption capacity. (2) 2.88-fold upregulation of AraE protein. (3) Protease and β-galactosidase increased by 6.05-fold and 50.31-fold, respectively. | [93] |
| B. subtilis 168 | (1) Overexpress the yjhG and yjhH genes. (2) Knock out ackA and ldh genes. (3) Introduce an auxiliary pathway composed of aldA, aceB and mdh genes | (1) Improved xylose absorption rate. (2) Fenamycin yield increased by 87%. | [92] |
| B. subtilis 1A751 | (1) Truncate the promoter PmalA. (2) Knock out the maltose hydrolysis gene malL/yvdK. | (1) Significantly increased the activity of promoter PmalA. (2) Improve the GFP expression. (3) High expression of luciferase and D-aminoacylase. (4) Luciferase and D-aminoacylase expression in this system is superior to that in the constitutive PhapII system. | [95] |
| B. subtilis CCTCCM 2016536 | Locus mutations are introduced in the region −2 to +10 of promoter PamyE. | The relative activity of D-allulose 3-epimerase was significantly enhanced, reaching 282.43% of the original strain. | [98] |
| B. amyliquefaciens TCCC 19030 | (1) Mutant PamyE promoter conserved sites: G3, C8, G9. (2) Break the cre sequence symmetry. (3) Adjustment of the relative distance between the cre sequence and the transcription start site to 30 bp. | (1) CcpA-CRE binding weakened; downstream gene transcription increased 3.92–5.46-fold. (2) Decrease the CCR effect and increase the PamyE strength. (3) GFP fluorescence intensity increased by 60.87%. | [99] |
| B. subtilis 168 | Introduction of A300W, A302W, L306W, and K308W mutations in the CcpA gene. | It cannot exert complete CCR/CCA effects on xynP (encode xylose transporter), ackA, and alsS (encodes the acetyllactate synthase). | [97,100] |
| B. amyloliquefaciens WH1 | Knock out the srfA gene. | (1) Biofilm formation is thin and fragile. (2) Unable to form spores and carbon metabolism disorder. (3) Downregulation of xylose metabolic gene xylB (xylulokinase) and galactose metabolic gene galK (galactokinase). High Expression of ccpA. | [101] |
| B. subtilis PY79 | (1) The light control system CcaSR v0.1 is constructed through the PCB production module, light-sensing module, and transcriptional output module. (2) Optimizing PCB/CcaS expression. (3) Optimizing output promoter activity. (4) Increases dynamic range. | (1) The photoresponsive light control system CcaSR v1.0 was successfully constructed in B. subtilis. (2) CcaSR v1.0 achieves more than 70 times activation and fast response dynamics. | [102] |
| B. subtilis 168/WB800 | (1) Screen glycerol-specific activation promoters. (2) Regulate the ratio of glucose to glycerol. (3) Overexpression of GlpP. | (1) Successfully developed and constructed an efficient glycerol-induced expression system (GIES) in B. subtilis. (2) The efficient expression of aspartate, nattokinase and serine protease was successfully achieved. | [77] |
| Protein | Optimization Process | Multiple/Yield U/mL | Reference |
|---|---|---|---|
| α-amylase AmyZ1 | (1) Screen out promoter PspoVG. | 952.6 (PspoVG) | [129] |
| (2) Build a double promoter. | 1139.8 (PspoVG-PspoVG) | ||
| (3) Truncating the 300 bp PspoVG sequence to obtain PspoVG2 (180 bp). | 1232.3 (PspoVG-PspoVG2) | ||
| (4) Introducing a C-to-T substitution in the AT box. | 1437.6 (PspoVG-PspoVG142) | ||
| (5) Optimization of signal peptide. | 1691.1 (Opt3) | ||
| Type I L-asparaginase | (1) Screen out promoter PyvyD. | 436.28 (PyvyD) | [130] |
| (2) Build a double promoter. | 502.11 (PaprE-PyvyD) | ||
| (3) Mutate the -35 and -10 regions of PyvyD. within the dual promoters PaprE-PyvyD. | 568.59 (PaprE-PyvyD-Mutant-3) | ||
| (4) Screen out RBS10. | 790.1 (PaprE-PyvyD-Mutant3-RBS10) | ||
| α-amylase AmyZ1 | (1) Screen out double promoter Pveg-ylb | 3687.7 (Pveg-ylb) | [72] |
| (2) Screen out signal peptide SPNucB | 4199.1 (Pveg-ylb-SPNucB) | ||
| (3) RBS optimization. | 4824.2 (SPNucB-RBS1) | ||
| Aldehyde dehydrogenases | (1) Screen out signal peptide SPyqzG. | 204.85 (PaprE-SPyqzG) | [135] |
| (2) Screen out promoter Pglv. | 254.82 (Pglv) | ||
| (3) Build a double promoter. | 268.26 (P43-Pglv) | ||
| L-asparaginase | (1) Screen out promoter P43 | 235.10 (P43) | [137] |
| (2) RBS sequence optimization. | 371.87 (P43-RBS207) | ||
| Bacilysin | Combine the classic strong SD sequence 5′-TAAGGAGG-3′ with the “ACAAACTC”-8nt interval sequence. | 80.3 (bacAstrongRBS) | [138] |
| GFPmut3 | Increase the short-interval sequence from 4 nt to 4–12 nt. | 4-fold change (7 nt) | [139] |
| β-glucuronidase | 27-fold change (9 nt) | ||
| Keratinase | (1) Screen signal peptides. | 84.3 × 103 (P43-SPdacB) | [140] |
| (2) Mutated RBS sequence. | 109.1 × 103 (P43-RBS16D12-SPdacB) | ||
| Thermo-alkaline β-mannanase | (1) Screen signal peptides. | 763 (PhpaII-SPlipA) | [141] |
| (2) Screen out promoter P43. | 908 (P43-SPlipA) | ||
| Glutaminase | (1) Screening of the optimal signal peptide from 173 candidate signal peptides. | 0.24 (ParpE-SPYndA) | [142] |
| (2) Screen out promoter PHpaII. | 4.52 (PHpaII-SPYndA) | ||
| (3) Build a double promoter. | 4.90 (PHpaII-ParpE -SPYndA) | ||
| Hyaluronate lyase | Screening signal peptides increases protein expression. | 1.86 × 104 (P43-SPabnA) | [26] |
| Alginate lyases | (1) Screen vectors and host bacteria. | 0.81 (B. subtilis WB600-pP43NMK) | [143] |
| (2) Screen out promoter PnprE. | 4.47 (PnrpE) | ||
| (3) Screen signal peptides. | 1.33 (SPvpr) | ||
| (4) Optimize the length and base composition of the RBS short-interval sequence. | 4.58 (RBS/NCS-1) | ||
| Upstream GFP/Downstream mCherry | (1) Screening terminators. | Up 2.2-fold change (GFP-TB5) | [144] |
| Down 28.4 change (TB5-mCherry) | |||
| (2) Establishes a double terminator. | Up 2.7-fold change (GFP-TH1.5b-TB5) | ||
| Down 30.5-fold change (TH1.5b-TB5-mCherry) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Zhang, P.; Tian, Z.; Huang, Y. Optimization Strategy of Expression Vectors and Regulatory Elements for Enhanced Protein Production in Bacillus subtilis. Int. J. Mol. Sci. 2025, 26, 10812. https://doi.org/10.3390/ijms262210812
Ye Z, Zhang P, Tian Z, Huang Y. Optimization Strategy of Expression Vectors and Regulatory Elements for Enhanced Protein Production in Bacillus subtilis. International Journal of Molecular Sciences. 2025; 26(22):10812. https://doi.org/10.3390/ijms262210812
Chicago/Turabian StyleYe, Ziru, Puyue Zhang, Zhong Tian, and Yong Huang. 2025. "Optimization Strategy of Expression Vectors and Regulatory Elements for Enhanced Protein Production in Bacillus subtilis" International Journal of Molecular Sciences 26, no. 22: 10812. https://doi.org/10.3390/ijms262210812
APA StyleYe, Z., Zhang, P., Tian, Z., & Huang, Y. (2025). Optimization Strategy of Expression Vectors and Regulatory Elements for Enhanced Protein Production in Bacillus subtilis. International Journal of Molecular Sciences, 26(22), 10812. https://doi.org/10.3390/ijms262210812






